Abstract
Peptides from the N-heptad repeat region of the HIV gp41 protein can inhibit viral fusion, but their potency is limited by a low tendency to form a trimeric coiled-coil. Accordingly, stabilization of N peptides by fusion with the stable coiled-coil IZ yields nanomolar inhibitors [Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187-11192]. Because the antiviral potency of IZN17 is limited by self-association equilibrium, we covalently stabilized the peptide by using interchain disulfide bonds. The resulting covalent trimer, (CCIZN17)3, has an extraordinary thermodynamic stability that translates into unprecedented antiviral potency: (CCIZN17)3 (i) inhibits fusion in a cell-cell fusion assay (IC50 = 260 pM); (ii)is the most potent fusion inhibitor described to date (IC50 = 40-380 pM) in a single-cycle infectivity assay against HIVHXB2, HIVNL4-3, and HIVMN-1; (iii) efficiently neutralizes acute viral infection in peripheral blood mononuclear cells; and (iv) displays a broad antiviral profile, being able to neutralize 100% of a large panel of HIV isolates, including R5, X4, and R5/X4 strains. In all of these assays, the potency of N-peptide inhibitor (CCIZN17)3 was equal to or more than the C-peptide inhibitor in clinical use, DP178 (also known as Enfuvirtide and Fuzeon). More importantly, we show that the two inhibitors, which have different targets in gp41, synergize when used in combination. These features make (CCIZN17)3 an attractive lead to develop as an antiviral drug, alone or in combination with DP178, as well as a promising immunogen to elicit a fusion-blocking neutralizing antibody response.
Keywords: fusion inhibitor
HIV type 1 (HIV-1) infects cells by fusing its membrane with the host cell membrane. The key player in viral entry into target cells is the envelope glycoprotein complex gp160, which is composed of the subunits gp120 and gp41. Before exposure to cellular receptors, the two subunits form a noncovalent complex that associates in a trimer to form spikes at the virus surface. Binding of gp120 to the host cellular receptors triggers a cascade of conformational changes in the envelope complex, culminating with the insertion of the transmembrane subunit gp41 into the host cell membrane. The mechanism of fusion involves two helical regions of gp41, an N-terminal heptad repeat (HR), HR1 (defined as N helix or N peptide in various studies), and a C-terminal HR, HR2 (defined as C helix or C peptide). HR1 and HR2 form a fusogenic (i.e., fusion-active) conformation called the “trimer-of-hairpins,” a structure common to the fusion mechanism of many enveloped viruses (1, 2) consisting of a bundle of six α-helices contributed by three gp41 monomers. Three C peptides, one per gp41 chain, pack in an antiparallel manner against a central three-stranded coiled-coil formed by the N regions of the same chains (3-6). It is generally accepted that fusion progresses by formation of an intermediate, a “prehairpin” conformation, that places the N-terminal fusion peptide near or in the target cell membrane, exposing the HR1 and HR2 regions (1). In this intermediate, both HR are vulnerable to binding by synthetic C and N peptides, which can thus inhibit viral infection by preventing formation of the fusogenic trimer-of-hairpins. C peptides are potent inhibitors of HIV-1, active at low nanomolar concentration (7, 8). One of them, DP178 or T-20, has been approved with the name Enfuvirtide (also known by its brand name Fuzeon) as the first member of a new class of antiretroviral drugs known as fusion inhibitors (9, 10).
Antiviral activity was also reported for N peptides. Two mechanisms were proposed: interference with trimer-of-hairpin formation by binding to the C peptides (1, 11, 12) or disruption of the inner coiled-coil by intercalating within the N helices (3, 13). Linear N peptides are generally far less potent than C peptides, possibly because in the absence of the latter they aggregate and are sequestered in nonproductive intermolecular assemblies. Accordingly, a correlation exists between the solution structure of N peptides and their antiviral activity (7, 8, 14).
Starting from this observation, very potent engineered peptides were described. Root et al. (15) designed the single-chain construct 5-helix, with alternating N and C helices (three and two, respectively) linked by interconnecting loops. 5-helix folds into a structure similar to the trimer-of hairpins, but with an unoccupied binding site for a C peptide. Accordingly, it binds a C peptide very efficiently, and inhibits HIV-1 at low nanomolar concentration. Louis et al. (16) designed a soluble trimeric coiled-coil, NCCG-gp41, with the N helix fused in helical phase to a minimal thermostable gp41 trimeric core and the smaller, soluble constructs N34CCG and N35CCG-N13, consisting of the internal trimeric gp41 coiled-coil, stabilized by intermolecular disulfide bonds (17). The increased stability translated to nanomolar inhibitory potency. Importantly, one of the constructs, N35CCG-N13, was able to elicit neutralizing antibodies (17), thus validating the prehairpin intermediate as a vaccine target.
Other low nanomolar N-peptide inhibitors were reported by Eckert and Kim (11). These inhibitors were chimeric molecules, consisting of a designed trimeric coiled-coil (18, 19) fused to a portion of the N helix. Eckert and Kim showed that the inhibitory activity was correlated to the stability of the coiled-coil structure.
Overall, these studies demonstrated that appropriate engineering of N peptides could increase their inhibitory potency by three orders of magnitude. Building on these observations, we describe here chimeric N peptides that combine the concept of fusion with a stable coiled-coil domain with the concept of covalent stabilization by using disulfide bonds. We show that these covalent chimeric constructs display antiviral potency in the picomolar range, with a remarkable breadth of neutralization against a large panel of HIV strains, including many primary isolates.
Materials and Methods
Peptide Synthesis and CD. All of the peptides in Table 1 were synthesized by solid-phase synthesis. For full details on peptide synthesis and CD, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Table 1. Sequences of HIV-1 gp41 N helix and derived chimeric peptides inhibitors.
| Peptide | Sequence |
|---|---|
| d a d a d a d a d a d a d | |
| N helix | QARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLK |
| N36 | SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL |
| N23 | IEAQQHLLQLTVWGIKQLQARIL |
| N17 | LLQLTVWGIKQLQARIL |
| IZN36 | IKKEIEAIKKEQEAIKKKIEAIEKEISGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL |
| IZN23 | IKKEIEAIKKEQEAIKKKIEAIEKEIEAQQHLLQLTVWGIKQLQARIL |
| IZN17 | IKKEIEAIKKEQEAIKKKIEAIEKLLQLTVWGIKQLQARIL |
| N17IZ | LLQLTVWGIKQLQARILAIKKEIEAIKKEQEAIKKKIEAI |
| CCIZN17 | CCGGIKKEIEAIKKEQEAIKKKIEAIEKLLQLTVWGIKQLQARIL |
| CCIZN23 | CCGGIKKEIEAIKKEQEAIKKKIEAIEKEIEAQQHLLQLTVWGIKQLQARIL |
| CCI10N17 | CCGGIKKKIEAIEKLLQLTVWGIKQLQARIL |
The HR1 sequence corresponds to residues 540-588 of HIV-HXB2, with alignment of corresponding positions a and d of the heptad repeats of the coiled-coil. Non-HIV residues are shown in italic; HIV residues are underlined. All peptides feature C-terminal carboxyamide and N-terminal acetyl, except IZN23 and CCIZN23 (which have N-terminal biotin).
HIV-1 Neutralization Assays. Three different assay formats were used. The first format was a single-cycle HIV-1 infectivity assay. P4-2/R5 cells (20, 21) are HeLa cells stably expressing human CD4 and CC-chemokine receptor 5 and harboring a β-galactosidase reporter gene driven by a tat-responsive fragment of the HIV-2 LTR. P4-2/R5 cells were maintained at 37°C and 5% CO2 in phenol red-free DMEM/10% FBS. For infectivity assays, cells were seeded in 96-well plates (Costar) at 2.5 × 103 cells per well and infected the following day with HIV-1 at a multiplicity of infection of 0.01 in the presence of peptides. Forty-eight hours later, cells were lysed and β-galactosidase activity was measured with a chemiluminescent substrate as described in ref. 22.
The second format was a spread assay using MT4 cells or peripheral blood mononuclear cells (PBMCs). Cells at 4 × 105 per ml (MT4 cells) or 106 per ml (PBMCs) were infected at a multiplicity of infection of 0.01, washed free of virus 24 h after infection and seeded into wells of the 96-well plates. Test peptides were diluted by two-fold serial dilutions and mixed with cells. Details of plate assembly for the (CCIZN17)3/DP178 combination experiment are given in Supporting Materials and Methods. Cultures were incubated at 37°C and 5% CO2 for 72 h and then assayed for viral production by a commercial p24 assay kit (Beckman Coulter).
The third format was a commercial phenotypic virus assay (ViroLogic, South San Francisco, CA) described in ref. 23.
HIV-1 Cell-Cell Fusion Assay. Inhibition of cell-cell fusion was measured by using a quantitative assay essentially as described in ref. 24. Briefly, inhibitors were titrated into cultures containing a mixture of SupT1 cells overexpressing cytoplasmic β-lactamase and HeLa cells expressing HXB2 gp160. HeLa cells were also loaded with the β-lactamase fluorescent substrate, CCF4-AM. Fusion was measured by quantifying CCF4 fluorescence in the blue and green channels, with the blue/green fluorescence ratio being proportional to the extent of fusion.
Results
Structure Activity Relationship of Chimeric N-Peptide Inhibitors. Our starting point was the study by Eckert and Kim (11). These authors compared the antiviral potency of chimeric N peptides consisting of the designed trimeric coiled-coil IZ (18, 19) fused to portions of the gp41 N helix, namely IZN17, IZN23, and IZN36 (Table 1). N17 is the portion of the N region comprising the gp41 hydrophobic pocket (6, 25). N23 and N36 include 6 or 19 (respectively) additional residues upstream to the N17 region (Table 1). In addition to providing an important set of contacts for the cognate C peptide during formation of the trimer-of-hairpin (6, 25), the N17 region contributes significantly to the stability of HR1 self-association, which is completely abolished by deletion of the hydrophobic pocket (26). IZN17, IZN23, and IZN36 all have similar inhibitory potency (11), suggesting that the antiviral activity of this class of chimeric N peptides is recapitulated in the N17 portion of the N helix, which we used for all our subsequent work.
To complement the data reported by Eckert and Kim (11), we investigated how the position of the IZ scaffold influences antiviral potency. We prepared the analog N17IZ, with the IZ domain C-terminal instead of N-terminal to the N17 sequence. An alanine was included between N17 and IZ to maintain the alternating a-to-g heptad repeat in frame (Table 1). Examination of the structure of the fusion-active 6-helical bundle suggests that, although IZN17 should be able to capture the N-terminal portion of the C helix, the C-terminal portion of the C helix would have to partially twist in order not to clash against the coiled-coil groove of IZ. By positioning the IZ domain in a different structural context, one could hope to reduce the steric hindrance to C-peptide binding, hence improving antiviral potency.
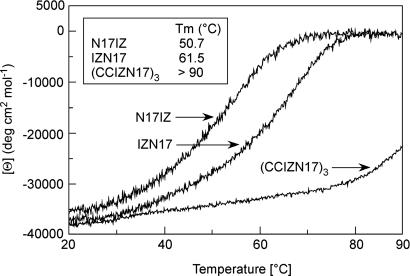
N17IZ is fully helical at 10 μM as its parent peptide IZN17 (Table 2; see also Fig. 6, which is published as supporting information on the PNAS web site). N17IZ is also very stable, with a melting temperature (Tm) of >90°C. In the presence of 2 M guanidinium chloride, N17IZ shows a cooperative thermal transition, with Tm = 50.7°C, whereas, for IZN17, Tm = 61.5°C (Table 2 and Fig. 1). Despite the preserved structural stability, N17IZ shows dramatically reduced antiviral activity, with IC50 = 800 nM, almost three orders of magnitude lower than IZN17 (Table 2). Structural studies have shown that the N-helix bundle extends only seven more residues at the C terminus of N17, followed by a region interconnecting the N and C helices (3). Our data suggest that a C-terminally positioned IZ domain may clash with some region of the fusion intermediate other than the C helix.
Table 2. Biophysical data and HIV-1 inhibitory activity of chimeric N peptides.
| Peptide | Helicity at 10 μM | Tm, °C | Tm 2 M GdnHCl, °C | IC50, nM |
|---|---|---|---|---|
| IZN17 | 99 | >90 | 61.5 | 1.26 |
| N17IZ | 96 | >90 | 50.7 | 799.94 |
| IZN23 | nd | >90 | nd | 2.33 |
| (CCIZN17)3 | 97 | >90 | >90 | 0.04 |
| (CCIZN23)3 | 100 | >90 | >90 | 0.31 |
| CCI10N17 | 65 | nd | nd | nd |
| (CCI10N17)3 | 85 | 60 | nd | 23.76 |
| 5-helix | nd | nd | nd | 9.93 |
| DP178 | nd | nd | nd | 1.14 |
Helicity was determined by CD spectroscopy. Tm is the midpoint of thermal denaturation transitions by CD spectroscopy. Shown is Tm determined alone and Tm in the presence of 2 M guanidinium chloride. IC50 values were determined from a single-cycle infectivity assay using HIV-HXB2. n.d., not done.
Fig. 1.
Thermal denaturation curves of IZN17, N17IZ, and (CCIZN17)3, as monitored by CD at 222 nm in 5 mM Hepes, pH 7.3/150 mM NaCl/2 M guanidinium hydrochloride. The melting temperatures (Tm), are 50.7°C, 61.5°C, and >90°C for N17IZ, IZN17, and (CCIZN17)3, respectively.
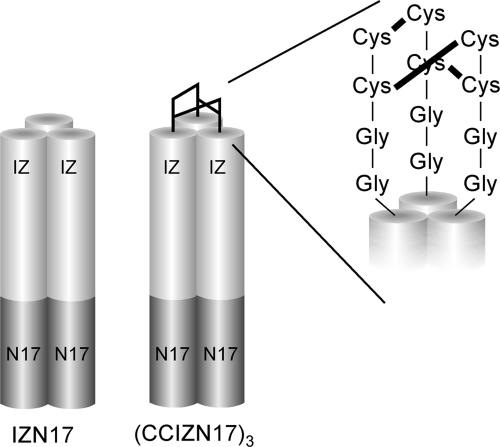
Covalently Trimeric Chimeric N-Peptide Inhibitors. The above data confirmed that the chimeric peptide IZN17 represented the best starting point for further improvement. We reasoned that, in this molecule, the presence of a coiled-coil structure depends on the monomer/trimer equilibrium in solution, and, thus, strictly depends on concentration. The observed antiviral potency of IZN17 should therefore result from the interplay between the binding constant of the trimer to the C peptide and the constant of oligomerization to form the coiled-coil. Rather than try to design more stable trimerization scaffolds to overcome this putative limitation, we designed a construct, CCIZN17, which can form a covalently stabilized coil-coil structure (Table 1 and Fig. 2). Similar to Louis et al. (16, 17), we introduced two cysteine residues in the monomeric IZN17 precursor to form three interchain disulfide bridges upon spontaneous assembly of the monomers into a trimer. Instead of introducing the cysteines inside the heptad repeat region of the coiled-coil as in the Louis et al. design (16, 17), we located a cysteine pair at the N terminus of the IZ domain that was spatially separated from the coiled-coil domain by two additional glycine residues. The glycine residues would endow the pair of cysteines with sufficiently high conformational freedom during the spontaneous formation of the interchain disulfide bonds.
Fig. 2.
Schematic model of the designed IZN17 and (CCIZN17)3 molecules. The IZN17 helices forming a homotrimeric coiled-coil are represented by cylinders. Each chimeric N peptide consists of an N-terminal designed trimeric coiled-coil (light gray) fused to a portion of the sequence from the N-helix region of gp41 (dark gray), namely N17 (residues 568-584 of HIVHXB2). The (CCIZN17)3 helices are covalently stabilized at the N terminus by three interchain disulfides between each pair of cysteines of each peptide chain. Only one of the possible combinations of the three disulfides is drawn.
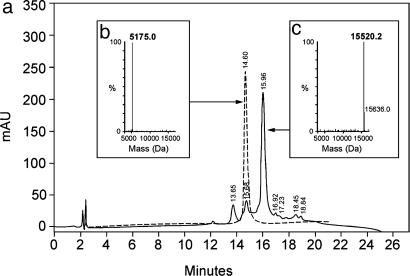
When dissolved at neutral pH, IZN17 folds spontaneously into a stable trimeric coiled-coil, with three peptide chains physically associated in parallel orientation (Fig. 2) (11). Similarly, three identical cysteine-containing molecules of CCIZN17 associate into a trimeric coiled-coil structure, and the resulting close proximity of the juxtaposed cysteine residues allows the formation of three intermolecular disulfide bonds under oxidizing conditions, as shown schematically in Fig. 2 and experimentally with HPLC-MS in Fig. 3.
Fig. 3.
Formation of the covalent trimer (CCIZN17)3 monitored by HPLC-MS. (a) Overlay of the chromatograms of the precursor CCIZN17 (dashed line) and of the oxidized (CCIZN17)3 after 18 h of incubation (solid line). (b and c) Hypermass reconstruction of the raw electrospray MS data of monomeric CCIZN17 (observed mass, 5,175.0 Da; calculated mass, 5,175.0 Da) (b) and the covalent trimer (CCIZN17)3, whose observed mass of 15,520.1 Da (calculated mass, 15,520.2 Da), is in agreement with the formation of three disulfide bonds (c).
The covalent trimer (CCIZN17)3 is fully helical, with a CD spectrum superimposable to that of IZN17 (Fig. 6). Thermal denaturation experiments show that, in the presence of 2 M guanidine hydrochloride, (CCIZN17)3 has a Tm that is >90°C, compared with a Tm of 61.5°C for IZN17 (Table 2 and Fig. 1). It appears, therefore, that covalent stabilization of the IZN17 trimer yields a molecule with an extraordinary thermodynamic stability.
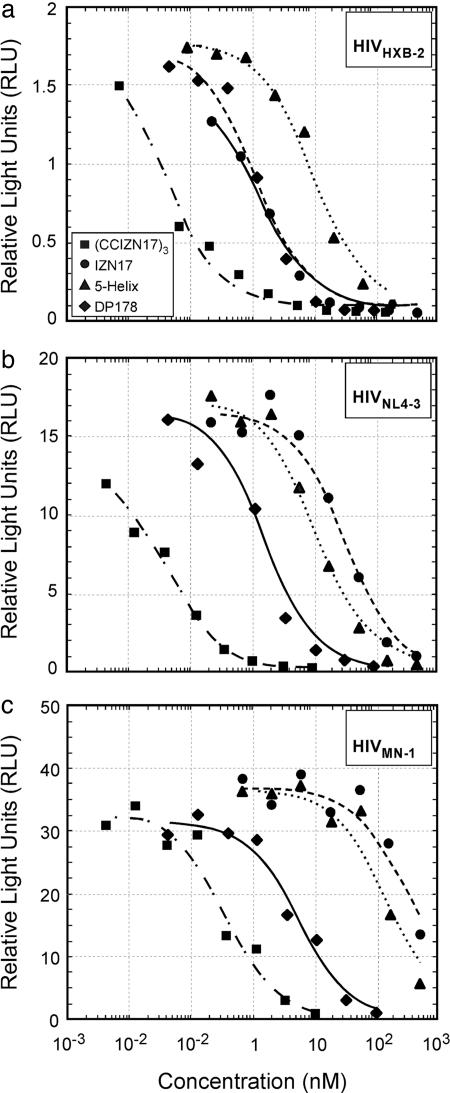
(CCIZN17)3 Is an Extremely Potent Inhibitor of Viral Infectivity. We next tested (CCIZN17)3 in a single-cycle infectivity assay using three viral isolates (HIVHXB2, HIVNL4-3, and HIVMN-1) and compared its activity with other N-peptide inhibitors and DP178, which is a C-peptide inhibitor (Table 2 and Fig. 4). (CCIZN17)3 displayed subnanomolar activity against all three strains (IC50 values of 0.04, 0.05, and 0.38 nM, respectively) thus being 30- to 700-fold more potent than the parent peptide IZN17 (IC50 values of 1.3, 33.7, and 394.7 nM, respectively). (CCIZN17)3 was also 250- to 400-fold more potent than 5-helix (IC50 values of 9.9, 12.0, and 159.4 nM, respectively). Its antiviral activity was also greater than the potent C-peptide drug DP178 (IC50 values of 1.2, 1.7, and 5.5 nM, respectively). Notably, (CCIZN17)3 potently blocks infection by the primary isolate HIV89.6 (IC50 = 5.13 nM), which is completely resistant to IZN17 and only poorly neutralized by 5-helix (IC50 = 213 nM).
Fig. 4.
Inhibition of infectivity of various HIV isolates. The calculated IC50 values for (CCIZN17)3, IZN17, 5-helix, and DP178 are (respectively) 0.04, 1.3, 9.9, and 1.2 nM (against HIVHXB2) (a); 0.046, 33.66, 12.0, and 1.7 nM (against HIVNL4-3)(b); and 0.38, 394.7, 159.4, and 5.5 nM (against HIVMN-1)(c).
(CCIZN17)3 was then tested in a PBMC infectivity assay. The results in Table 3 are shown as IC95 (nM) for the three primary isolates HIV89.6, HIVBaL, and HIVJR-FL and the lab-adapted virus, HIVH9IIIb. The assay was performed in PBMCs from two different healthy donors, and results were in good agreement. (CCIZN17)3 potently neutralized three of four viruses, with an IC95 of one order of magnitude lower than that of IZN17, and it was also somewhat more potent than DP178. The primary isolate HIV89.6 was not inhibited by IZN17, barely inhibited by (CCIZN17)3, and only weakly inhibited by DP178 (Table 3).
Table 3. Comparative inhibition of infection of PBMCs from two different donors by (CCIZN17)3, IZN17, and DP178.
| IC95, nM
|
||||||||
|---|---|---|---|---|---|---|---|---|
| HIV-189.6
|
HIV-1BaL
|
HIV-1H9IIIb
|
HIV-1JR-FL
|
|||||
| Inhibitor | D1 | D2 | D1 | D2 | D1 | D2 | D1 | D2 |
| (CCIZN17)3 | >500 | 500 | 63 | 63 | 15 | 8 | 63 | 63 |
| IZN17 | >500 | >1000 | >500 | 500 | 250 | 125 | 500 | 250 |
| DP178 | 500 | 250 | >500 | 250 | 63 | 15 | 250 | 125 |
The IC95 is the concentration of test compound that inhibited virus growth by at least 95% compared with untreated infected control cultures. D1, assay run in PBMC from donor 1; D2, assay run in PBMC from donor 2.
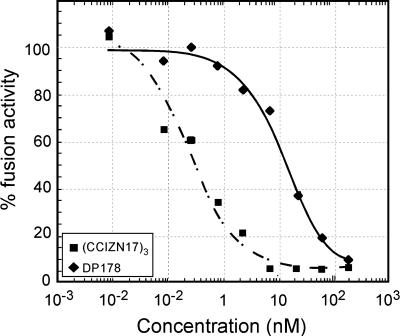
(CCIZN17)3 Is a Picomolar Inhibitor of HIV Fusion. We next examined the ability of (CCIZN17)3 to interfere with the fusion process in a cell-cell fusion assay and compared its activity to DP178. (CCIZN17)3 potently inhibits entry at subnanomolar concentrations (IC50 = 260 pM), showing a 55-fold higher potency than DP178 (IC50 = 14.5 nM) (Fig. 5).
Fig. 5.
Inhibition of HIV fusion in a cell-cell fusion assay. The calculated IC50 values for (CCIZN17)3 and DP178 are 0.26 nM and 14.5 nM, respectively.
(CCIZN17)3 Displays a Broad Antiviral Profile. To determine the breadth of neutralization of (CCIZN17)3, the peptide was tested in a commercial phenotypic virus assay (ViroLogic) (23) against a large panel of HIV isolates (Table 4, which is published as supporting information on the PNAS web site). The panel included R5, X4, and R5/X4 isolates. Remarkably, (CCIZN17)3 was able to neutralize 100% of the strains tested, with potencies between 3.9 and 202 nM. This variable susceptibility to inhibition is comparable with what had been observed for the DP178: among 14 viral isolates tested, susceptibility varied over an ≈30-fold range (24). It is now being recognized that resistance to fusion inhibitors is complex (27, 28) and can derive from multiple factors other than the sequence of the gp41 HR (29, 30). Accordingly, the strains in Table 4, which are more resistant to (CCIZN17)3, are also more resistant to known neutralizing antibodies to gp120 and gp41 (M.D.M. and R.G., unpublished data).
Combination of (CCIZN17)3 with the C-Peptide Inhibitor DP178. Because (CCIZN17)3 and DP178 target different regions of the prehairpin intermediate and because DP178 does not include the HR2 region, which binds to the hydrophobic pocket of N17 (6), the two inhibitors should work independently and could in principle be administered in combination. Combination therapy has been shown to be particularly effective in minimizing the emergence of resistant viruses, which are quickly selected during the course of therapy with a single drug (31). Therefore, we performed an experiment in which the two fusion inhibitors were administrated separately and in combination to assess their individual and joint effects in a spread assay in MT4 cells. The joint effect is defined as antagonistic, additive, or synergistic when the response to the combination is respectively worse, the same, or better than the expected additive response, respectively.
For (CCIZN17)3 and DP178, the combination showed a strong synergistic effect at most of the dose combinations and an additive effect at the marginal dose combinations, i.e., when the concentration of one inhibitor was too low for a detectable inhibition or high enough to provide almost full inhibition by itself (Table 5, which is published as supporting information on the PNAS web site).
Other Cysteine-Capped IZN Inhibitors. The strategy of covalent stabilization by interchain disulfide bridge formation was then applied to other chimerae. We first prepared the covalent trimer of IZN23, (CCIZN23)3 (Table 1), which was readily formed upon incubation at neutral pH of the precursor CCIZN23 (see Supporting Materials and Methods). This molecule is also very stable, with Tm > 90°C in 2 M guanidinium hydrochloride. In the single-cycle infectivity assay, IZN23 showed an IC50 of 2.3 nM against HIVHXB2, whereas (CCIZN23)3 showed an IC50 of 0.31 nM (Table 2). Thus, also in this case the covalently stabilized trimeric (CCIZN23)3 is more potent than the parent noncovalent inhibitor, IZN23. However the gain in potency is only 7-fold, confirming that covalent stabilization is optimally applied to the N17 portion of the N helix, which recapitulates most of the binding energy required for fusion inhibition.
IZ Is Indispensable for the Antiviral Activity of Covalently Chimeric N Peptides. We finally investigated whether covalent stabilization might compensate for reduction in the favorable packing interactions provided by a shortened IZ coiled-coil. Thus, we designed (CCI10N17)3, a chimeric construct comprising the N17 residues and only 10 of the original 24 IZ residues (I10) (Table 1). Upon incubation at neutral pH, the precursor CCI10N17 oxidized to form the covalent trimer (CCI10N17)3 but with a yield of only 40%, the remainder being dimeric and tetrameric species. When the precursor CCI10N17 was analyzed by CD, it was found to be only 61% helical at 15 μM concentration, unlike IZN17 which is fully helical under the same conditions (Fig. 7, which is published as supporting information on the PNAS web site). This result confirms that the scaffold length is critical for the formation of non-covalently-stabilized, chimeric N-peptide coiled-coils. Moreover, the covalent trimer (CCI10N17)3 is only 85% helical (Fig. 7), with a thermal denaturation curve showing poor cooperativity, and a Tm of only 60°C (Table 2). Overall, the data indicate that, in this case, covalent trimerization via a cysteine cap provides only marginal stabilization to the helical structure (from 61% to 85%) and, thus, that a longer scaffold is essential to nucleate a stable trimer. Consistent with previous observations, (CCI10N17)3 is a much less potent HIV inhibitor than (CCIZN17)3, with an IC50 of only 19 nM against HIVHXB2 (Table 2).
Discussion
When suitably stabilized in a coiled-coil conformation, N peptides are potent inhibitors of HIV-1 entry, with IC50 values in the low nanomolar range (11, 15-17). In this study, we focused our attention on the potent chimeric IZN17 peptide described by Eckert and Kim (11). The N17 sequence is the portion of the N region that comprises the gp41 hydrophobic pocket (6, 23), the region within HR1 that provides the most important set of contacts for binding of the cognate HR2 domain. Accordingly, within the IZN peptide series, IZN17 recapitulates all of the requirements for inhibitory potency, because addition of more gp41 residues to produce IZN23 and IZN36 does not result in better inhibitors (11).
To design a more potent inhibitor, we focused on N17 and found that the location of the scaffolding IZ domain relative to N17 (N- or C-terminal) has a dramatic impact on antiviral activity, as shown by the 800-fold loss in potency of N17IZ with respect to IZN17, despite the comparable structural stability (Table 2). We then addressed the main potential limitation of this inhibitor, i.e., the self-association equilibrium. The antiviral activity of the molecule depends on the presence of a trimeric coiled-coil structure, which is necessarily concentration-dependent. We thus designed a construct, CCIZN17, which can form a coiled-coil structure covalently stabilized by interchain disulfide bonds. Previously, disulfide-stabilized coiled-coil trimers of gp41 (16, 17) showed increased helical content with respect to N peptides that translated to nanomolar antiviral activity. Our covalent scaffolded trimeric (CCIZN17)3 construct forms a highly stable fully helical structure that very efficiently inhibits cell-cell fusion. In a single-cycle infectivity assay against various HIV isolates, (CCIZN17)3 shows much increased potency with respect to IZN17, with subnanomolar antiviral activity. Moreover, its activity is one order of magnitude higher than IZN17 in a PBMC infectivity assay. These data suggest that the maximum inhibitory potency displayed by the latter was indeed limited by its trimerization equilibrium in solution, which is confirmed by the gain in potency of another peptide in the IZN series, IZN23, when stabilized with a cysteine cap to yield (CCIZN23)3.
All of the elements of (CCIZN17)3 appear to synergize for optimal activity. The IZ domain cannot be shortened, as shown by the analog (CCI10N17)3. In fact, (CCI10N17)3, comprising only 10 of 24 residues of IZ, is much less stable and accordingly much less potent, with an IC50 of only 19 nM in the single-cycle infectivity assay.
In addition to showing extremely high antiviral potency, (CCIZN17)3 displays a remarkable breadth of neutralization against a large panel of HIV isolates, being able to neutralize 100% of the strains tested in a commercial phenotypic virus assay. These properties compare favorably with the recently approved C-peptide inhibitor DP178 (also known as Enfuvirtide and Fuzeon). In a variety of assays, (CCIZN17)3 was equally or more potent than DP178. More importantly, we were able to demonstrate that the two inhibitors, which have different targets in gp41, synergize when used in combination. The use of two potent fusion inhibitors in combination should minimize the risk that viruses resistant to DP178 (10) or (CCIZN17)3 would emerge. Taken together, these features make (CCIZN17)3 an attractive lead to develop as an antiviral drug, alone or in combination with DP178.
In addition, the cysteine-capped inhibitors and (CCIZN17)3 in particular likely represent faithful mimetics of at least one of the possible conformations of gp41 coiled-coil during the HIV fusion process. As such, cysteine-capped inhibitors and (CCIZN17)3 may also find use as selector molecules in screening campaigns for small-molecule fusion inhibitors (26, 32-34) and as immunogens to elicit a neutralizing antibody response targeting the HIV fusion intermediate(s) (17, 35, 36). The data of Louis et al. (17) argue well for the latter strategy.
Supplementary Material
Acknowledgments
We thank Gennaro Ciliberto, Riccardo Cortese, Daria J. Hazuda, and John Shiver for thoughtful discussions throughout the project and critical review of the manuscript; Fabio Talamo for analytical MS; and Manuela Emili for artwork.
Author contributions: E.B., M.F., P.I., X.S.H., M.D.M., R.G., and A.P. designed research; E.B., M.F., P.I., R.H., A.V.C., X.S.H., W.A.S., M.D.M., and R.G. performed research; E.B., R.G., and A.P. analyzed data; and E.B. and A.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HIV-1, HIV type 1; HR, heptad repeat; PBMC, peripheral blood mononuclear cell.
References
- 1.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777-810. [DOI] [PubMed] [Google Scholar]
- 2.Colman, P. M. & Lawrence, M. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 309-319. [DOI] [PubMed] [Google Scholar]
- 3.Caffrey, M., Cai, M., Kaufman, J., Stahl, S. J., Wingfield, P. T., Covell, D. G., Gronenborn, A. M. & Clore, G. M. (1998) EMBO J. 17, 4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan, K., Liu, J., Wang, J., Shen, S. & Lu, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426-430. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 7.Wild, C., Greenwell, T., Shugars, D., Rimsky-Clarke, L. & Matthews, T. (1995) AIDS Res. Hum. Retroviruses 11, 323-325. [DOI] [PubMed] [Google Scholar]
- 8.Wild, C., Oas, T., McDanal, C., Bolognesi, D. & Matthews, T. (1992) Proc. Natl. Acad. Sci. USA 89, 10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaBonte, J., Lebbos, J. & Kirkpatrick, P. (2003) Nat. Rev. Drug Discovery 2, 345-346. [DOI] [PubMed] [Google Scholar]
- 10.Matthews, T., Salgo, M., Greenberg, M., Chung, J., DeMasi, R. & Bolognesi, D. (2004) Nat. Rev. Drug Discovery 3, 215-225. [DOI] [PubMed] [Google Scholar]
- 11.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. (1999) Cell 99, 103-115. [DOI] [PubMed] [Google Scholar]
- 13.Bewley, C. A., Louis, J. M., Ghirlando, R. & Clore, G. M. (2002) J. Biol. Chem. 277, 14238-14245. [DOI] [PubMed] [Google Scholar]
- 14.Wild, C., Dubay, J. W., Greenwell, T., Baird, T., Jr., Oas, T. G., McDanal, C., Hunter, E. & Matthews, T. (1994) Proc. Natl. Acad. Sci. USA 91, 12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884-888. [DOI] [PubMed] [Google Scholar]
- 16.Louis, J. M., Bewley, C. A. & Clore, M. G. (2001) J. Biol. Chem. 276, 29485-29489. [DOI] [PubMed] [Google Scholar]
- 17.Louis, J. M., Nesheiwat, I., Chang, L., Clore, M. G. & Bewley, C. A. (2003) J. Biol. Chem. 278, 20278-20285. [DOI] [PubMed] [Google Scholar]
- 18.Eckert, D. M., Malashkevich, V. N. & Kim, P. S. (1998) J. Mol. Biol. 284, 859-865. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki, K., Hiroaki, H., Kohda, D. & Tanaka, T. (1998) Protein Eng. 11, 1051-1055. [DOI] [PubMed] [Google Scholar]
- 20.Charneau, P., Alizon, M. & Clavel, F. (1992) J. Virol. 66, 2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., Di Marzio, P., Marmon, S., Sutton, R. E., Hill, C. M., et al. (1996) Nature 381, 661-666. [DOI] [PubMed] [Google Scholar]
- 22.Joyce, J. G., Hurni, W. M., Bogusky, M. J., Garsky, V. M., Liang, X., Citron, M. P., Danzeisen, R. C., Miller, M. D., Shiver, J. W., Keller, P. M. (2002) J. Biol. Chem. 277, 45811-45820. [DOI] [PubMed] [Google Scholar]
- 23.Richman, D. D., Wrin, T., Little, S. J. & Petropoulos, C. J. (2003) Proc. Natl. Acad. Sci. USA 100, 4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lineberger, J. E., Danzeisen, R., Hazuda, D. J., Simon, A. J. & Miller, M. D. (2002) J. Virol. 76, 3522-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan, D. C., Chutkowsky, C. T. & Kim, P. S. (1998) Proc. Natl. Acad. Sci. USA 95, 15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer, J. J., Hasan, A., Wilson, K. L., White, J. M., Matthews, T. J. & Delmedico, M. K. (2003) Biochemistry 42, 4945-4953. [DOI] [PubMed] [Google Scholar]
- 27.Derdeyn, C. A., Decker, J. M., Sfakianos, J. N., Zhang, Z., O'Brien, W. A., Ratner, L., Shaw, G. M. & Hunter, E. (2001) J. Virol. 75, 8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, M. D. & Hazuda, D. J. (2004) Drug Res. Updates 7, 89-95. [DOI] [PubMed] [Google Scholar]
- 29.Derdeyn, C. A., Decker, J. M., Sfakianos, J. N., Wu, X., O'Brien, W. A., Ratner, L., Kappes, J. C., Shaw, G. M. & Hunter, E. (2000) J. Virol. 74, 8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves, J. D., Gallo, S. A., Ahmad, N., Miamidian, J. L., Harvey, P. E., Sharron, M., Pohlmann, S., Sfakianos, J. N., Derdeyn, C. A., Blumenthal, R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulick, R. M., Mellors, J. M., Havlir, D., Eron, J. J., Gonzalez, C., McMahon, D., Jonas, L., Meibohm, A., Holder, D., Schleif, W. A., et al. (1998) J. Am. Med. Assoc. 280, 35-41. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S. & Jiang, S. (2004) Curr. Pharm. Des. 10, 1827-1843. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, S., Lin, K., Zhang, L. & Debnath, A. K. (1999) J. Virol. Methods 80, 85-96. [DOI] [PubMed] [Google Scholar]
- 34.Naicker, K. P., Jiang, S., Lu, H., Ni, J., Boyer-Chatenet, L., Wang, L. X. & Debnath, A. K. (2004) Bioorg. Med. Chem. 12, 1215-1220. [DOI] [PubMed] [Google Scholar]
- 35.Burton, D. R., Desrosiers, R. C., Doms, R. W., Koff, W. C., Kwong, P. D., Moore, J. P., Nabel, G. J., Sodroski, J., Wilson, I. A. & Wyatt, R. T. (2004) Nat. Immunol. 5, 233-236. [DOI] [PubMed] [Google Scholar]
- 36.Sundaram, R., Lynch, M. P., Rawale, S. V., Sun, Y., Kazanji, M. & Kaumaya, P.T.P. (2004) J. Biol. Chem. 279, 24141-24151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.