Abstract
The roles of positive directional selection (selective sweeps) and negative selection (background selection) in shaping the genome-wide distribution of genetic variation in humans remain largely unknown. Here, we optimize the parameter values of a model of the removal of deleterious mutations (background selection) to observed levels of human polymorphism, controlling for mutation rate heterogeneity by using interspecific divergence. A point of “best fit” was found between background-selection predictions and estimates of human effective population sizes, with reasonable parameter estimates whose uncertainty was assessed by bootstrapping. The results suggest that the purging of deleterious alleles has had some influence on shaping levels of human variation, although the effects may be subtle over the majority of the human genome. A significant relationship was found between background-selection predictions and measures of skew in the allele frequency distribution. The genome-wide action of selection (positive and/or negative) is required to explain this observation.
Levels of human nucleotide polymorphism are positively correlated with the physical density of genetic recombination in humans (Nachman et al. 1998; Przeworski et al. 2000; Nachman 2001; Hellmann et al. 2003). This correlation also exists in many other eukaryotic species including Drosophila (e.g., Begun and Aquadro 1992; for reviews, see Andolfatto 2001; Aquadro et al. 2001; Schlötterer 2002). Two principal nonexclusive classes of hypotheses posed to explain this observation are (1) genetic recombination is, or is correlated with, a mutagenic process (Lercher and Hurst 2002; Waterston et al. 2002; Hardison et al. 2003; Hellmann et al. 2003); and (2) recombination allows increased independence from the effects of widespread diversity reducing selection (for reviews, see Andolfatto 2001; Aquadro et al. 2001). The primary selective processes hypothesized to explain the apparent reduction of variation in regions of low recombination are (2a) background selection associated with the ongoing selective removal of new deleterious mutations from the population and (2b) hitchhiking associated with positive directional selection.
Background selection is related to the classical concept of purging a mutational load (Haldane 1937; Muller 1950; Crow 1958; Charlesworth et al. 1993). Assuming a uniform distribution of deleterious mutations across the genome, regions of lower recombination will have an increased probability of linkage between neutral and deleterious variants; therefore, the probability of the removal of neutral polymorphism from the population is increased (but see also Palsson and Pamilo 1999). Background selection thus predicts reductions in levels of variation in regions of rarer crossing over. The effects of background selection can be summarized as the fraction of neutral variation remaining (f0) after the reducing effects of selection on linked deleterious mutants, which is often thought of as a regional reduction in the effective population size (Ne). The predicted amount of neutral polymorphism removed from a population increases with the deleterious mutation rate (u). The amount of variation removed also increases with the reduction of the strength of selection against heterozygous deleterious mutants (sh) because of persistence in the population, but ultimate removal, of weakly deleterious alleles (Kimura et al. 1963; Crow and Simmons 1983; Charlesworth et al. 1993). Indeed, the ability to provide some degree of independence among loci in order to more efficiently purge deleterious alleles has been considered one of the primary reasons for the evolution of multiple chromosomes and meiotic recombination (e.g., Felsenstein 1974; Kondrashov 1988; Charlesworth 1990; Antezana and Hudson 1997). Furthermore, to the extent that deleterious mutations are known to be a frequent occurrence, the process of background selection has been proposed as the appropriate null selective model to reject in favor of hitchhiking in explaining the correlation between diversity and recombination (Charlesworth et al. 1993; Stephan 1995; Hamblin and Aquadro 1996).
Hitchhiking refers to the rapid increase in frequency of genetic variants linked to a positively selected rare allele. This process is predicted to reduce genetic polymorphism, the size and amount determined primarily by the strength of selection and local rates of recombination (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Durrett and Schweinsberg 2004). Apparent instances of hitchhiking are reported for several loci in humans, based on deviations from both flanking levels of variation and the expectation of a steady-state allele frequency distribution (for reviews, see Aquadro et al. 2001; Bamshad and Wooding 2003). However, positive selection cannot reasonably explain all of the changes in genetic variation in humans because the required positively selected mutation rate would have to equal or exceed the neutral mutation rate (Andolfatto 2001).
Hellmann et al. (2003) found that changes in mutation rate, correlated with recombination rates, are sufficiently large enough to account for the genome-wide positive correlation between changes in polymorphism and rates of recombination in humans. However, mutation rate changes associated with recombination can only explain 6% of the total variation in polymorphism. Humans appear to have a high deleterious mutation rate (e.g., Eyre-Walker and Keightley 1999; Nachman and Crowell 2000), and genome-wide integrated genetic-physical maps are available (Kong et al. 2002); thus, it is natural to ask how much, if any, of the remaining variation in polymorphism along the chromosomes, after correcting for mutation rate heterogeneity, can be explained by background selection.
A great deal of uncertainty surrounds the parameter values necessary for creating baseline background-selection predictions along the human chromosomes. Therefore, in this report, we optimize the parameters of a background-selection model to observed levels of human polymorphism using a weighted least-squares regression and estimate the uncertainty associated with our parameter estimates by bootstrapping. We control for mutation rate heterogeneity by using human-chimpanzee divergence to estimate an effective population size (N̂e) for each genic region. The parameter estimates of these predictions are of interest in that they allow estimates of the fraction of the genome under constraint (i.e., purifying selection) and of the strength of selection against deleterious mutants, as well as affect how we interpret skewed allele frequency spectrums. The predicted levels of variation can be used as a null hypothesis representing the genome-wide effects of neutral and deleterious variation. Locus-specific departures can be evaluated against this prediction as candidates for positive selection. Additionally, these predictions can be used to select regions of the genome that maximize or minimize the region-specific effective population size in order to study various selective and demographic processes.
Results
We estimated effective population sizes (N̂e) of 126 autosomal loci (SeattleSNPs 2005, http://pga.gs.washington.edu;) using average nucleotide heterozygosity (π) and human-chimpanzee levels of nucleotide divergence (d, equation 6 in Methods; Supplemental Table 1). The parameters of a background-selection model (the deleterious mutation rate u, the strength of selection against heterozygous deleterious mutants sh, and the effective population size without selection N0) (slightly modified from Hudson and Kaplan 1995) were varied over a grid of parameter values, incorporating physical locations and changes in rates of recombination across the majority of the human genome (Kong et al. 2002). The point of “best fit” between the estimated and expected Ne (i.e., the maximum variance that can be explained by the model) as measured by r2 was recorded (equations 7 and 8 in Methods). After this optimization, a maximum correlation between the nucleotide polymorphism and divergence data and model predictions was found with a per generation deleterious mutation rate of û/Mb = 0.0016 (genomic Û = 10.2 = 2 × 0.0016û/Mb × 3200 Mb), a relative strength of selection against heterozygous deleterious mutants of ŝĥ = 0.018 and an effective population size before selection of N̂0 = 32,800 (Fig. 1). Predictions of the gene-specific effective population sizes were reduced from strict neutral levels (N̂0) to effective population sizes of 30,800 to 11,800 for the loci included (Supplemental Table 1). This range of population sizes leads to as much as a 62% reduction in polymorphism among the observed loci predicted by background-selection alone. However, at these best-fit values, the model of background selection can explain only r2 = 7.9% of the actual variation in effective population size estimates among loci (equation 8 in Methods). The correlation between observed and predicted Ne values is not statistically significant (determined by nonparametric bootstrapping, P = 0.28). However, we consider the observed relationship to be biologically meaningful for three reasons. First, bootstrapping is typically conservative because chance outliers (and perhaps loci affected by other forms of selection) in the total data set are overrepresented in a subset of the pseudo-samples (Efron and Tibshirani 1993). Second, reasonable parameter values are found that are consistent with previous literature and/or theoretical expectations (see below). Third, significant correlations between background-selection predictions and distortions in the allelic spectrum were found that require a role of selection to explain (also below).
Figure 1.
Plots of the bootstrapping distribution of the estimated deleterious mutation rate (û/Mb), strength of selection (ŝĥ), and the effective population size without selection (N̂0). The best-fit value for all the data is contained in the weight of the outcomes. û and ŝĥ appear to be positively correlated (i.e., as the deleterious mutation rate increases, selection strength must also increase to maintain a similar outcome). û and N̂0 also appear to be correlated (as the deleterious mutation rate increases, the effective population size must also increase to maintain a similar outcome). A small number of points that were widely dispersed below ŝĥ = 0.00001 and/or û/Mb = 0.00001 are not included in the plot.
Assessing parameter uncertainty by bootstrapping
In order to estimate the uncertainty associated with our parameter estimates, we carried out nonparametric bootstrapping, which produced a “cloud” of points clustered around the original estimate (Fig. 1). Confidence intervals can be estimated from the bootstrapping outcomes using the “reflection” method by removing the same number of outcomes from the upper and lower edges of the bootstrapping distribution (Efron and Tibshirani 1993). The 90% confidence interval for the per generation deleterious mutation rate is 7.7 × 10-5 to 4.7 × 10-3 per single-copy megabase or 0.49 to 30 per diploid genome complement (2 × 3200 Mb). This is consistent with previous lower-bound estimates of the genomic deleterious mutation rate based on relative ratios of nonsynonymous to synonymous substitutions among humans and other primates (e.g., Û = 1.6-3.1 [Eyre-Walker and Keightley 1999]; Û = 1.5-4.0 [Nachman and Crowell 2000]) and is higher than the deleterious mutation rate estimated from amino-acid-altering protein electrophoretic band morph mutations (Û = 0.4 [Neel et al. 1988; Keightley and Eyre-Walker 1999]).
The 90% confidence interval of sh, the strength of selection against deleterious mutations in the heterozygote is 2.0 × 10-5 to 0.17. Values of sh are not predicted to be less than (2Ne)-1 if selection is to have any appreciable effect on a per nucleotide basis since below this point the force of stochastic drift in changing allele frequencies is greater than the selective differences (Kimura 1955). The lower bound for our estimate of sh is very close to this theoretical boundary.
The 90% confidence interval of N0, the effective population size without the reducing effect of background selection, is 28,700 to 51,000 diploid individuals. In general, the effective population size before the reducing effects of background selection is expected to be larger than the effective population sizes estimated from observed data. These values are larger than effective population sizes traditionally estimated for humans (e.g., 10,000 individuals [Li and Sadler 1991]; 14,600 [Nachman et al. 1998]) and suggest that many Ne values in the literature may at least be partially affected by diversity reducing selection. However, we make some assumptions in order to account for the expected contribution to divergence of polymorphism in the common ancestor of humans and chimpanzees (equation 6 in Methods), which will affect Ne and N0 estimates.
Identifying locus-specific departures
Estimated gene-specific effective population sizes along the human autosomes (Fig. 2) are, as expected, generally reduced on average near the physical centers of the chromosomes and maximized near the edges. A few of the autosomes contain regions of dramatic expected reductions in effective population size, portions of the interiors of Chromosomes 1, 9, 11, and 16; and the p-edge of Chromosome 13, which tend to be heterochromatic areas near the centromeres. Of interest are loci like ABO (ABO blood group; MIM 110300, http://www.ncbi.nlm.nih.gov/omim/), which has also been found to have signals of selection robust to simple demographic assumptions (Akey et al. 2004). ABO has a very high estimated effective population size (91,200) and a large excess of intermediate frequency alleles (Tajima's D = 2.06) consistent with balancing selection and a possible role of this antigen in disease susceptibility (e.g., cholera [Glass et al. 1985]; norwalk virus [Hutson et al. 2002]). Also of interest, as candidates for hitchhiking events, are loci that deviate below (or have less variation than) predictions based on background selection and divergence (e.g., DCN, FGA, ICAM1, IL5, PPARA, PROS1, PROZ, SMP1, THBD, TNFAIP1). Of these, DCN (decorin; MIM 125255) has also been found to have signals of selection robust to simple demographic assumptions (Akey et al. 2004), and is associated with renal disease (De Cosmo et al. 2002). IL5 (Interleukin 5; MIM 147850) is a member of the T-helper 2 (Th2) interleukin immune defense cluster on Chromosome 5 (e.g., Brombacher 2000), and positive selection has been reported for other members of this group (IL4 [Rockman et al. 2003]; IL13 [Tarazona-Santos and Tishkoff 2004]). ICAM1 (Intercellular adhesion molecule 1; MIM 147840) is a Plasmodium falciparum cell adhesion receptor (Berendt et al. 1989), a rhinovirus receptor (e.g., Bella et al. 1998), and plays a role in both septic shock (Xu et al. 1994) and autoimmunity (Bullard et al. 1997). ICAM1 has also been identified as a gene undergoing significantly accelerated amino acid replacements along the human lineage (Clark et al. 2003). Similarly, DCN and TNFAIP1 (tumor necrosis factor α induced protein 1; MIM 191161) appear to be rapidly evolving along the chimpanzee lineage (Clark et al. 2003). Genes undergoing rapid evolution may deviate below background-selection predictions because they have undergone a recent selective sweep and/or because divergence has been overestimated owing to an excess of selected fixations. Finally, a few gene regions have paradoxical deviations, typified by MC1R (melanocortin 1 receptor; MIM 155555), which has an excess of intermediate frequency alleles, but low levels of variation compared to divergence, which is not consistent with a simple model of either balancing selection or a selective sweep, but may reflect a partial selective sweep (see also Harding et al. 2000).
Figure 2.
Predicted (lines) and estimated (circles) effective population size estimates (Ne) along the human autosomes under the model of background selection (equation 2). The upper and lower 90% bootstrapping-based confidence intervals are solely for the background-selection estimates. The deviations of individual gene regions depend on evolutionary variance and their individual sampling properties. Filled black circles correspond to positive Tajima's D values, filled gray circles correspond to Tajima's D values between 0 and -1; filled white circles correspond to Tajima's D values <-1.
These 126 loci were chosen for the SeattleSNPs resequencing study largely because of their medical interest; many are involved in human disease interactions with clear fitness consequences (e.g., ABO and cholera [Glass et al. 1985]; IL4 and HIV-1 progression [Valentin et al. 1998]; CSF2 and pneumonia [LeVine et al. 1999]). Therefore, it is not difficult to imagine positive selection acting at some fraction of these loci. Individual gene regions affected by forces other than background selection may deviate from the level of variation predicted by fitting the background-selection model to the rest of the data. Inclusion of these positively selected loci in the data set may detract from the proportion of variation accounted for by the background-selection model, as measured by r2. As an ad hoc exploration to identify positive selection in the data, we tabulated how often each locus was included in the highest 10% of bootstrapping outcomes when ordered by r2 values and asked: Are there significant underrepresentations in the collection of gene regions that seem to best fit background-selection predictions. A goodness-of-fit test between observed counts and expectations from a Poisson distribution of mean 100 rejected the null distribution (Supplemental Fig. 1) (cells were pooled to satisfy Cochran's [1954] guidelines; χ2 = 27.75, df = 14, P < 0.025). The most underrepresented loci in the top 10% (ordered by r2) of bootstrapping replicates are VEGF (vascular endothelial growth factor; MIM 192240; P = 1.16 × 10-19 with a Bonferroni correction) and LTA (Lymphotoxin-α; MIM 153440; P = 6.92 × 10-12 with a Bonferroni correction). These two loci are 15.2 cM apart on Chromosome 6 and deviate above the background-selection predictions, and LTA has a positive Tajima's D estimate consistent with balancing selection (Fig. 2; Supplemental Table 1). The genetic grouping of VEGF and LTA suggests errors in the local recombination rate estimates, and the very large effective population size estimates suggest errors in the mutation rate estimates, might be responsible for their underrepresentation. However, LTA is found near the major histocompatibility complex (MHC) on Chromosome 6 (Jongeneel et al. 1991) and has an immune regulation function (Chin et al. 2003), thus it may, in fact, be affected by some form of balancing selection. Curiously, in addition to being a vascular growth factor (Ferrara and Henzel 1989), VEGF is inhibited by the dopamine neurotransmitter (Basu et al. 2001); is neurotrophic, neuroprotective, and neurogenic (Jin et al. 2002 and the references therein); may affect neurocognitive function by promoting glucose passage across the blood-brain barrier during acute hypoglycemia (Dantz et al. 2002); and appears to be responsible for increased neurogenesis and improved cognitive response to enriched environments and learning tasks: “VEGF may be a key mediator linking the environment to neurogenesis, learning and memory” (Cao et al. 2004, p. 832). The excess of variation compared to divergence and an excess of rare alleles found at VEGF is not consistent with simple models of balancing selection or selective sweeps, but may be consistent with a model of diversifying selection in humans. One of the loci that we identified above as having less variation than predicted by background selection also has a tendency to be underrepresented in these replicates (ICAM1, P = 0.69, 0.0055 with and without a Bonferroni correction, respectively), consistent with putative hitchhiking. PROS1 (Protein S; MIM 176880) is a clear outlier that is awkward to explain. When included, PROS1 appears to increase the amount of variation that can be explained by background selection. However, PROS1 was identified above as a gene region that deviated below background-selection predictions and has a strongly negative Tajima's D value (-1.44). Alternatively, the low level of variation and skewed allelic spectra toward an excess of rare alleles at PROS1 may be entirely consistent with the effects of background selection, as described below.
Comparing background-selection predictions to the allelic spectra
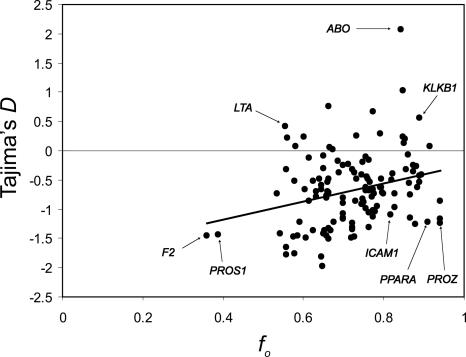
Simulation studies have reported distortions in the allele frequency distribution associated with the removal of weakly deleterious mutations, particularly in populations with effective sizes as small as that of modern humans (Charlesworth et al. 1993, 1995; Fu 1997; Tachida 2000; Gordo et al. 2002; Williamson and Orive 2002). We find that f̂0 (the fraction of remaining neutral variation after background selection) is positively correlated with Tajima's D values (r2 = 0.074, P = 0.002) (Fig. 3; Tajima 1989). This is similar to the correlation between Tajima's D and the recombination rate recently reported by Stajich and Hahn (2005). There is a concern that this correlation may arise from, or be exaggerated by, sharing of the same π values between Tajima's D and f̂0 calculations. To address this, the loci were divided, using a random function, into two subsamples with an equal chance of being included in each subsample. The first subsample was used to reoptimize the model parameters identically to the method used for the full data set. The resulting f̂0 estimates were calculated for all loci. The second sample was used to test for a correlation between D and f̂0. Therefore, in this second treatment, none of the data from genic regions whose π values were used to calculate N̂e and optimize f̂0 were used in the comparison between the models f̂0 predictions and the Tajima's D estimates. Essentially the same result, a significant positive correlation, was again found (r2 = 0.103, P = 0.009). These results indicate that, overall, areas predicted to be affected more by background selection are also increasingly skewed from an expected steady-state allele frequency distribution toward an excess of rare alleles. This is not unexpected given the parameter values we have estimated (ŝĥ = 0.018 and N̂0 = 32,800), but the skew can be problematic for discriminating between the genome-wide effects of positive and negative selection in humans.
Figure 3.
A plot of Tajima's D versus f̂0 predictions. The observed frequency distribution is increasingly skewed toward an excess of rare alleles as predicted variation is reduced (assuming background selection). This could be a result of either positive or negative selection or both. The correlation between Tajima's D and f̂0 remains significant even if the apparent outliers (ABO, F2, and PROS1) are selectively removed (r2 = 0.045, P = 0.019). Note, if a linear extrapolation is made from the best-fit regression line to f0 = 1, in an effort to account for the effects of selection, there is little to no negative skew (D ≈ 0) predicted for humans, consistent with a nearly constant ancestral population size.
Discussion
The process of background selection as modeled can explain 7.9% of the variation in observed nucleotide polymorphism among autosomal gene regions across the human genome, after adjustments for variation in mutation rates. Comparisons of the range of locus-specific effective population sizes to their corresponding expectation suggests that the role of the removal of deleterious alleles in shaping levels of human variation can be subtle over the majority of autosomal loci (Fig. 2), as predicted for mammalian chromosomes (Nordborg et al. 1996). This is in contrast to findings in Drosophila, where a large amount of observed variation is consistent with background selection (Hudson and Kaplan 1995; Charlesworth 1996; Hamblin and Aquadro 1996). However, this does not necessarily mean that other deterministic factors like positive selection have a large effect on standing variation in humans. We are faced with the high evolutionary variance of nucleotide heterozygosity (π) in modern humans (Tajima 1983; Nei 1987), variance of lineage coalescence in the common human-chimpanzee ancestor, and uncertainty of the relative size and age of this common ancestor (e.g., Takahata et al. 1995), as well as the possibility of positive selection affecting divergence estimates, particularly when regulatory elements might be included (e.g., Wray et al. 2003). Thus, inferences about any single locus as outliers from background-selection predictions should be verified with further data collection and analysis (e.g., by measuring changes in polymorphism at flanking regions, divergence estimates from additional species, and explicit tests of positive selection).
It should also be noted that we assume a uniform distribution of deleterious mutations for each physical segment of the genome. Known genes are nonrandomly distributed along a chromosome; however, the majority of interspecific conserved sequences appear to be nongenic (Shabalina et al. 2001; Dermitzakis et al. 2002; Waterston et al. 2002). The unequal distributions of nongenic and genic conservation appear to largely cancel each other out and yield a relatively uniform (at least as a first approximation) distribution of conserved nucleotides (Dermitzakis et al. 2002). However, considering changes in rates of purifying selection as a function of interspecific nucleotide conservation rather than physical distance, should also improve background-selection predictions.
The tolerance of a high deleterious mutation rate
A diploid genomic deleterious mutation rate (Û) of 10.2 represents a very large per generation mutational load for humans (e.g., Haldane 1937; Crow 1958; Kimura and Maruyama 1966; Kondrashov 2001). However, when the proportion of the human genome under functional constraint (estimated as 5% by comparisons to mouse) (Waterston et al. 2002) and the genomic mutation rate (estimated as 175 new mutations per generation) (Nachman and Crowell 2000) is considered, then a high U is predicted (in this example Û = 8.75).
Both synergistic epistatic fitness between deleterious alleles (Kimura and Maruyama 1966; Crow and Kimura 1978; Charlesworth 1990; Kondrashov 1994) and inbreeding (Glémin 2003) can more efficiently purge the genome of deleterious alleles by removing multiple alleles at a rate greater than expected under independent fitness effects with random mating and may help resolve this paradox. Furthermore, the removal of gametes carrying deleterious alleles (e.g., during atresia) (Gougeon 1996) may also allow an increased tolerance of a high per generation deleterious mutation rate and is more efficient at purging deleterious alleles because each gamete has only one genomic copy.
Effective population size estimates
To the extent that selection may vary the effective population size along the chromosome, different areas of the genome may contain different information about the history of a species since the time to the coalescence of sample lineages is a function of the effective population size (Kingman 1982). Loci in regions of low predicted Ne, such as the center of Chromosome 1, where the effects of stochastic drift are predicted to be magnified, will be expected to have greater differences in allele frequencies and may provide more information on the recent demographic structure of modern humans. Conversely, regions of high predicted Ne, such as the ends of most of the autosomes, may have maximal segregating lineage depth and provide information on more ancient demographic processes. In this case, gene regions with high f̂0 can have an expected Ne as large as 30,000 diploid individuals and deviate little from neutral predictions (although this prediction is sensitive to some assumptions made in our divergence estimate; see Methods). In a sample of sufficient size, the expected time to a most recent common ancestor is expected to be 4Ne generations (Kingman 1982). With an average generation time of 25 yr, this corresponds to three million years ago (Mya). Therefore, by selecting loci with high f̂0, it should be possible to study lineages that predate the emergence of Homo sapiens within the last 200,000 yr (White et al. 2003; McDougall et al. 2005) and perhaps even predate the origin of the genus Homo ∼2.3 Mya (Kimbel et al. 1996).
Background selection's effect on divergence
Can the correlation between rates of recombination and divergence observed by Hellmann et al. (2003), and explained as neutral changes in mutation rates, also be explained by background selection acting in the common ancestral species of humans and baboons? Assuming an average generation time of 15 yr and a common ancestor 25 Mya (Goodman et al. 1998; Yoder and Yang 2000), humans and Old World monkeys are separated by 3.3 million generations (2g). Background selection may have had a larger effect in this common ancestor considering that baboons have lower rates of recombination (and thus more linkage between deleterious and neutral alleles) than humans (Rogers et al. 2000). However, even with a 10-fold difference in effective population sizes along the chromosomes due to background selection, an interlocus maximum effective population size of 240,000 diploid individuals (Ne,max) is required in the common ancestor to entirely explain the expected 20% change in divergence associated with recombination rates:
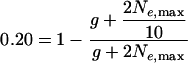 |
(1) |
(note, the scaled mutation rate 2μ cancels out and the expected contribution to divergence 2gμ of the coalescence of two lineages in a common ancestor is 4Neμ). This is larger than the effective population sizes typically estimated in primates (e.g., Chen and Li 2001; Chiarello and de Melo 2001; Storz et al. 2002; Yang 2002; Li et al. 2003; Wall 2003). However, Satta et al. (2004) estimate an N̂e of 105 over much of primate evolution, and Takahata and Satta (1997) estimate an N̂e of 106 in the Oligocene. Thus, while background selection may be unlikely to be entirely responsible for the patterns observed by Hellmann et al. (2003), it is also hard to rule out a nontrivial contribution of background selection to changes in divergence between species.
Patterns of skewed allele frequency spectra
The traditional treatment of background selection, and the model we use, assumes efficient selection; deleterious mutants are removed fast enough to make no contribution to heterozygosity (Charlesworth et al. 1993). However, this assumption may break down with smaller effective population sizes, weaker selective coefficients, and nonmultiplicative mutant interactions. With human parameters, an Ne on the order of 25,000 diploid individuals and sh values approaching 0.01 or less, analyses and simulations of the process of background selection predict that deleterious mutants can persist and rise to high enough frequencies to begin to contribute to sample heterozygosity and to co-segregate (Charlesworth et al. 1995). Ultimately these mutants will likely be removed, and they tend to be young and to contribute mutations as rare alleles in external lineages (Williamson and Orive 2002). Furthermore, interference among linked mutants can also reduce the efficiency of selection in quickly removing these mutants (Hill and Robertson 1966; Felsenstein 1974). The result is an ongoing population expansion, where a subset of “healthy” chromosomes ultimately contributes to all future chromosomes, and, in a population sample, a class of chromosomes are held at a lower than expected sample frequency due to linked deleterious mutants that are inefficiently purged. This results in an apparent non-neutral pattern, particularly in regions of low recombination (Charlesworth et al. 1995; Tachida 2000; but see also Przeworski et al. 1999). Indeed, we found a significant positive correlation between Tajima's D and levels of variation predicted by background selection for the 126 SeattleSNPs loci (r2 = 0.068, P = 0.0058) (Fig. 3). This suggests that regional rare-skewed allele frequency distributions alone are not conclusive indicators of positive selection and that the effects of background selection, in humans, cannot be simply thought of as a reduction in the effective population size.
The selection of candidate loci that have undergone positive selection might be refined in light of the potential effects of background selection on allele frequency distributions (Fig. 3). For example, PROS1 has a low level of variation, compared to divergence, and an excess of rare alleles, but is very close to background-selection predictions (both in terms of levels of variation [Fig. 2] and allele frequency distribution [Fig. 3]). Therefore, PROS1 may not be as promising a candidate region for the presence of a selective sweep as PPARA (peroxisome proliferators-activated receptor-α; MIM 170998) and PROZ (protein Z; MIM 176895), which have slightly higher levels of variation and less skewed allelic distributions (Figs. 2 and 3), but deviate much more from Tajima's D values seen at high f̂0 values. Similarly, KLKB1 (prekallikrein deficiency; MIM 229000) may not be as likely a candidate for balancing selection as LTA, which deviates more from the negative Tajima's D value expected at lower f̂0 values (Fig. 3).
The correlation between Tajima's D and f̂0 could be due to selective sweeps with hitchhiking. However, Kitano et al. (2003) find a correlation in skewed allele frequency distributions from 10 X-linked genes in both humans and chimpanzees. This finding suggests that allele frequency skews are, to some degree, a static regional phenomena preserved between species. This may be difficult to explain under a hitchhiking model, where individual signals of positive selection are predicted to be intermittent and short-lived (Simonsen et al. 1995; Przeworski 2002). The allele frequency distortion correlation with f̂0 could also arise from an interaction of the reduction in effective population size due to background selection and other selective and demographic factors. For example, the human population size is expanding; the coalescent distribution in regions of reduced effective population size is more influenced by recent events; therefore, regions of lower f0 may reflect a greater signal of the expansion of modern humans. Also consider that regions of low f0 are predicted to experience accelerated genetic drift (due to a smaller effective population size) compared to regions of higher f0. If a sample is composed of members from recently structured populations, larger differences in rare allele frequencies among subpopulations may result in a sampled excess of rare alleles in areas of low f0 (Ptak and Przeworski 2002; Hammer et al. 2003).
Conclusions
We predict a null level of DNA sequence polymorphism expected across the human autosomes based on a simple model of background selection. Background-selection predictions may help refine candidate loci influenced by positive selection and identify gene regions informative in studying recent or ancient demographic patterns. The results of Hellmann et al. (2003) indicate that neutral mutation rates may be the main determinant of genome-wide variation, but a genome-wide role of selection in humans is required to explain the correlation between background-selection predictions and skews in the allele frequency spectrum.
Methods
Background-selection model
We use the background selection with recombination derivation of Hudson and Kaplan (1995). Because we correct for differences in mutation rates (see below), we replace average pairwise nucleotide heterozygosity (π) (Nei and Li 1979) in the original Hudson and Kaplan (1995) equation with effective population size, Ne. The expected effective population size under this model is calculated as follows:
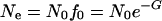 |
(2) |
where
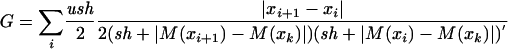 |
(3) |
N0 is the effective population size without the effects of background selection, f0 is the probability of not being linked to (and removed by) a deleterious mutant, u is the deleterious mutation rate per physical unit (in this case we used megabase pairs), sh is the selective coefficient against the deleterious mutants (s) multiplied by the degree of dominance (h), x refers to the physical position being considered, and M refers to the genetic map position in morgans of physical position x. For each locus of interest (k) the probability of being linked to, and removed by, a deleterious mutant is the sum of all the probabilities contributed by each physical segment (i to i + 1) along the chromosome. The distribution of linked deleterious mutants is assumed to be Poisson; thus the probability of the mutation-free class is given by e-G. The model assumes that selection is efficient so that mutations are removed quickly enough not to contribute to observable sample polymorphism and that mutant alleles are at low enough frequency to always be present in the heterozygote form (Hudson and Kaplan 1995).
Human polymorphism data
Publicly available estimates of DNA sequence variation for 126 autosomal gene regions (SeattleSNPs 2005, http://pga.gs.washington.edu) assayed in combined African-American (24 individuals) and European-American CEPH (23 individuals) population samples were used as a basis for levels of human genetic polymorphism (Supplemental Table 1). Because we are interested in patterns general to humans; because, relative to many other organisms, there is a high degree of shared polymorphism and evolutionary history in humans; and because these samples, in all likelihood, already consist of individuals admixed among different genetically structured human populations (e.g., Parra et al. 1998), we chose to combine the African-American and European-American samples in an effort to reduce sample variance. Only noncoding regions (averaging a total of 16.5 kb in length) were used in the analysis (i.e., 3′- and 5′-flanking and intron sites). We chose to exclude the X- and Y-chromosomes and focus on the autosomes in the present report because (1) more multilocus data from a common set of DNA samples are available for the autosomes; (2) the autosomes make up the majority of the genome; and (3) selection, mutation, and effective population size parameters are expected to be different between the sex chromosomes and the autosomes because of the X-chromosome's hemizygosity in males (coupled with effective sex ratio uncertainty) (Charlesworth 2001), the Y-chromosome's single-copy nature and absence in females, and a higher mutation rate in males (for review, see Li et al. 2002).
Background selection is predicted to have more of an effect on average pairwise nucleotide heterozygosity (π) (Nei and Li 1979) than the number of segregating sites in a sample (S) (Watterson 1975), particularly when selection is weak and effective population size, Ne, is small (Charlesworth et al. 1993). Therefore we used total π estimates from each gene region (including coding and noncoding sites) as our principal measure of genetic polymorphism.
In an ideal Wright-Fisher mutation drift equilibrium population of constant size, under the infinite sites model, π is an unbiased, but inconsistent (i.e., maintains a finite variance under infinite sampling) (Tajima 1983; Nei 1987), estimator of the population mutation parameter θ (Haldane 1939; Kimura 1968). For autosomal genes in an ideal diploid population, θ =4Neμ, where Ne is the diploid effective population size and μ is the mutation rate per generation.
Controlling for mutation rate heterogeneity, estimating Ne
Possible differences in the proportion of nucleotides under functional constraint as well as increased mutation rates in regions of higher recombination and higher GC content require differences in effective mutation rates to be accounted for in order to combine gene regions for analysis. We correct for mutation rate differences among gene regions by using average pairwise DNA divergence between the humans and a chimpanzee, Pan troglodytes, sample (SeattleSNPs 2005, http://pga.gs.washington.edu;). Polymorphism in the ancestral population of humans and chimpanzees could have made a substantial contribution to the observed interspecific divergence (d), because the division between the two species is recent (4-6 Mya or ∼250,000 generations, g) (e.g., Gavan 1953; Nishida et al. 1990; Hill and Hurtado 1996; Stauffer et al. 2001; Burnet et al. 2002; Wall 2003), and the common ancestor between humans and chimpanzees before this time may have had a large effective population size (see below), resulting in more ancient sequence divergence dates (e.g., 5.5-11 Mya) (Bailey et al. 1991; Goodman et al. 1998; Kumar and Hedges 1998; Huelsenbeck et al. 2000; Arnason and Janke 2002; Hasegawa et al. 2003; Yang and Yoder 2003). Also, the ancestral human-chimpanzee speciation may have occurred over a considerable period of time, adding to the variation in these estimates (Osada and Wu 2005). The ancestral contribution to divergence is expected to be θa =4Naμ, where θa and Na are the corresponding ancestral values. We correct divergence by using an ancestral constant, a, that is equal to the ratio of ancestral to modern effective population sizes. a is multiplied by the modern human θ estimate, based on π, and this product (aπ) is subtracted from the species divergence to yield a divergence value corrected for ancestral polymorphism (d - aπ). This yields an estimate of the modern effective population size, N̂e, for each gene region based on π:
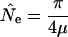 |
(4) |
where
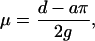 |
(5) |
g is the number of generations since species division (here we use g = 250,000), and 2g represents the total number of generations between the two species. This simplifies to:
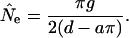 |
(6) |
Estimates of the effective population size of the immediate common ancestor to humans and chimpanzees range from 1-2 (Yang 2002) to 4-10 (Takahata and Satta 1997, estimated separately from the Oligocene value cite above; Chen and Li 2001; Wall 2003) times larger than modern humans. These latter values are almost surely overestimates because an ancestral correction as large as four removes all the divergence between the species for some loci and leads to negative modern effective population size estimates (i.e., the human to chimpanzee species divergence is predicted to be entirely contained by lineages in the common ancestor). Because estimating the effective population size of the human-chimpanzee common ancestor is not a central goal of this report, we optimized a values with the full data set at the beginning of the analysis. The best-fit value, a = 2.0, was then used for all subsequent bootstrapping analyses. The particular value of a used should not bias the organization of levels of variation along the chromosome (which could affect û and ŝĥ), but a will affect N̂0.
Use of integrated maps
We used the integrated genetic and physical maps reported by deCODE, which result from 1257 meioses, 5136 markers, and 4690 recombination rate interval estimates (Kong et al. 2002). Sex-averaged genetic positions were used for all the autosomes. Each SeattleSNPs (2003, http://pga.gs.washington.edu;) locus was located in the April 2003 freeze of the UCSC Human Genome Browser (http://genome.ucsc.edu/; Kent et al. 2002), and assigned the genetic and physical position of the nearest marker reported in deCODE's map.
Least-squares optimization procedure, comparing estimated and expected Ne
By computer algorithm, values of u and sh were systematically varied over a range of 1 × 10-5 to 1. Each resulting f0 prediction was fit, by the weighted least-squared error described below, to the observed DNA polymorphism data by a one-dimensional optimization of N0. The best-fit minimum weighted squared error was found for a particular coordinate of u, sh, and N0 in this range and reported.
Although the chromosomes between gene regions were sampled in common in the SeattleSNPs data set, the lengths sequenced, local recombination rates, and levels of nucleotide variation differed among loci and affect the expected variance of the sample π estimate. In order to appropriately take these differences into account, the expected variance, V, was estimated for each datum, under its corresponding sampling parameters, by the calculated variance of π values resulting from 10,000 coalescent simulations using the MS program of Hudson (2002). The expected distribution of π is not symmetrical (Tajima 1983; Nei 1987); therefore, separate upper and lower variances were calculated for each datum according to standard statistical methods. The squared errors between the background-selection prediction and the observed value were weighted by each datum's expected upper or lower variance estimate, corresponding to the individual deviation, in order to find the best-fit parameter configuration by weighted regression (e.g., Neter et al. 1996; cf. Andolfatto and Przeworski 2001).
Several of the SeattleSNPs loci shared the same, physically nearest deCODE marker. To avoid a single location's influence from being overrepresented in the optimization procedure, the variance of each individual SeattleSNPs locus was multiplied by the number of loci (l) that shared the same deCODE position. This approach proportionally downweights the contribution of clustered loci to the sum of the squared errors (SSE). Thus, for each locus i:
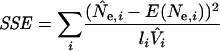 |
(7) |
where, for the i-th locus, N̂e,i is the estimated effective population size (equation 6), E(Ne,i) is the expected effective population size according to background selection (equation 2), li is the number of loci that share the same deCODE position, and V̂i is the estimated sample variance. Note, this is a constrained regression with the slope of the regression line fixed at 1 and the intercept 0. The r2 value, the proportion of variance that can be accounted for by the model predictions, is calculated as follows:
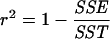 |
(8) |
where
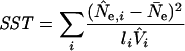 |
(9) |
and N̄e is the mean N̂e value.
Bootstrapping procedure
Nonparametric bootstrapping was used to assess the general robustness of the parameter values (Efron and Tibshirani 1993). Random sampling of the loci with replacement generated pseudo-samples of the same size as the original data set. For each pseudo-sample, the weighted regression optimization described above was repeated and the best-fit parameter values were found and reported. This procedure assumes that each locus is genetically far enough apart to reflect independent outcomes of the evolutionary process. For the majority of loci included in each pseudo-sample, this assumption is easily met. If we consider an expectation of 100 recombination events in the coalescent history between two loci on a single chromosome (assuming an ideal population of constant size) as sufficiently independent,
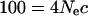 |
(10) |
where c is the per generation recombinant fraction, and an effective population size of only 10,000 diploid individuals, then genetic distances as small as one-quarter of a centimorgan are effectively independent:
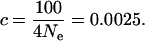 |
(11) |
The average genetic distance between adjacent markers in the deCODE map is 0.7 cM, and the contribution of the subset of loci that map to the same marker is down-weighted so that the total contribution from each marker is equivalent to a single locus, as described above.
Supplementary Material
Acknowledgments
We thank Vanessa Bauer DuMont, Yuseob Kim, Guy Reeves, Rasmus Nielsen, Bret Payseur, Molly Przeworski, Wolfgang Stephan, and three anonymous reviewers for helpful discussion and suggestions. We also thank Deborah Nickerson and the SeattleSNPs project for making these data publicly available. This work was supported in part by grants from Sigma Xi (to F.A.R.), National Institutes of Health grant GM36431 (to C.F.A), and National Science Foundation grant DMS-0201037 (to Richard Durrett, C.F.A., and Rasmus Nielsen).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3413205.
References
- Akey, J.M., Eberle, M.A., Rieder, M.J., Carlson, C.S., Shriver, M.D., Nickerson, D.A., and Kruglyak, L. 2004. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2: 1591-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P. 2001. Adaptive hitchhiking effects on genome variability. Curr. Opin. Genet. Dev. 11: 635-641. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P. and Przeworski, M. 2001. Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics 158: 657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antezana, M.A. and Hudson, R.R. 1997. Before crossing over: The advantages of eukaryotic sex in genomes lacking chiasmatic recombination. Genet. Res. 70: 7-25. [DOI] [PubMed] [Google Scholar]
- Aquadro, C.F., Bauer DuMont, V., and Reed, F.A. 2001. Genome-wide variation in the human and fruitfly: A comparison. Curr. Opin. Genet. Dev. 11: 627-634. [DOI] [PubMed] [Google Scholar]
- Arnason, U. and Janke, A. 2002. Mitogenomic analyses of eutherian relationships. Cytogenet. Genome Res. 96: 20-32. [DOI] [PubMed] [Google Scholar]
- Bailey, W.J., Fitch, D.H.A., Tagle, D.A., Czelusniak, J., Slightom, J.L., and Goodman, M. 1991. Molecular evolution of the ψη-globin gene locus: Gibbon phylogeny and the hominoid slowdown. Mol. Biol. Evol. 8: 155-184. [DOI] [PubMed] [Google Scholar]
- Bamshad, M. and Wooding, S.P. 2003. Signatures of natural selection in the human genome. Nat. Rev. Genet. 4: 99-111. [DOI] [PubMed] [Google Scholar]
- Basu, S., Nagy, J.A., Pal, S., Vasile, E., Eckelhoefer, I.A., Bliss, V.S., Manseau, E.J., Dasgupta, P.S., Dvorak, H.F., and Mukhopadhyay, D. 2001. The neurotransmitter dopamine inhibits angiogenesis induced by vascular permeability factor/vascular endothelial growth factor. Nat. Med. 7: 569-574. [DOI] [PubMed] [Google Scholar]
- Begun, D.J. and Aquadro, C.F. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519-520. [DOI] [PubMed] [Google Scholar]
- Bella, J., Kolatkar, P.R., Marlor, C.W., Greve, J.M., and Rossmann, M.G. 1998. The structure of the two amino-terminal domains of human ICAM-1 suggest how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. 95: 4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendt, A.R., Simmons, D.L., Tansey, J., Newbold, C.I., and Marsh, K. 1989. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 341: 57-59. [DOI] [PubMed] [Google Scholar]
- Brombacher, F. 2000. The role of interleukin-13 in infectious diseases and allergy. BioEssays 22: 646-656. [DOI] [PubMed] [Google Scholar]
- Bullard, D.C., King, P.D., Hicks, M.J., Dupont, B., Beaudet, A.L., and Elkon, K.B. 1997. Intercellular Adhesion Molecule-1 deficiency protects MRL/MpJ-Faslpr mice from early lethality. J. Immunol. 159: 2058-2067. [PubMed] [Google Scholar]
- Burnet, M., Guy, F., Pilbeam, D., Mackaye, H.T., Likius, A., Ahounta, D., Beauvilain, A., Blondel, C., Bocherens, H., Boisserie, J.R., et al. 2002. A new hominid from the Upper Miocene of Chad, Central Africa. Nature 418: 145-151. [DOI] [PubMed] [Google Scholar]
- Cao, L., Jiao, X., Zuzga, D.S., Liu, Y., Fong, D.M., Young, D., and During, M.J. 2004. VEGF links hippocampal activity with neurogenesis, learning and memory. Nat. Genet. 36: 827-835. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B. 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55: 199-221. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Background-selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. Camb. 68: 131-149. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 68: 131-149. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., Morgan, M.T., and Charlesworth, D. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., Charlesworth, B., and Morgan, M.T. 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F. and Li, W. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarello, A.G. and de Melo, F.R. 2001. Primate population densities and sizes in Atlantic forest remnants of northern Espírito Santo, Brazil. Int. J. Primatol. 22: 379-396. [Google Scholar]
- Chin, R.K., Lo, J.C., Kim, O., Blink, S.E., Christiansen, P.A., Peterson, P., Wang, Y., Ware, C., and Fu, Y.-X. 2003. Lymphotoxin pathway directs thymic Aire expression. Nat. Immun. 4: 1121-1127. [DOI] [PubMed] [Google Scholar]
- Clark, A.G., Glanowski, S., Nielsen, R., Thomas, P.D., Kejariwal, A., Todd, M.A., Tanenbaum, D.M., Civello, D., Lu, F., Murphy, B., et al. 2003. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302: 1960-1963. [DOI] [PubMed] [Google Scholar]
- Cochran, W.G. 1954. Some methods for strengthening the common χ2 test. Biometrics 10: 417-451. [Google Scholar]
- Crow, J.F. 1958. Some possibilities for measuring selection intensities in man. Hum. Biol. 30: 1-13. [PubMed] [Google Scholar]
- Crow, J.F. and Kimura, M. 1978. Efficiency of truncation selection. Proc. Natl. Acad. Sci. 76: 396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J.F. and Simmons, M.J. 1983. The mutation load in Drosophila. In The genetics and biology of Drosophila (eds. M. Ashburner, et al.), pp. 1-35. Academic Press, London.
- Dantz, D., Bewersdorf, J., Fruehwald-Schultes, B., Kern, W., Jelkmann, W., Born, J., Fehm, H.L., and Peters, A. 2002. Vascular endothelial growth factor: A novel endocrine defensive response to hypoglycemia. J. Clin. Endocr. Metab. 87: 835-840. [DOI] [PubMed] [Google Scholar]
- De Cosmo, S., Tassi, V., Thomas, S., Piras, G.P., Trevisan, R., Cavallo Perin, P., Bacci, S., Zucaro, L., Cisternino, C., Trischitta, V., et al. 2002. The Decorin gene 179 allelic variant is associated with a slower progression of renal disease in patients with type 1 diabetes. Nephron 92: 72-76. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Butcher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578-582. [DOI] [PubMed] [Google Scholar]
- Durrett, R. and Schweinsberg, J. 2004. Approximating selective sweeps. Theor. Pop. Biol. 66: 129-138. [DOI] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R.J. 1993. An introduction to the bootstrap. Chapman & Hall, London.
- Eyre-Walker, A. and Keightley, P.D. 1999. High genomic deleterious mutation rates in hominids. Nature 397: 344-347. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1974. The evolutionary advantage of recombination. Genetics 78: 737-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara, N. and Henzel, W.J. 1989. Pituitary follicular cells secrete a novel heparin-binding growth factor specific for vascular endothelial cells. Biochem. Biophys. Res. Commun. 161: 851-858. [DOI] [PubMed] [Google Scholar]
- Fu, Y.-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavan, J.A. 1953. Growth and development of the chimpanzee; a longitudinal and comparative study. Hum. Biol. 25: 93-143. [PubMed] [Google Scholar]
- Glass, R.I., Holmgren, J., Haley, C.E., Khan, M.R., Svennerholm, A.M., Stoll, B.J., Belayet Hossain, K.M., Black, R.E., Yunus, M., and Barua, D. 1985. Predisposition for cholera of individuals with O blood group. Possible evolutionary significance. Am. J. Epidemiol. 121: 791-796. [DOI] [PubMed] [Google Scholar]
- Glémin, S. 2003. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57: 2678-2687. [DOI] [PubMed] [Google Scholar]
- Goodman, M., Porter, C.A., Czelusniak, J., Page, S.L., Schneider, H., Shoshani, J., Gunnell, G., and Groves, C.P. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9: 585-598. [DOI] [PubMed] [Google Scholar]
- Gordo, I., Navarro, A., and Charlesworth, B. 2002. Muller's ratchet and the pattern of variation at a neutral locus. Genetics 161: 835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon, A. 1996. Regulation of ovarian follicular development in primates: Facts and hypotheses. Endocr. Rev. 17: 121-155. [DOI] [PubMed] [Google Scholar]
- Haldane, J.B.S. 1937. The effect of variation on fitness. Am. Naturalist 71: 337-349. [Google Scholar]
- ____. 1939. The equilibrium between mutation and random extinction. Ann. Eugen. 9: 400-405. [Google Scholar]
- Hamblin, M.T. and Aquadro, C.F. 1996. High nucleotide sequence variation in a region of low recombination in Drosophila simulans is consistent with the background-selection model. Mol. Biol. Evol. 13: 1133-1140. [DOI] [PubMed] [Google Scholar]
- Hammer, M.F., Blackmer, F., Garrigan, D., Nachman, M.W., and Wilder, J.A. 2003. Human population structure and its effects on sampling Y chromosome sequence variation. Genetics 164: 1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, R.M., Healy, E., Ray, A.J., Ellis, N.S., Flanagan, N., Todd, C., Dixon, C., Sajantila, A., Jackson, I.J., Birch-Machin, M.A., et al. 2000. Evidence for variable selective pressures at MC1R. Am. J. Hum. Genet. 66: 1351-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, M., Thorne, J.L., and Kishino, H. 2003. Time scale of eutherian evolution estimated without assuming a constant rate of molecular evolution. Genes Genet. Syst. 78: 267-283. [DOI] [PubMed] [Google Scholar]
- Hellmann, I., Ebersberger, I., Ptak, S.E., Pääbo, S., and Przeworski, M. 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. and Hurtado, A.M. 1996. Ache life history: The ecology and demography of a foraging people. Aldone de Gruyter, New York.
- Hill, W.G. and Robertson, A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269-294. [PubMed] [Google Scholar]
- Hudson, R.R. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337-338. [DOI] [PubMed] [Google Scholar]
- Hudson, R.R. and Kaplan, N.L. 1995. Deleterious background-selection with recombination. Genetics 141: 1605-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., Larget, B., and Swofford, D. 2000. A compound Poisson process for relaxing the molecular clock. Genetics 154: 1879-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson, A.M., Atmar, R.L., Graham, D.Y., and Estes, M.K. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185: 1335-1337. [DOI] [PubMed] [Google Scholar]
- Jin, K., Zhu, Y., Sun, Y., Mao, X.O., Xie, L., and Greenberg, D.A. 2002. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc. Natl. Acad. Sci. 99: 11946-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel, C.V., Briant, L., Udalova, I.A., Sevin, A., Nedospasov, S.A., and Cambon-Thomsen, A. 1991. Extensive genetic polymorphism in the human tumor necrosis factor region and relation to extended HLA haplotypes. Proc. Natl. Acad. Sci. 88: 9717-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N.L., Hudson, R.R., and Langley, C.H. 1989. The “hitchhiking effect” revisited. Genetics 123: 887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P.D. and Eyre-Walker, A. 1999. Terumi Mukai and the riddle of deleterious mutation rates. Genetics 153: 515-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome. Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbel, W.H., Walter, R.C., Johanson, D.C., Reed, K.E., Aronson, J.L., Assefa, Z., Marean, C.W., Eck, G.G., Robe, R., Hovers, E., et al. 1996. Late Pliocene Homo and Oldowan tolls from the Hadar Formation (Kada Hadar member), Ethiopia. J. Hum. Evol. 31: 549-561. [Google Scholar]
- Kimura, M. 1955. Stochastic processes and distribution of gene frequencies under natural selection. Cold Spring Harbor Symp. Quant. Biol. 20: 33-53. [DOI] [PubMed] [Google Scholar]
- ____. 1968. Evolutionary rate at the molecular level. Nature 217: 624-626. [DOI] [PubMed] [Google Scholar]
- Kimura, M. and Maruyama, T. 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., Maruyama, T., and Crow, J.F. 1963. The mutation load in small populations. Genetics 48: 1303-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman, J.F.C. 1982. The coalescent. Stochastic Processes Appl. 13: 235-248. [Google Scholar]
- Kitano, T., Schwarz, C., Nickel, B., and Pääbo, S. 2003. Gene diversity patterns at 10 X-chromosomal loci in humans and chimpanzees. Mol. Biol. Evol. 20: 1281-1289. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A.S. 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435-440. [DOI] [PubMed] [Google Scholar]
- ____. 1994. Muller's ratchet under epistatic selection. Genetics 136: 1469-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Sex and U. Trends Genet. 17: 75-77. [DOI] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241-247. [DOI] [PubMed] [Google Scholar]
- Kumar, S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J. and Hurst, L.D. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337-340. [DOI] [PubMed] [Google Scholar]
- LeVine, A.M., Reed, J.A., Kurak, K.E., Cianciolo, E., and Whitsett, J.A. 1999. GM-CSF-deficient mice are susceptible to pulmonary group B streptococcal infection. J. Clin. Invest. 103: 563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H. and Sadler, L.A. 1991. Low nucleotide diversity in man. Genetics 129: 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H., Yi, S.J., and Makova, K. 2002. Male-driven evolution. Curr. Opin. Genet. Dev. 12: 650-656. [DOI] [PubMed] [Google Scholar]
- Li, H., Meng, S.-J., Men, Z.-M., Fu, Y.-X., and Zhang, Y.-P. 2003. Genetic diversity and population history of golden monkeys (Rhinopithecus roxellana). Genetics 164: 269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J. and Haigh, J. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23-35. [PubMed] [Google Scholar]
- McDougall, I., Brown, F.H., and Fleagle, J.G. 2005. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature 433: 733-736. [DOI] [PubMed] [Google Scholar]
- Muller, H.J. 1950. Our load of mutations. Am. J. Hum. Genet. 2: 111-176. [PMC free article] [PubMed] [Google Scholar]
- Nachman, M.W. 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17: 481-485. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W. and Crowell, S.L. 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M.W., Bauer, V.L., Crowell, S.L., and Aquadro, C.F. 1998. DNA variability and recombination rates at X-linked loci in humans. Genetics 150: 1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel, J.V., Satoh, C., Goriki, K., Asakawa, J., Fujita, M., Takahashi, N., Kageoka, T., and Hazama, R. 1988. Search for mutations altering protein change and/or function in children of atomic bomb survivors: Final report. Am. J. Hum. Genet. 42: 663-676. [PMC free article] [PubMed] [Google Scholar]
- Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York.
- Nei, M. and Li, W.H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. 76: 5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neter, J., Kutner, M.H., Nachtsheim, C.J., and Wasserman, W. 1996. 10.1 Unequal error variances remedial measures—Weighted least squares. In Applied linear statistical models (eds. R.T. Hercher et al.), pp. 400-409. Richard D. Irwin, Inc., Chicago.
- Nishida, T., Takasaki, H., and Takahata, Y. 1990. Demography and reproductive profiles. In Chimpanzees of the Mahale Mountains (ed. T. Nishida), pp. 63-97. Tokyo University Press, Tokyo.
- Nordborg, M., Charlesworth, B., and Charlesworth, D. 1996. The effect of recombination on background selection. Genet. Res. 67: 159-174. [DOI] [PubMed] [Google Scholar]
- Osada, N. and Wu, C.-I. 2005. Inferring the mode of speciation from genomic data: A study of the great apes. Genetics 169: 259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson, S. and Pamilo, P. 1999. The effects of deleterious mutations on linked, neutral variation in small populations. Genetics 153: 475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, E.L., Marcini, A., Akey, J., Martinson, J., Batzer, M.A., Cooper, R., Forrester, T., Allison, D.B., Deka, R., Ferrell, R.E., et al. 1998. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 63: 1839-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M. 2002. The signature of positive selection at randomly chosen loci. Genetics 160: 1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M., Charlesworth, B., and Wall, J.D. 1999. Genealogies and weak purifying selection. Mol. Biol. Evol. 16: 246-252. [DOI] [PubMed] [Google Scholar]
- Przeworski, M., Hudson, R.R., and Di Rienzo, A. 2000. Adjusting the focus on human variation. Trends Genet. 16: 296-302. [DOI] [PubMed] [Google Scholar]
- Ptak, S.E. and Przeworski, M. 2002. Evidence for population growth in humans is confounded by fine-scaled population structure. Trends Genet. 18: 559-563. [DOI] [PubMed] [Google Scholar]
- Rockman, M.V., Hahn, M.W., Soranzo, N., Goldstein, D.B., and Wray, G.A. 2003. Positive selection on a human-specific transcription factor binding site regulating IL4 expression. Curr. Biol. 13: 2118-2123. [DOI] [PubMed] [Google Scholar]
- Rogers, J., Mahaney, M.C., Witte, S.M., Nair, S., Newman, D., Wedel, S., Rodriguez, L.A., Rice, K.S., Silfer, S.H., Perelygin, A., et al. 2000. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics 67: 237-247. [DOI] [PubMed] [Google Scholar]
- Satta, Y., Hickerson, M., Watanabe, H., O'hUigin, C., and Klein, J. 2004. Ancestral population sizes and species divergence times in the primate lineage on the basis of intron and BAC end sequences. J. Mol. Evol. 59: 478-487. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C. 2002. Towards a molecular characterization of adaptation in local populations. Curr. Opin. Genet. Dev. 12: 683-687. [DOI] [PubMed] [Google Scholar]
- Shabalina, S.A., Ogurtsov, A.Y., Kondrashov, V.A., and Kondrashov, A.S. 2001. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 17: 373-376. [DOI] [PubMed] [Google Scholar]
- Simonsen, K.L., Churchill, G.A., and Aquadro, C.F. 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141: 413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J.E. and Hahn, M.W. 2005. Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 22: 63-73. [DOI] [PubMed] [Google Scholar]
- Stauffer, R.L., Walker, A., Ryder, O.A., Lyons-Weiler, M., and Hedges, S.B. 2001. Human and ape molecular clocks and constraints on paleontological hypotheses. J. Hered. 92: 469-474. [DOI] [PubMed] [Google Scholar]
- Stephan, W. 1995. An improved method for estimating the rate of fixation of favorable mutations based on DNA polymorphism data. Mol. Biol. Evol. 12: 959-962. [DOI] [PubMed] [Google Scholar]
- Stephan, W., Wiehe, T.H.E., and Lenz, M.W. 1992. The effect of strongly selected substitutions on neutral polymorphism—Analytical results based on diffusion-theory. Theor. Popul. Biol. 41: 237-254. [Google Scholar]
- Storz, J.F., Ramakrishnan, U., and Alberts, S.C. 2002. Genetic effective size of a wild primate population: Influence of current and historical demography. Evolution 56: 817-829. [DOI] [PubMed] [Google Scholar]
- Tachida, H. 2000. Molecular evolution in a multistate nearly neutral mutation model. J. Mol. Evol. 50: 69-81. [DOI] [PubMed] [Google Scholar]
- Tajima, F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N. and Satta, Y. 1997. Evolution of the primate lineage leading to modern humans: Phylogenetic and demographic inferences from DNA sequences. Proc. Natl. Acad. Sci. 94: 4811-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., Satta, Y., and Klein, J. 1995. Divergence time and population size in the lineage leading to modern humans. Theor. Popul. Biol. 48: 198-221. [DOI] [PubMed] [Google Scholar]
- Tarazona-Santos, E. and Tishkoff, S.A. 2004. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes Immun. 6: 53-65. [DOI] [PubMed] [Google Scholar]
- Valentin, A., Lu, W., Rosati, M., Schneider, R., Albert, J., Karlsson, A., and Pavlakis, G.N. 1998. Dual effect of interleukin 4 on HIV-1 expression: Implications for viral phenotypic switch and disease progression. Proc. Natl. Acad. Sci. 95: 8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J.D. 2003. Estimating ancestral population sizes and divergence times. Genetics 163: 395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Argawala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Watterson, G.A. 1975. On the number of segregating sites in genetical models without recombination. Theor. Pop. Biol. 7: 256-276. [DOI] [PubMed] [Google Scholar]
- White, T.D., Asfaw, B., DeGusta, D., Gilbert, H., Richards, G.D., Suwa, G., and Howell, F.C. 2003. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423: 742-747. [DOI] [PubMed] [Google Scholar]
- Williamson, S. and Orive, M.E. 2002. The genealogy of a sequence subject to purifying selection at multiple sites. Mol. Biol. Evol. 19: 1376-1384. [DOI] [PubMed] [Google Scholar]
- Wray, G.A., Hahn, M.W., Abouheif, E., Balhoff, J.P., Pizer, M., Rockman, M.V., and Romano, L. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377-1419. [DOI] [PubMed] [Google Scholar]
- Xu, H., Gonzalo, J.A., St Pierre, Y., Williams, I.R., Kupper, T.S., Cotran, R.S., and Springer, T.A. 1994. Leukocytosis and resistance to septic shock in intercellular adhesion molecule-1-deficient mice. J. Exp. Med. 180: 95-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. 2002. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162: 1811-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Yoder, A.D. 2003. Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene loci and calibration points, with application to a radiation of cute-looking mouse lemur species. Syst. Biol. 52: 705-716. [DOI] [PubMed] [Google Scholar]
- Yoder, A.D. and Yang, Z. 2000. Estimation of primate speciation dates using local molecular clocks. Mol. Biol. Evol. 17: 1081-1090. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/; UCSC Human Genome Browser.
- http://pga.gs.washington.edu; SeattleSNPs, NHLBI Program for Genomic Applications, UW-FHCRC, Seattle, WA, March 26, 2005.
- http://www.ncbi.nlm.nih.gov/omim/; Online Mendelian Inheritance in Man (OMIM).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.