Abstract
Arabidopsis thaliana contains four DICER-LIKE (DCL) genes with specialized functions in small RNA biogenesis for RNA interference-related processes. A mutant with defects in DCL4 was identified and analyzed for microRNA- and endogenous, small interfering RNA (siRNA)-related functions. The dcl4-2 mutant contained normal or near-normal levels of microRNAs (21 nt) and heterochromatin-associated siRNAs (24 nt). In contrast, this mutant lacked each of three families of 21-nt trans-acting siRNAs (ta-siRNAs) and possessed elevated levels of ta-siRNA target transcripts. The dcl4-2 mutant resembled an rna-dependent RNA polymerase 6 mutant in that both mutants lacked ta-siRNAs and displayed heterochronic defects in which vegetative phase change was accelerated. Double mutant analyses with dcl2-1, dcl3-1, and dcl4-2 alleles revealed hierarchical redundancy among DCL activities, leading to alternative processing of ta-siRNA precursors in the absence of DCL4. These data support the concept that plants have specialized and compartmentalized DCL functions for biogenesis of distinct small RNA classes.
Eukaryotes contain small RNA-dependent pathways that negatively regulate gene expression at the transcriptional or posttranscriptional level (1). Small RNAs categorized as microRNAs (miRNAs) or small interfering RNAs (siRNAs) arise from imperfectly base-paired foldback structures or from dsRNA precursors, respectively (1, 2). Small RNAs associate with factors, such as ARGONAUTE (AGO) proteins, in effector complexes to guide target RNA cleavage, translational repression, or chromatin modification (3). miRNA primary transcripts arise from genetically defined PolII units, whereas dsRNA precursors for siRNAs may arise from bidirectional transcription of a locus, transcription of extended inverted duplications, or the activity of an RNA-dependent RNA polymerase (RDR) on a suitable RNA template (1, 2). RNaseIII-type enzymes, termed Drosha and Dicer (DCR) in animals or DCR-LIKE (DCL) in plants, catalyze processing of miRNA and siRNA precursors to 21- to 24-nt duplexes (2, 4). Loading of effector complexes with miRNAs or siRNAs involves asymmetric strand selection from the duplex based on the thermodynamic properties of each end (5, 6).
Although gene families encoding common components (AGO or DCR/DCL) of the small RNA biogenesis and effector complex machineries are conserved across eukaryotic kingdoms, small RNA pathways have become diversified and specialized between and within kingdoms (4). This diversification and specialization is most evident in plants, such as Arabidopsis thaliana, which contain 4 DCL, 10 AGO, and at least 3 functional RDR genes (4). miRNAs form through multiple processing steps catalyzed by DCL1 and require AGO1 to guide cleavage (or nondegradative repression in some cases) of target mRNAs in trans (7-11). Several classes of siRNAs form through distinct pathways. Heterochromatin-associated siRNAs (predominantly 24-nt) form through the activities of RDR2, RNA polymerase IV, and DCL3 and require AGO4 for activity to direct or reinforce cytosine methylation of DNA and histone H3 methylation at Lys-9 (12-16). Formation of posttranscriptionally active siRNAs from exogenous (viral and transgenic) sources may involve RDR1 or RDR6 and, for some viruses, DCL2 (4). Endogenous, trans-acting siRNAs (ta-siRNAs) arise from PolII genes and function like miRNAs to guide cleavage of target mRNAs (17-19). ta-siRNAs require RDR6 and suppressor of gene silencing 3 (SGS3) (of unknown biochemical function) for precursor formation (18, 19). ta-siRNA formation also requires DCL1, although the specific role of DCL1 may be indirect (17-19). The DCL activity that catalyzes processing of dsRNA precursors for ta-siRNAs is unclear based on previous studies. All known classes of endogenous small RNAs in Arabidopsis require HEN1, an RNA methyltransferase that modifies the 3′ end of miRNAs and siRNAs (20). Among the four DCLs in Arabidopsis, DCL4 is the conspicuous protein for which no activities were assigned previously.
The biogenesis pathway for ta-siRNAs involves site-specific cleavage of primary transcripts guided by a miRNA (17). Two miRNAs, miR173 and miR390, function in this capacity in Arabidopsis (17). The processed transcript is then converted to dsRNA through the activities of RDR6 and SGS3 (17-19). A DCL activity then catalyzes siRNA duplex formation in 21-nt increments, starting from the processed end of the precursor. Active ta-siRNAs, therefore, are accurately phased with respect to the miRNA-guided cleavage site (17).
A. thaliana has three known families of ta-siRNA-encoding genes, designated TAS1, TAS2, and TAS3 (17-19). The TAS1 family is composed of three genes that encode a closely related set of ta-siRNAs [for example, siR255/siR480(+)] that target four mRNAs encoding proteins of unknown function (17-19). TAS2-derived ta-siRNAs (for example, siR1511) targets a set of mRNAs encoding pentatricopeptide repeat proteins (17, 18). The TAS3 locus specifies two ta-siRNAs that target a set of mRNAs for several Auxin response factors (ARFs), including ARF3 (ETTIN) and ARF4 (17, 21). Arabidopsis mutants with defects in RDR6 and SGS3 lack ta-siRNAs and exhibit accelerated transition from juvenile to adult phase during vegetative development (18, 19), suggesting that ta-siRNAs regulate developmental timing, presumably through regulation of ta-siRNA target genes.
In this study, we identified a role for DCL4 in ta-siRNA biogenesis. dcl4 mutant plants exhibit phase-change phenotypes that resemble those of rdr6 mutants, specifically lack ta-siRNAs, and accumulate elevated levels of ta-siRNA target mRNAs. These results indicate that DCL4 functions to process ta-siRNA precursors in a distinct small RNA biogenesis pathway.
Materials and Methods
Plant Materials. The dcl1-7 allele was described in ref. 22. The dcl1-7 mutant line used in this study, which was created by introducing the original dcl1-7 allele into the Col-0 background, was kindly provided by R. Scott Poethig (University of Pennsylvania, Philadelphia) (18). Mutant lines containing dcl2-1, dcl3-1, and rdr6-15 alleles were described in refs. 14 and 23. The dcl4-2 mutant contains a T-DNA [portion of the Ti (tumor-inducing) plasmid] insertion between nucleotides 9005 and 9046 from the start codon (Genomanalyse im Biologischen System Pflanze seed line GABI160G05). This mutant was backcrossed to Col-0, and homozygous lines were selected after two generations. The double mutant lines with dcl2dcl3, and dcl2dcl4 alleles were generated by standard genetic crosses. Homozygous mutant genotypes in each line were confirmed by allele-specific PCR assays after two generations.
RNA Preparation and Blot Assays. RNA was extracted from plant leaf or inflorescence tissues by using TRIzol reagent (Invitrogen), followed by purification with RNA/DNA Midi columns (Qiagen, Valencia, CA).
For detection of DCL4-specific mRNA, total RNA (20 μg) from inflorescence tissue was resolved by formaldehyde denaturing 1.5% agarose gel electrophoresis. Blot hybridization was done in PerfectHyb Plus buffer (Sigma) with 32P-radiolabeled probes prepared from PCR-amplified cDNA fragments. The RNaseIII domain probe (RIII; ORF nucleotides 3360-4006) was amplified from cloned cDNA with primers DCL4-RIII-F (5′-GCTTGAGGTGCTTGGTGATGCATT-3′) and DCL4-RIII-R (5′-CAGCATCCCCAAGAAACTCCAA-3′). The PCR primers for probe A (nucleotides 4183-5109) were BamHI/NcoI-DCL4-4183F (5′-GAGAGGATCCATGGAGGATTATACCAATTTCCT-3′) and KpnI/SacI-DCL4-5109R (5′-GAGAGGTACCGAGCTCAGCAAAGGAATCCAGAAT-3′). The PCR primers for probe B (nucleotides 4846-5109) were DCL4-g9119F (5′-GGACCAAGCAGCAAAACCGCAAA-3′) and DCL4-R (5′-TCAGCAAAGGAATCCAGAATGCTT-3′). The tyrosine aminotransferase probe for the control hybridization was described in ref. 24.
For detection of small RNAs, quantitative small RNA blot assays were done with synthetic RNA oligonucleotide (Dharmacon Research, Layfayette, CO) as standard, with the exception of the AtSN1 siRNA blot, for which labeled in vitro transcripts instead of an end-labeled oligonucleotide was used as a probe. Briefly, for each small RNA blot, total RNA (12.5 μg per lane) samples in triplicates were run in parallel with a synthetic RNA oligonucleotide standard (two lanes; 0.5 and 2.5 fmol per lane, respectively). Gel electrophoresis and blot hybridization were done as described in ref. 23. Blot signals were measured with an Instant Imager, and statistical analysis was done by standard t test.
Analysis of DCL4 cDNA. The coding region of DCL4 was cloned by using a standard RT-PCR procedure. Superscript III (Invitrogen) and oligo(dT) primer were used for first-strand cDNA synthesis, and primers attB1-DCL4-F (5′-GGGGACAAGT T TGTACAAAAA AGCAGGCT TGATGCGTGACGAAGTTGACTTGAGCTT-3′) and attB2-DCL4-R_NoStop (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGCAAAGGAATCCAGAATGCTTGAGGCACCATA-3′) were used to amplify the DCL4-specific sequence with PfuUltra high-fidelity DNA polymerase (Stratagene). The PCR products were gel-purified and cloned in pDONR207 vector by using Gateway technology (Invitrogen) to create pENTR-DCL4_NoStop. Three clones were randomly selected for complete sequence analysis. To determine end sequences of DCL4 transcripts, RNA ligase-mediated 5′ and 3′ RACE was done as described in ref. 25. Gene-specific primers were DCL4-g64R (5′-CTCTGTCTCGCTTCCCCAAAAGCTT-3′) and DCL4-g36R (5′-GGGAATGGTCAAGCTCAAGTCAACTT-3′) for 5′ RACE, and DCL4-g9379F (5′-GATGCGCCCAATATGACATTGGAAT-3′) and DCL4-g9442F (5′-GAGCACGCTGCCCAAGCTGCTATAT-3′) for 3′ RACE. A total of 25 and 15 clones were sequenced for 5′ and 3′ RACE, respectively. The DCL4 transcript sequences were deposited in the GenBank database (accession no. DQ118423).
Expression Profiling. RNA was extracted from rosette leaf (leaves 6-8) and inflorescence (stages 1-12) tissues from six plants per genotype (Col-0 control, dcl1-7, dcl4-2, and rdr6-15) per replicate sample. All microarray analyses were done with ATH1 arrays (Affymetrix, Santa Clara, CA) by using three biological replicates grown under identical conditions. RNA isolation, labeling, and hybridization were done as described in ref. 17. Raw intensity values were normalized by using rma express (26) and imported into genespring 7 (Agilent Technologies, Palo Alto, CA) for analysis. Principal Components Analysis was done with a set of 92 and 7 validated or high-quality-predicted miRNA and ta-siRNA target genes, respectively, represented on the ATH1 array (17). Significantly coaffected genes were identified by the sam method (27), with a false discovery rate of 0.05. In three independent analyses, genes exhibiting significant up-regulation in dcl1-7, dcl4-2, and rdr6-15 in either tissue type were identified.
Results
Arabidopsis DCL4 Encodes All Highly Conserved Domains Found in Other DCR Family Members. Members of the DCR family in eukaryotes share highly conserved structural features, including a helicase domain, dual RNase III-like domains, dsRNA-binding motif(s), and a centrally located Piwi/Ago/Zwille (PAZ) domain (22, 28). Previous descriptions of domain structure of the four Arabidopsis DCL proteins indicated that DCL4 was unusual because of the lack of a PAZ domain (22). We analyzed the DCL4 mRNA by RT-PCR cloning and sequencing and by mapping the 5′ and 3′ UTRs with RNA ligase-mediated RACE procedures. The cloned cDNA revealed a 5,109-bp ORF, which contrasted with the previous prediction of a 5,031-bp ORF. This discrepancy was apparently due to inaccurate predictions at multiple exon/intron junctions, including those flanking exons 6, 9, 17, and 18, and introns 5, 8, 16, and 17 in the revised annotation (Fig. 1A Upper), resulting in a predicted protein with internal deletions and insertions. The previous annotation lacked 101 bp (3,000-3,100 nt from the start codon) encompassing the coding region for part of the PAZ domain. With this revision, DCL4 is predicted to encode a protein of 1,702 residues with a central PAZ domain in addition to all other conserved domains (Fig. 1 A Lower).
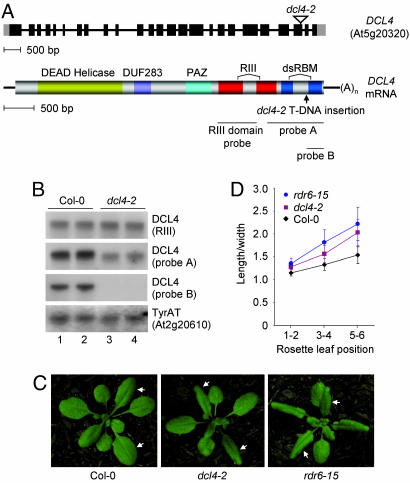
Fig. 1.
Characterization of Arabidopsis dcl4-2 mutant. (A) Organization of DCL4 genomic DNA (Upper) and mRNA (Lower). In the genomic diagram, exons and introns are illustrated as black bars and lines, respectively, and 5′ and 3′ UTRs are shown as gray bars. The T-DNA insertion site in dcl4-2 is indicated. Regions encoding conserved domains are indicated by colors in the mRNA diagram. The arrowhead indicates the dcl4-2 insertion point. Locations of cDNA-derived probe sequences are indicated by thin lines. dsRBM, dsRNA binding motif; DUF283, domain of unknown function 283. (B) RNA blot analyses of dcl4-2 mRNA. Blot assays with duplicate samples from inflorescence tissue of wild-type (lanes 1 and 2) and mutant (lanes 3 and 4) plants were done by using three DCL4-specific or tyrosine aminotransferase (TyrAT) probes. (C) Phenotypes of early rosette leaves of dcl4-2, rdr6-15, and wild-type plants. Arrowheads indicate rosette leaves 5 and 6. (D) Leaf growth (length/width) in rosette leaves of dcl4-2, rdr6-15, and wild-type plants. Mean and SD are shown for each data point (n = 40).
The 5′ RACE assay revealed two alternative transcription start sites that would give rise to transcripts with 5′ UTRs of 198 or 55 nt (Fig. 1 A and Fig. 6, which is published as supporting information on the PNAS web site). The 3′ RACE assay revealed two polyadenylation sites, which would define mRNAs with 3′ UTRs of 231 or 278 nt (Figs. 1 A and 6). The upstream polyadenylation site was supported by an EST clone (GenBank accession no. AV547573) containing precisely the same 3′ UTR sequence.
Identification of the dcl4-2 Mutant. To explore the function of Arabidopsis DCL4, we identified a mutant line (dcl4-2; Genomanalyse im Biologischen System Pf lanze seed line GABI160G05) harboring a T-DNA insertion beginning after nucleotide 9005 from the DCL4 start codon. This insertion should result in termination of the wild-type sequence before the coding region for the second dsRNA binding motif (Fig. 1 A). A series of RNA blot assays was done to characterize the dcl4-2 transcript. Probes corresponding to sequences upstream of the insertion site (RIII probe and probe A) detected wild-type DCL4 and mutant dcl4-2 transcripts of similar size (Fig. 1B). A probe (probe B) specific for sequences downstream of the insertion site detected the wild-type transcript but not the dcl4-2 mutant transcript (Fig. 1B). These results suggest that the dcl4-2 transcript is likely chimeric with partial DCL4- and T-DNA-specific sequences but not with sequences encoding the second dsRNA-binding motif. A 3′ RACE analysis confirmed the chimeric nature of the dcl4-2 transcript and revealed a 192-nt segment of T-DNA-derived sequence (Fig. 6).
Homozygous dcl4-2 mutant plants (Col-0 background) had elongated and downwardly curled rosette leaves, which are phenotypes also associated with rdr6 mutants (Fig. 1C) (18). The length/width ratios of early rosette leaves from the dcl4-2 and rdr6-15 mutant plants were significantly (P < 0.001) higher than those of equivalent leaves in Col-0 plants (Fig. 1D), although the phenotype of the dcl4-2 mutant was weaker than the rdr6-15 phenotype (Fig. 1 C and D). In addition, precocious production of abaxial trichomes in early rosette leaves was also detected in dcl4-2 and rdr6-15 mutants (data not shown). The elongated leaf and early abaxial trichome phenotypes are hallmarks of accelerated juvenile-to-adult phase change. In fact, a role for RDR6 and the ta-siRNA pathway in vegetative phase change was shown by Peragine et al. (18). The phenotypic similarity between the dcl4-2 and rdr6-15 mutants, therefore, suggests that DCL4 functions in the ta-siRNA pathway to affect vegetative phase change.
Expression Profiling of miRNA and ta-siRNA Targets in the dcl4-2 Mutant. Given the phenotypic similarity between the dcl4-2 and rdr6-15 mutants (Fig. 1 C and D), we hypothesized that DCL4 functions specifically in the ta-siRNA pathway, perhaps as the enzyme that catalyzes ta-siRNA formation from dsRNA precursors. If this hypothesis were true, then dcl4-2 and rdr6-15 mutants should have similar effects on ta-siRNAs and ta-siRNA target genes. In previous studies, ta-siRNAs accumulated to low levels and ta-siRNA target transcripts accumulated to relatively high levels in rdr6 mutant plants (17-19). Arabidopsis transcript levels were measured by expression profiling in two tissue types of Col-0, dcl1-7, dcl4-2, and rdr6-15 plants. The dcl1-7 mutant was included to distinguish genes that were regulated by miRNAs (primarily affected in dcl1-7) versus ta-siRNAs (affected in rdr6-15 and dcl1-7).
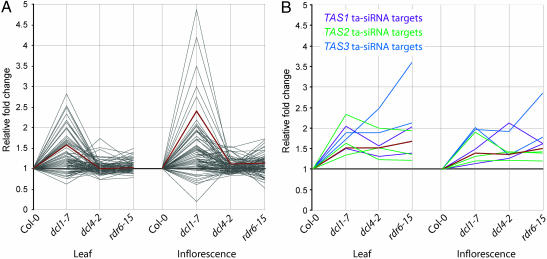
As shown previously, miRNA target gene transcripts as a group were generally up-regulated in dcl1-7. This gene set, however, was largely unaffected in dcl4-2 and rdr6-15 (Fig. 2A). This finding was confirmed by principal components analysis, in which at least 60% of the variation in both tissue types was explained by an eigenvector that was elevated specifically in dcl1-7 (Fig. 2 A). These data suggest that DCL4 is not required for miRNA activity. In contrast, all predicted and validated ta-siRNA targets that were represented on the array (seven targets; see Table 1, which is published as supporting information on the PNAS web site) were significantly up-regulated in at least one tissue type in dcl1-7, dcl4-2, and rdr6-15 plants. Coregulation of ta-siRNA target mRNAs was supported strongly by principal components analysis in both of the tissues analyzed (Fig. 2B). In total, only nine genes were significantly up-regulated in all three mutants (false discovery rate = 0.05; sam method) (Table 1). Because the dcl4-2 mutant affected ta-siRNA but not miRNA targets, it is likely to be required for ta-siRNA processing downstream of miRNA-guided cleavage of primary ta-siRNA transcripts. Among transcripts from TAS1, TAS2, and TAS3, only TAS2 [Arabidopsis Genome Initiative (AGI) no. At2g39680) transcript was represented in the ATH1 array. This transcript was up-regulated by 1.3- to 3.3-fold in each mutant, although a significant difference was detected only in the rdr6-15 plants (data not shown).
Fig. 2.
Expression profiling of miRNA and ta-siRNA target genes in dcl1-7, dcl4-2, and rdr6-15 mutant lines. Mean fold-change values were calculated for miRNA (A) and ta-siRNA (B) target transcripts in Col-0 (1.0 by definition) and mutant plants. The primary principal components analysis eigenvectors that accounted for >60% or >90% of the variation with miRNA and ta-siRNA targets, respectively, are shown by the red lines. Targets for ta-siRNAs from TAS1 (purple), TAS2 (green), and TAS3 (blue) are color-coded. Note that only seven of nine predicted or validated ta-siRNA targets are represented on the array (Table 1).
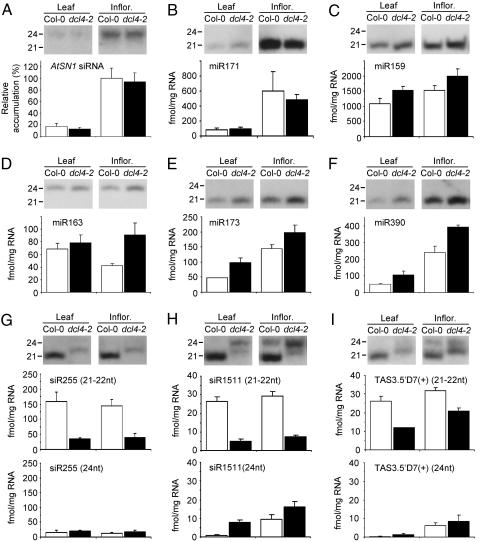
Biogenesis of Endogenous ta-siRNAs Is Impaired in dcl4-2 Plants. The requirement for DCL4 in biogenesis of endogenous ta-siRNAs was tested by using dcl4-2 mutant plants and quantitative RNA blot assays. DCL3- and RDR2-dependent heterochromatin-associated, AtSN1-derived siRNAs (24 nt) and DCL1-dependent miRNAs were also tested. AtSN1 siRNAs accumulated to levels that were similar in wild-type and dcl4-2 plants (Fig. 3A). Among five miRNAs tested, none accumulated to significantly lower levels in dcl4-2 compared with wild-type plants (Fig. 3 B-F). Curiously, several miRNAs, including miR159, miR163, miR173, and miR390, accumulated to slightly but significantly (P < 0.05) higher levels in leaves or inflorescences of dcl4-2 plants. These data indicate that DCL4 is unlikely to be required for heterochromatin-associated siRNA or miRNA biogenesis. The slight increase in miRNA levels in dcl4-2 plants may reflect a pleiotropic, indirect effect of loss of DCL4.
Fig. 3.
RNA blot assays for small RNAs in dcl4-2 plants. Triplicate RNA samples from leaf and inflorescence (Inflor.) tissues of dcl4-2 mutant and wild-type plants were analyzed. A representative blot image and mean (±SD) small RNA levels (fmol/mg total RNA based on quantitative standards) are presented in columns in each panel. Small RNAs tested were AtSN1 siRNAs (A), miR171 (B), miR159 (C), miR163 (D), miR173 (E), and miR390 (F) and ta-siRNAs siR255 (G), siR1511(H), and TAS3.5′D7(+) (I). In blot assays for ta-siRNAs, signals from the 21- plus 22-nt zone and the 24-nt zone were measured and presented in separate graphs. Note that it was not possible to accurately measure 22-nt ta-siRNA forms in the presence of high levels of 21-nt forms.
In contrast, each of three 21-nt ta-siRNAs [siR255, siR1511, and TAS3.5′D7(+)] accumulated to significantly lower (P < 0.003) or undetectable levels in each tissue tested in dcl4-2 plants (Fig. 3 G-I). These ta-siRNAs arise from each of the three known ta-siRNA gene families (TAS1, TAS2, and TAS3, respectively). Loss of ta-siRNA signal was specific for 21-nt ta-siRNA species. Low levels of 24-nt siR255-related and siR1511-related RNA were detected in wild-type plant tissues (as noted in ref. 17) and to similar or significantly elevated levels in dcl4-2 tissues (Fig. 3 G-I). Additionally, elevated levels of 22-nt species related to siR255, siR1511, and TAS3.5′D7(+) were detected in dcl4-2 plants (Fig. 3 G-I). Although we were unable to accurately measure the abundance of the 22-nt RNAs separate from the 21-nt ta-siRNAs because of overlapping signals, visual inspection of RNA blot data clearly indicated that the 22-nt species accumulated predominantly in dcl4-2 plants (Fig. 3 G-I). These data indicate that DCL4 is required for biogenesis of 21-nt ta-siRNAs but that alternatively processed forms of different sizes or structure arise in the absence of DCL4.
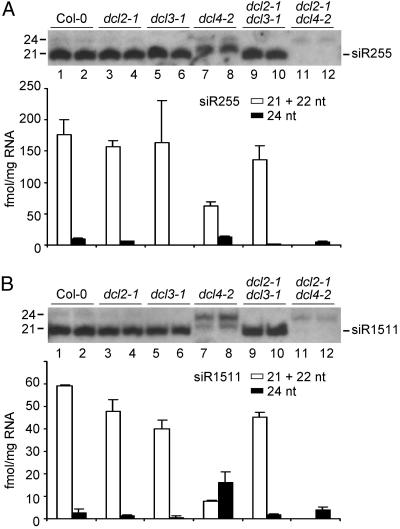
The basis for multiple-sized (22- and 24-nt) siR255- and siR1511-related species was investigated by single and double mutant analysis using dcl2-1, dcl3-1, and dcl4-2 alleles (Fig. 4). Consistent with previous TAS3 ta-siRNA results, the 24-nt forms of siR255 and siR1511 were lost in dcl3-1 and dcl2-1dcl3-1 double mutant plants (Fig. 4) (17). These data reinforce the conclusion that DCL3 functions as the 24-nt small-RNA-generating enzyme in Arabidopsis (12-15). The 22-nt siR255 and siR1511 forms that accumulated in dcl4-2 plants were lost specifically in dcl2-1dcl4-2 plants (Fig. 4), which indicates clearly that the 22-nt forms are DCL2 products rather than small RNAs formed by an aberrant dcl4 activity.
Fig. 4.
RNA blot assays for ta-siRNAs in single and double mutant plants. Duplicate total RNA samples from inflorescence tissue of mutant and wild-type plants were analyzed for siR255 (A) and siR1511 (B) RNAs. In each panel, a blot image and a bar graph showing signal intensity from the 21- plus 22-nt (open bars) and 24-nt (filled bars) zones are presented.
Discussion
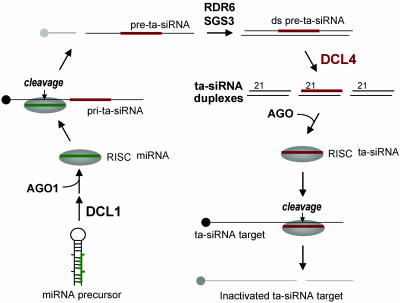
The DCL4 gene was shown to be required for ta-siRNA biogenesis and activity. Loss of DCL4 activity had no negative effect on miRNA or heterochromatic siRNA accumulation, indicating that DCL4 has a relatively specialized function within the ta-siRNA pathway. In contrast, loss-of-function dcl1 mutants affected miRNA and ta-siRNA formation (17-19). Given the results presented here, it is now clear how to interpret these observations. DCL1 is required for miRNA biogenesis, and miRNAs (miR173 and miR390) are necessary for site-specific processing of primary ta-siRNA transcripts (17) (Fig. 5). This processing defines an end structure that, after RDR6- and SGS3-directed conversion to dsRNA, provides a start point for DCL4-mediated processing and formation of ta-siRNA duplexes. Thus, loss of DCL1 function affects miRNA and ta-siRNA biogenesis, whereas loss of DCL4 affects only ta-siRNA formation (Fig. 5). The dcl4-2 mutant exhibited vegetative phase change defects that resembled those observed in rdr6 mutants, which confirms the initial conclusions of Peragine et al. (18) about the involvement of Arabidopsis ta-siRNAs in phase change. This involvement likely reflects the regulatory role of ta-siRNAs on target genes that control phase change.
Fig. 5.
Model for trans-acting siRNA biogenesis in Arabidopsis. RISC, RNA-induced silencing complex.
Given that the effective ta-siRNAs are phased in 21-nt increments, deviation from 21-nt spacing between cleavage events would result in formation of siRNAs with suboptimal complementarity with target sequences. Thus, it is worth considering whether the alternatively processed (DCL2- and DCL3-dependent) 22- and 24-nt ta-siRNA-related species that form in the absence of DCL4 are functional. The relatively weak phenotype of dcl4-2 may have been due to partial redundancy of functions among the four DCL family members. However, these alternatively processed forms would have been offset from the authentic processing phase to varying extents, depending on the DCL product size and position of the active ta-siRNA sequence relative to the phasing site. We predict, therefore, that the alternatively processed siRNAs had relatively low or no targeting activity on ta-siRNA targets.
With the findings reported here, we conclude that the three known branches of endogenous RNA interference-related processes (miRNA, ta-siRNA, and heterochromatic siRNA) are associated with specialized DCL functions. The ta-siRNA-specific DCL4 is likely associated with RDR6 and SGS3. One possibility is that DCL4 functions in a coordinated complex with RDR6 and SGS3 to convert suitable, perhaps aberrant, RNA templates to dsRNA and catalyze siRNA processing. However, the determinants governing specificity for this pathway remain unclear. Importantly, RDR6 and SGS3 were also identified through genetic screens for Arabidopsis mutants with defects in sense transgene-induced RNA silencing (29, 30). These two factors were also shown to be required for RNA silencing induced by a DNA virus and some RNA viruses (29-31). Conceivably, RNA targets for sense transgene- or virus-induced RNA silencing possess features that resemble endogenous RDR6 templates (ta-siRNA precursors). These features might include the lack of a cap or polyadenylated tail, which would resemble the miRNA-guided cleavage products of ta-siRNA primary transcripts (Fig. 5). Whether DCL4 is necessary for sense transgene and antiviral silencing remains to be determined.
Phylogenetic analysis of DCR/DCL genes in plants, animals, and fungi indicate that all members of this class possess a monophyletic origin and that proliferation of family members occurred after kingdoms split (Z.X. and J.C.C., unpublished observations). We logically assume that the specialization of RNA interference branches in plants involved the functionalization of DCL family members with unique specificities. Diversification of DCL and other RNA interference functions in plants also involved acquisition of regulatory control mechanisms, such as the targeting of DCL1 and AGO mRNAs by plant-specific miRNAs (11, 32). Thus, we suggest that acquisition of DCL functions associated with distinct RNA interference branches in plants occurred during, not before, evolution of the plant lineage.
Note Added in Proof. While this paper was under review, Gasciolli et al. (33) reported similar results on the function of DCL4 in trans-acting siRNA production.
Supplementary Material
Acknowledgments
We thank R. Scott Poethig for the dcl1-7 allele in Col-0, Anne-Marie Girard for assistance with microarray experiments, Mark Dasenko for sequencing, and Krissy Remple and Noah Fahlgren for assistance with morphological measurements. We also thank the members of the J.C.C. laboratory for advice, discussion, and technical support. This work was supported by National Science Foundation Grant MCB-0209836, National Institutes of Health Grant AI43288, and U.S. Department of Agriculture Grant 2005-35319-15280.
Author contributions: Z.X., E.A., and J.C.C. designed research; Z.X., E.A., and A.W. performed research; Z.X., E.A., and J.C.C. analyzed data; and Z.X., E.A., and J.C.C. wrote the paper.
Abbreviations: DCR, Dicer; DCL, DCR-LIKE; miRNA, microRNA; siRNA, small interfering RNA; ta-siRNA, trans-acting siRNA; RDR, RNA-dependent RNA polymerase; AGO, ARGONAUTE; SGS3, suppressor of gene silencing 3.
Data deposition: The cDNA sequence for DCL4 has been deposited in the GenBank database (accession no. DQ118423). Microarray data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO accession no. GSE3011).
References
- 1.Finnegan, E. J. & Matzke, M. A. (2003) J. Cell Sci. 116, 4689-4693. [DOI] [PubMed] [Google Scholar]
- 2.Bartel, D. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 3.Denli, A. M. & Hannon, G. J. (2003) Trends Biochem. Sci. 28, 196-201. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe, D. (2004) Nature 431, 356-363. [DOI] [PubMed] [Google Scholar]
- 5.Khvorova, A., Reynolds, A. & Jayasena, S. D. (2003) Cell 115, 209-216. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N. & Zamore, P. D. (2003) Cell 115, 199-208. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B. & Bartel, D. P. (2002) Genes Dev. 16, 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park, W., Li, J., Song, R., Messing, J. & Chen, X. (2002) Curr. Biol. 12, 1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurihara, Y. & Watanabe, Y. (2004) Proc. Natl. Acad. Sci. USA 101, 12753-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidner, C. A. & Martienssen, R. A. (2004) Nature 428, 81-84. [DOI] [PubMed] [Google Scholar]
- 11.Vaucheret, H., Vazquez, F., Crete, P. & Bartel, D. P. (2004) Genes Dev. 18, 1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herr, A. J., Jensen, M. B., Dalmay, T. & Baulcombe, D. C. (2005) Science 308, 118-120. [DOI] [PubMed] [Google Scholar]
- 13.Onodera, Y., Haag, J. R., Ream, T., Nunes, P. C., Pontes, O. & Pikaard, C. S. (2005) Cell 120, 613-622. [DOI] [PubMed] [Google Scholar]
- 14.Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., Jacobsen, S. E. & Carrington, J. C. (2004) PLoS Biol. 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. (2002) EMBO J. 21, 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zilberman, D., Cao, X. & Jacobsen, S. E. (2003) Science 299, 716-719. [DOI] [PubMed] [Google Scholar]
- 17.Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. (2005) Cell 121, 207-221. [DOI] [PubMed] [Google Scholar]
- 18.Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L. & Poethig, R. S. (2004) Genes Dev. 18, 2368-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., Hilbert, J. L., Bartel, D. P. & Crete, P. (2004) Mol. Cell 16, 69-79. [DOI] [PubMed] [Google Scholar]
- 20.Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., Steward, R. & Chen, X. (2005) Science 307, 932-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, L., Carles, C. C., Osmont, K. S. & Fletcher, J. C. (2005) Proc. Natl. Acad. Sci. USA. 102, 9703-9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauer, S. E., Jacobsen, S. E., Meinke, D. W. & Ray, A. (2002) Trends Plant Sci. 7, 487-491. [DOI] [PubMed] [Google Scholar]
- 23.Allen, E., Xie, Z., Gustafson, A. M., Sung, G. H., Spatafora, J. W. & Carrington, J. C. (2004) Nat. Genet. 36, 1282-1290. [DOI] [PubMed] [Google Scholar]
- 24.Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A. & Carrington, J. C. (2003) Dev. Cell 4, 205-217. [DOI] [PubMed] [Google Scholar]
- 25.Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S. A. & Carrington, J. C. (2005) Plant Physiol., 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed]
- 26.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185-193. [DOI] [PubMed] [Google Scholar]
- 27.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 29.Dalmay, T., Hamilton, A., Rudd, S., Angell, S. & Baulcombe, D. C. (2000) Cell 101, 543-553. [DOI] [PubMed] [Google Scholar]
- 30.Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., Jouette, D., Lacombe, A. M., Nikic, S., Picault, N., et al. (2000) Cell 101, 533-542. [DOI] [PubMed] [Google Scholar]
- 31.Muangsan, N., Beclin, C., Vaucheret, H. & Robertson, D. (2004) Plant J. 38, 1004-1014. [DOI] [PubMed] [Google Scholar]
- 32.Xie, Z., Kasschau, K. D. & Carrington, J. C. (2003) Curr. Biol. 13, 784-789. [DOI] [PubMed] [Google Scholar]
- 33.Gasciolli, V., Mallory, A. C., Bartel, D. P. & Vaucheret, H. (2005) Curr. Biol., 10.1016/S0960982205006743. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.