Abstract
Since the associations between Helicobacter pylori genotype and disease differ in Asia and the West, we investigated the correlation between HP0638, encoding an outer membrane protein, and potential markers of virulence (cagA, vacA, and iceA). For 109 strains from nine countries, the status of cagA, vacA, and iceA was determined by PCR and/or a line probe assay. We also studied 18 strains from 8 patients (parents and 6 daughters) from a Dutch family and paired strains collected on average 8 years apart from 11 patients. When the HP0638 signal sequences were amplified by PCR and DNA sequence determinations were performed, 89 (96%) of 93 cagA-positive strains had HP0638 in frame, versus none (0%) of 16 cagA-negative strains (P < 0.001). Among strains in which HP0638 was in frame, a six-CT dinucleotide repeat pattern was dominant in Western countries (23 of 33 strains [70%]), while a pattern of three CT repeats with another CT after four T’s (3 + 1-CT-repeat pattern) was dominant in East Asia (31 of 46 strains [67%]); however, specific CT repeat patterns did not correlate with clinical outcome. HP0638 phylogenetic trees also showed geographic characters. The HP0638 frame status and CT dinucleotide repeat patterns were identical for 9 of 11 pairs of strains obtained on average 8 years apart from individuals and the 15 strains obtained from the mother and all six daughters. Thus, HP0638 frame status and cagA status are strongly correlated. The CT dinucleotide repeat pattern in the putative HP0638 signal sequence has geographic characters and appears stable in particular patients and families over a period of years. Analysis of HP0638 CT polymorphisms may serve as a new typing system to discriminate H. pylori isolates for epidemiological purposes.
Helicobacter pylori, a microaerophilic, gram-negative bacterium that colonizes the human stomach, is involved in the pathogenesis of peptic ulcer disease and gastric cancer (GCA) (8, 9). H. pylori is highly genetically diverse, with isolates being easily distinguishable by various fingerprinting techniques (2, 3, 32) or by sequencing of representative gene segments (16). Extensive interstrain gene transfer and recombination are important causes of such diversity (1, 18, 31).
Recently, several H. pylori genes related to the risk of disease were identified (22). The cytotoxin-associated gene (cagA) is a marker for the cag island, whose presence is associated with a more severe clinical outcome (9, 12, 13, 15, 20, 36). The cag island genes encode proteins that enhance the virulence of the strain, for example, by increasing host cell cytokine production (5, 12, 35) and by altering protein tyrosine phosphorylation (24). A protein that induces vacuoles in epithelial cells is encoded by vacA (14, 21, 29, 33). vacA is present in all H. pylori strains and contains at least two variable regions (6). The s region (encoding the signal peptide) exists as s1 (including s1a, s1b, and s1c) or s2 allelic types (39). The m region (middle) occurs as m1 or m2 allelic types. Most vacA s1 strains are cagA positive, and most vacA s2 strains are cagA negative (6), even though these two genetic elements do not have any physical linkage on the H. pylori chromosome (10). The vacA and cagA DNA sequences in strains from the United States and Europe differ from those of strains in China and Japan (7, 17, 26, 38, 39, 41, 46). For example, in Western countries, most s1 strains are s1a or s1b, while about 80% of the s1 strains from East Asia are s1c (39). The allelic groupings of babA and babB, other outer membrane protein genes, are independent of one another, and each allelic group has geographic variations (28). cagA-positive strains frequently are vacA s1, vacA m1, and iceA1. The iceA1 genotype also is associated with vacA s1 and vacA m1 (42).
Several previous studies addressed whether H. pylori clusters geographically by examining sequences of housekeeping genes (1), ureAB (11), and cagA (23, 41, 45); these reports indicated that H. pylori sequences have geographic characters. However, Asian and Western strains remain difficult to distinguish, and further genetic markers are needed to reliably differentiate strains from different regions of the world.
Gram-negative bacterial outer membranes mediate the interaction with the surrounding environment. For H. pylori to survive and persist in the gastric milieu, specific adaptations involving its outer membrane proteins would be expected. The expression of HP0638, encoding the OipA outer membrane protein, has been reported to correlate with interleukin 8 (IL-8) induction (44). Variations in dinucleotide repeats located in the region encoding the signal sequence determine whether or not the complete open reading frame (ORF) is in frame. Since the associations between H. pylori genotype and disease differ in Asia and the West, we hypothesized that the expression of HP0638 might correlate with that of other genes associated with inflammation, such as cagA, and that geographically based alleles might exist. We also sought to determine whether HP0638 frame status is stable in isolates from individuals over time and among members of a family.
MATERIALS AND METHODS
Bacterial strains.
We studied H. pylori isolates from 109 patients undergoing upper gastrointestinal tract endoscopy in six different areas (38 in the United States, 13 in Europe, 4 in Colombia, 38 in Japan, 8 in China, and 8 in India) (Table 1). For each patient, clinical status had been determined as described previously (5). From the 109 patients, biopsy specimens were homogenized with a glass rod and incubated on brucella agar plates with 10% newborn calf serum (BS agar; Intergen) for 5 to 7 days at 37°C in a 5% CO2 atmosphere. A single colony was picked and streaked for isolation on a BS agar plate for 3 days, and H. pylori strains were identified by Gram staining and catalase, oxidase, and urease assays. We also examined a group of 18 strains previously collected from 8 patients (parents and 6 adult daughters) from one extended Dutch family (37) and paired strains collected on average 8 years apart from 11 patients as described previously (19).
TABLE 1.
Characteristics of the 109 H. pylori strains studied
| Origin | Clinical diagnosis | No. of strains |
|---|---|---|
| Asian countries (n = 54) | ||
| Japan | DU | 9 |
| GU | 16 | |
| GCA | 6 | |
| NUD | 7 | |
| China | DU | 4 |
| GCA | 4 | |
| India | DU | 1 |
| NUD | 7 | |
| Western countries (n = 55) | ||
| United States | ||
| Caucasian | DU | 10 |
| GU | 1 | |
| NUD | 17 | |
| African-Americans | DU | 3 |
| GU | 2 | |
| NUD | 5 | |
| Europe | DU | 4 |
| GU | 1 | |
| NUD | 8 | |
| Colombia | DU | 1 |
| GU | 3 |
Assessment of H. pylori cagA, vacA, and iceA1 status.
A chloroform-phenol extraction method was used to obtain DNA from the H. pylori isolates as described previously (43). Analyses for the presence of cagA, vacA s- and m-region allelic types, and iceA subtypes were done by PCR with established primers (6, 36, 40). PCRs were performed by standard methods with a reaction volume of 50 μl containing 0.5 U of Taq (Qiagen), 1.5 mM MgCl2, and 200 ng of each primer. The PCR protocol (30 cycles) included a denaturing step at 94°C for 1 min, annealing at 5°C below the predicted melting temperature of the primers for 1 min, and extension at 72°C for 1 min/kb of amplification product. cagA and vacA status also had been confirmed by reverse hybridization with a line probe assay (40).
Assessment of HP0638 status.
According to the complete genome sequence of H. pylori strains 26695 and J99 (4, 34), CT dinucleotide repeats are located in the region encoding the signal sequence of HP0638. The region of the signal sequence of HP0638 including the repeats was amplified by PCR with primers AN9260 (5′-CAAGCGCTTAACAGATAGGC-3′) and AN9261 (5′AAGGCGTTTTCTGCTGAAGC-3′) (44). PCR products were purified by using a QiaQuick PCR purification kit and a QiaQuick gel extraction kit (both kits from Qiagen); directly sequenced on both strands by using an automated Applied Biosystems, Inc., sequencer at the New York University Medical Center DNA Sequencing Core Facility; and analyzed by using Sequencer 3.1.1 (Gene Code Corp., Inc., Ann Arbor, Mich.).
Phylogenetic analysis.
Alignments of HP0638 nucleotides 1 to 150 derived from the PCR with primer pair AN9260-AN9261 were created by using GCG Pileup (Wisconsin Package version 9.1), and phylograms were constructed.
RESULTS
cagA, vacA, and HP0638 status of H. pylori strains from different geographic areas.
The 109 strains studied could be divided into dichotomous groups by cagA status (positive, 93; negative, 16), vacA s allelic type status (s1, 95; s2, 14), vacA m allelic type status (m1, 80; m2, 29), and iceA status (iceA1, 61; iceA2, 48). HP0638 in-frame status was more correlated with cagA-positive (100%; P < 0.001), vacA s1 (100%; P < 0.001), vacA m1 (83%; P < 0.001), and iceA1 (62%; P < 0.01) than HP0638 out-of-frame status (cagA positive, 20%; vacA s1, 20%; vacA m1, 30%; and iceA1, 30%). HP0638 was in frame in all 54 strains from East Asia and India and in 35 (64%) of the 55 strains from western countries, essentially mirroring cagA status (Table 2). Of the 109 patients studied, those carrying strains in which HP0638 was in frame had the more severe diseases (duodenal ulcer [DU], 30%; gastric ulcer [GU], 25%; nonulcer dyspepsia [NUD], 34%; and GCA, 11%) than did those with strains in which HP0638 was out of frame (DU, 25%; GU, 5%; and NUD, 70%). Since all 54 strains from Asian patients both were cagA positive and had HP0638 in frame, no analysis of independent relationships with clinical findings was possible. For the 55 patients from western countries, the effects of cagA positivity, HP0638 frame status, and clinical outcome also could not be dissociated (Table 2). All 16 cagA-negative strains had vacA s2 alleles, and all 93 cagA-positive strains had s1 alleles. Among the 93 cagA-positive strains, 54 Asian strains had the s1c (43) or s1a (11) allele, whereas 39 western strains had the s1a (14) or s1b (25) allele.
TABLE 2.
Variations in HP0638 CT repeat region sequences in 109 H. pylori strains from six geographic regions
| Origin | cagA genotypea | No. of CT repeats | HP0638 CT repeat region sequence | Frame status | Total | No. of isolates by diagnosis
|
|||
|---|---|---|---|---|---|---|---|---|---|
| DU | GU | NUD | GCA | ||||||
| Asian countries | |||||||||
| Japan (n = 38) | + | 1 + 1 + 1 | CTTTCTGTCTTTCTCGTT | In | 3 | 2 | 1 | 0 | 0 |
| 1 + 1 + 2 | CTAACTTTCTTT(CT)2CGTT | In | 1 | 0 | 1 | 0 | 0 | ||
| 1 + 3 | CTTTCTGT(CT)3CGTT | In | 2 | 0 | 2 | 0 | 0 | ||
| 2 + 1 | CTTT(CT)2TTTTCTCGTT | In | 1 | 0 | 0 | 1 | 0 | ||
| 3 + 1 | CTAA(CT)3TTTTCTCGTT | In | 26 | 6 | 10 | 6 | 4 | ||
| 3 + 2 | CTAA(CT)3TT(CT)2CGTT | In | 1 | 0 | 1 | 0 | 0 | ||
| 5 | CTTTTACTAA(CT)5TTCGTTb | In | 4 | 1 | 1 | 0 | 2 | ||
| China (n = 8) | + | 2 + 1 | CTTTTA(CT)2TTCTGTCT | In | 1 | 1 | 0 | 0 | 0 |
| 3 | CTAA(CT)3TTTTATCGTT | In | 1 | 1 | 0 | 0 | 0 | ||
| 3 + 1 | CTAA(CT)3TTTTCTCGTT | In | 5 | 2 | 0 | 0 | 3 | ||
| 3 + 2 | CTAA(CT)3TT(CT)2CGTT | In | 1 | 0 | 0 | 0 | 1 | ||
| India (n = 8) | + | 1 + 1 + 1 | CTTTCTGTCTTTCTCGTT | In | 1 | 0 | 0 | 1 | 0 |
| 2 + 3 | CTAA(CT)2TT(CT)3CGTT | In | 1 | 0 | 0 | 1 | 0 | ||
| 3 + 1 | CTAA(CT)3TTTTCTCGTT | In | 2 | 0 | 0 | 2 | 0 | ||
| 6 | CTTACTAA(CT)6CGTT | In | 4 | 1 | 0 | 3 | 0 | ||
| Total | + | In | 54 | 14 | 16 | 14 | 10 | ||
| Western countries | |||||||||
| Europe (n = 13) | + | 6 | CTTACTAA(CT)6CGTT | In | 5 | 2 | 1 | 2 | 0 |
| 8 | CTTA(CT)8CGTTc | In | 1 | 0 | 0 | 1 | 0 | ||
| 9 | CTTACTAA(CT)9CGTTd | In | 2 | 1 | 0 | 1 | 0 | ||
| 7 | CTTA(CT)7CGTT | Out | 1 | 0 | 0 | 1 | 0 | ||
| 8 | CTTACTAA(CT)8CGTTc | Out | 1 | 0 | 0 | 1 | 0 | ||
| − | 9 | CTTA(CT)9CGTTd | Out | 1 | 0 | 0 | 1 | 0 | |
| 10 | CTTTTA(CT)10CGTT | Out | 2 | 1 | 0 | 1 | 0 | ||
| United States | |||||||||
| Caucasians (n = 28) | + | 6 | CTTACTAA(CT)6CGTT | In | 13 | 7 | 0 | 6 | 0 |
| 9 | CTTACTAA(CT)9CGTTd | In | 3 | 0 | 1 | 2 | 0 | ||
| 4 | ATAA(CT)4CGTT | Out | 1 | 0 | 0 | 1 | 0 | ||
| 12 | CTTA(CT)12CGTTe | Out | 1 | 1 | 0 | 0 | 0 | ||
| − | 5 | CTTACTAA(CT)5CGTTb | Out | 3 | 0 | 0 | 3 | 0 | |
| 7 | CTTACTAA(CT)7CGTT | Out | 5 | 1 | 0 | 4 | 0 | ||
| 8 | CTTACTAA(CT)8CGTTc | Out | 1 | 0 | 0 | 1 | 0 | ||
| 9 | CTTA(CT)9CGTTd | Out | 1 | 1 | 0 | 0 | 0 | ||
| African-Americans (n = 10) | + | 5 | CTTACTAACC(CT)5CGTTb | In | 2 | 1 | 0 | 1 | 0 |
| 6 | CTTACTAA(CT)6CGTT | In | 5 | 2 | 1 | 2 | 0 | ||
| 9 | CTTACTAA(CT)9CGTTd | In | 1 | 0 | 1 | 0 | 0 | ||
| 12 | CTTACTAA(CT)12CGTTe | In | 1 | 0 | 0 | 1 | 0 | ||
| − | 7 | CTTACTAA(CT)7CGTT | Out | 1 | 0 | 0 | 1 | 0 | |
| Colombia (n = 4) | + | 6 | CTTACTAA(CT)6CGTT | In | 1 | 0 | 1 | 0 | 0 |
| 9 | CTTACTAA(CT)9CGTTd | In | 1 | 0 | 1 | 0 | 0 | ||
| − | 8 | CTTACTAA(CT)8CGTTc | Out | 2 | 1 | 1 | 0 | 0 | |
| Total | + | In | 35 | 13 | 6 | 16 | 0 | ||
| Out | 4 | 1 | 0 | 3 | 0 | ||||
| − | Out | 16 | 4 | 1 | 11 | 0 | |||
+, positive; −, negative.
ORF with five CT repeats is in frame due to insertion of TT sequence 6 bp upstream of CT repeats or insertion of CC sequence immediately upstream of CT repeats.
ORF with eight CT repeats is in frame due to deletion of CTAA sequence immediately upstream of CT repeats.
ORF with nine CT repeats is out of frame due to deletion of CTAA sequence immediately upstream of CT repeats.
ORF with 12 CT repeats is out of frame due to deletion of CTAA sequence immediately upstream of CT repeats.
Analysis of HP0638 variants and vacA s-region allelic types.
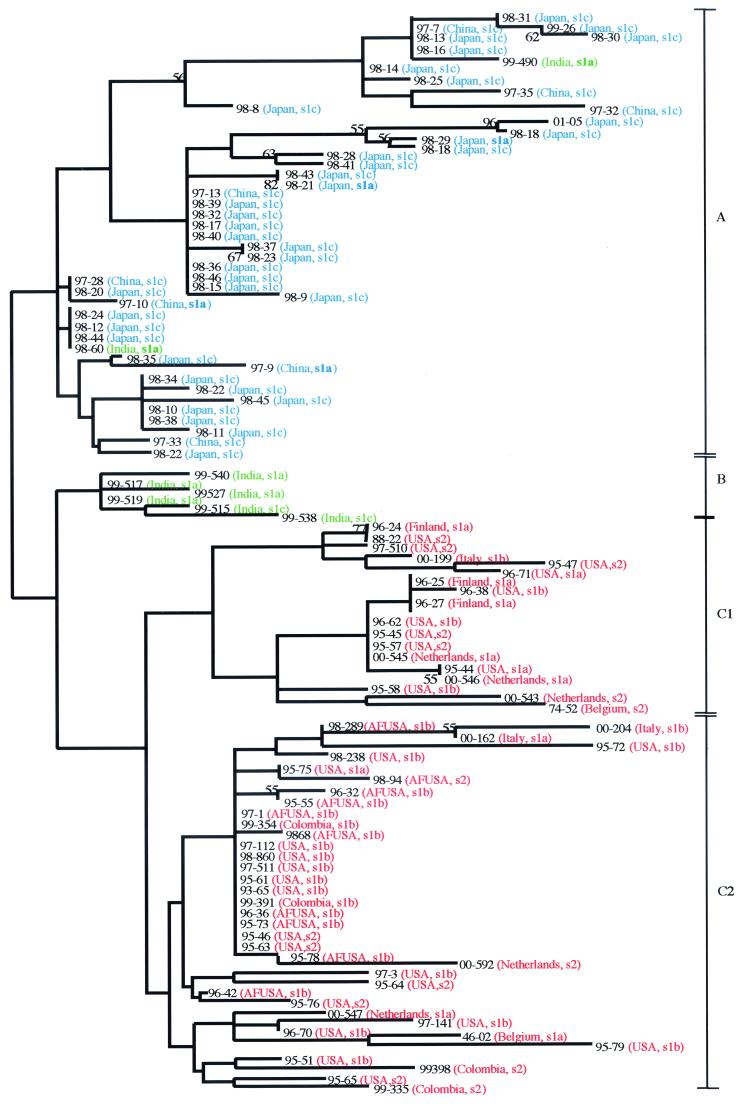
The sequence heterogeneity of nucleotides 1 to 150 of the HP0638 PCR products was examined. From each sample, the PCR amplicon was of the expected size. Phylogenetic analysis of these sequences revealed the existence of three major groupings based on geographic origin (Fig. 1). Strains from East Asia appeared exclusively in group A, six of the eight Indian strains constituted group B, and Western strains comprised group C. Group C was further subdivided into subgroups C1 and C2. C1 consisted of strains from Caucasian individuals only, whereas strains from African-Americans appeared only in subgroup C2. Phylogenetic analysis of the HP0638 fragment retained geographic characters even when the CT repeat region was removed (data not shown).
FIG. 1.
Phylogram of H. pylori strains based on the first 150 bp of the HP0638 ORF. Regions from 109 strains were aligned by using GCG Pileup, subjected to phylogenetic analysis by using Paup 4.0b2, and subjected to neighbor-joining analysis based on Kimura’s two-parameter model distance matrices (25). Isolates from East Asia (blue) and western countries (red) segregate into two major branches (A and C). Branch C can be divided into two subbranches (C1 and C2); all strains from African-Americans (AFUSA) are on subbranch C2. vacA s-region allelic types also are shown. Branch B, which is paraphyletic to the other two major branches, is composed of strains from the Indian subcontinent (green).
CT dinucleotide repeats in the signal peptide coding region of HP0638.
CT repeat patterns differed between the groups of strains based on their geographic origin (Tables 2 and 3). Among the 46 strains isolated from East Asia, all had ≤5 CT repeats (Table 3, values in boldface), and the pattern of three CT repeats with another after four T’s (3 + 1-CT-repeat pattern) was the most common (67%). Conversely, among the 35 strains in which HP0638 was in frame and which were isolated from western countries, all but 2 had ≥6 CT repeats (Table 3, values in boldface), and the six-CT-repeat pattern was the most common (24 of 35 strains [69%]). The strains from India had mixed CT repeat patterns (Tables 2 and 3). Twenty strains in which HP0638 was out of frame showed variable CT repeat patterns. There was no correlation between specific CT repeat patterns and clinical outcome (DU, GU, NUD, or GCA) (Table 2). In strains with both in-frame and out-of-frame HP0638, 5-, 8-, 9-, or 12-CT-repeat patterns were found (Tables 2 and 3). In strains in which a TT sequence was present 6 bp upstream of the CT repeats or a CC sequence was present immediately upstream of the CT repeats, HP0638 was in frame, even when five CT repeats were present. For strains with CTAA deleted immediately upstream of the CT repeats, frame status depended on the number of repeats (8, in frame; 9 or 12, out of frame) (Table 2).
TABLE 3.
Comparison of variations in HP0638 CT repeat region sequences among the 109 H. pylori strains, grouped by HP0638 frame status
| HP0638 status | No. (%) of cagA-positive strains | No. of CT repeats | No. of strains by geographic regiona
|
|||
|---|---|---|---|---|---|---|
| East Asia (n = 46) | India (n = 8) | Western countries (n = 55) | Total (n = 109) | |||
| In frame (n = 89) | 89 (100) | 3 + 1 | 31 | 2 | 0 | 33 |
| <4 (other than 3 + 1) | 11 | 2 | 0 | 13 | ||
| 5 | 4 | 0 | 2 | 6 | ||
| 6 | 0 | 4 | 24 | 28 | ||
| 8 | 0 | 0 | 1 | 1 | ||
| 9 | 0 | 0 | 7 | 7 | ||
| 12 | 0 | 0 | 1 | 1 | ||
| Total | 46 | 8 | 35 | 89 | ||
| Out of frame (n = 20) | 4 (20) | 4 | 0 | 0 | 1 | 1 |
| 5 | 0 | 0 | 3 | 3 | ||
| 7 | 0 | 0 | 7 | 7 | ||
| 8 | 0 | 0 | 4 | 4 | ||
| 9 | 0 | 0 | 2 | 2 | ||
| 10 | 0 | 0 | 2 | 2 | ||
| 12 | 0 | 0 | 1 | 1 | ||
| Total | 0 | 0 | 20 | 20 | ||
For values shown in boldface, the distribution of CT repeats was significantly (P < 0.001) different between East Asia isolates (all 46 had ≤5 repeats) and Western isolates (33 of 35 had ≥6 repeats).
HP0638 status of strains from an extended family.
Next, we examined 18 strains from an extended Dutch family, including parents and six adult daughters (Table 4). Since the 15 strains from the mother and from her daughters all had been shown to have nearly identical randomly amplified polymorphic DNA patterns, they were believed to be from the same origin, whereas the strains from the father clearly differed (37). Each of the 15 strains from the mother or daughters showed the same unique 5 + 2-CT-repeat pattern, and HP0638 was in frame in all. In contrast, the father’s strains showed a six- or seven-CT-repeat pattern, and all had HP0638 out of frame (Table 4).
TABLE 4.
Variations in HP0638 CT repeat region sequences among members of an extended family in The Netherlandsa
| Strain designation | Patient age (yr)b | Family relationship | cagA status | No. of CT repeats | HP0638 CT repeat region sequence | Frame status |
|---|---|---|---|---|---|---|
| 1a | 60 | Father | − | 6 | CTCTTA(CT)6CGTTTTG | Out |
| 1b | − | 7 | CTCTTA(CT)7CGTTTTG | Out | ||
| 1c | − | 7 | CTCTTA(CT)7CGTTTTG | Out | ||
| 2a | 55 | Mother | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 2b | − | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 3a | 35 | Daughter 1 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 4a | 32 | Daughter 2 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 4b | − | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 5a | 29 | Daughter 3 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 5d | − | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 6a | 26 | Daughter 4 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 6b | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 6c | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 7a | 25 | Daughter 5 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 7b | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 8a | 24 | Daughter 6 | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
| 8b | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In | ||
| 8c | + | 5 + 2 | CTTTTA(CT)5TT(CT)2CGTT | In |
Strain designations, family relationships, and cagA status (−, negative; +, positive) were as previously described (37).
Age on date when H. pylori culture was obtained.
HP0638 status of paired isolates obtained years apart from the same host.
Next, we examined HP0638 frame status for pairs of H. pylori isolates obtained an average 8 years apart from each of 11 persons (Table 5). Nine of 11 strain pairs (patients 3 to 13) were highly related to one another, based on recA sequence analysis and on randomly amplified polymorphic DNA and amplified fragment length polymorphism analyses (19). In two (22%) of the nine patients, the number of CT repeats differed between the first and second isolates and, in both patients, HP0638 was out of frame in the first isolate. In only one patient (patient 5) did the HP0638 frame status differ between the two isolates; despite this difference, both isolates were cagA negative. In patient 9, the numbers of CT repeats were eight in the first isolate and seven in the second, but in both isolates HP0638 was out of frame.
TABLE 5.
Variations in HP0638 CT repeat region sequences in paired H. pylori strains obtained 7 to 10 years apart from the same hosta
| Strain designation | Yr apart |
H. pylori genotype
|
No. of CT repeats | HP0638 CT repeats region sequence | Frame status | ||||
|---|---|---|---|---|---|---|---|---|---|
| cagA |
vacA
|
iceA | IS605 | ||||||
| s type | m type | ||||||||
| 1A | 8.8 | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In |
| 1B | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In | |
| 2A | 7.2 | − | s2 | m2 | 2 | − | 6 | CTAA(CT)6CGTT | In |
| 2B | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In | |
| 3A | 7.9 | − | s2 | m2 | 2 | + | 10 | CTAA(CT)10CGTT | Out |
| 3B | − | s2 | m2 | 2 | + | 10 | CTAA(CT)10CGTT | Out | |
| 4A | 8.7 | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In |
| 4B | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In | |
| 5A | 7.0 | − | s2 | m2 | 2 | − | 7 | CTAA(CT)7CGTT | Out |
| 5B | − | s2 | m2 | 2 | − | 6 | CTAA(CT)6CGTT | In | |
| 7A | 7.0 | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In |
| 7B | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In | |
| 8A | 9.8 | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In |
| 8B | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In | |
| 9A | 8.8 | − | s2 | m2 | 2 | − | 8 | CTAA(CT)8CGTT | Out |
| 9B | − | s2 | m2 | 2 | − | 7 | CTAA(CT)7CGTT | Out | |
| 10A | 10.2 | + | s1b | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In |
| 10B | + | s1b | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In | |
| 11A | 7.8 | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In |
| 11B | + | s1a | m1 | 2 | − | 6 | CTAA(CT)6CGTT | In | |
| 13A | 7.4 | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In |
| 13B | + | s1a | m1 | 1 | + | 6 | CTAA(CT)6CGTT | In | |
Strain designations and genotypes (+, positive; −, negative) were as described previously (19).
DISCUSSION
Although H. pylori has worldwide distribution (39, 41), the important differences in the vacA s1 alleles (39) and in the 5′ region of cagA (41), among other polymorphisms, strongly indicate that certain strains predominate in specific geographic areas. Despite evidence suggesting such geographic characters for H. pylori (1, 11, 23, 45), it remains difficult to clearly stratify H. pylori strains into geographic groupings. Therefore, we examined additional strain-specific markers that clearly differentiate H. pylori strains. We found clear geographic differences in HP0638 sequences for the first 150 nucleotides (Fig. 1), and we found geographic characters even for the CT repeat patterns in the signal sequence region (Table 3). HP0638 frame status correlated strongly with cagA and vacA status; all of the strains in which HP0638 was in frame were cagA positive and vacA s1, whereas most of the strains in which HP0638 was out of frame were cagA negative (80%) and vacA s2 (70%). In our study, HP0638 frame status was most closely correlated with cagA status, suggesting that cagA positivity could affect selection for in-frame status. Further studies are needed to assess the correlation between HP0638 status and cagA positivity.
That the isolates from all the daughters in the extended Dutch family shared the same 5 + 2-CT-repeat pattern and HP0638 in-frame status with the isolates from the mother but not the father further supports the presumption that the daughters acquired their H. pylori isolates from their mother, confirming and extending previous findings (37). These findings also suggest the stability of the HP0638 frame status and CT repeat pattern under in vivo conditions over the presumed combined 150 person-years of colonization. The stability of HP0638 sequences in this setting encourages us to examine other epidemiologically related strains to assess variability and to determine the underlying causes of variability. The CT repeat differences observed in the HP0638 signal sequence regions in two of nine paired strains from the same hosts (Table 4) are consistent with the presumed high frequency of slipped-strand mispairing associated with dinucleotide repeats (27) and further support the concept that the H. pylori population in an individual host represents a mixture of closely related clonal variants, or “quasispecies” (19). That the 206-nucleotide sequence within recA was identical within each pair (19) indicates that the strains from each patient had a similar origin and suggests that the CT repeat region may be hypervariable. Alternatively, it is possible that the numbers of HP0638 CT repeats in the paired strains changed as a result of in vitro passage. Arguing against that hypothesis is the finding that the 15 related isolates obtained from the extended family were all identical for frame status and number of CT repeats (Table 5). Further long-term in vitro passages and animal models will help elucidate whether in vivo variation or in vitro passage accounts for changes in HP0638 frame status in H. pylori.
Of the 109 patients studied, those carrying strains in which HP0638 was in frame had more severe diseases than did those with strains in which HP0638 was out of frame (NUD, [70%]) (Table 3). Among the strains of each group, with HP0638 either in frame or out of frame, there was no correlation between specific CT repeat patterns and clinical outcome. Thus, among HP0638 polymorphisms, only HP0638 frame status affected clinical status. However, the correlation of HP0638 frame status with cagA status was so strong (Table 2) that it was not possible to establish the relationship of HP0638 status to pathogenicity. Yamaoka et al. (44) reported that HP0638 mutants induced significantly lower IL-8 levels from AGS gastric epithelial cells than did their wild-type parent strains. However, in a recent study (5a), HP0638 disruption did not affect IL-8 production or CagA tyrosine phosphorylation status in AGS gastric epithelial cells. Akanuma et al. [M. Akanuma, K. Ogura, S. Maeda, Y. Mitsuno, Y. Hirata, H. Yoshida, Y. Shiratori, and M. Omata, Abstr. Gastroenterol. 120(Suppl. 1):A-100, 2001] also reported that HP0638 mutants induced IL-8 levels similar to those induced by their wild-type parent strains but that HP0638 is important for gastric H. pylori infection of gerbils. In vitro experiments indicate that IL-8 is induced in epithelial cells after viable H. pylori organisms attach (30); outer membrane proteins would be good candidates for the unknown proinflammatory virulence factors. Further study of the role of HP0638 as well as other outer membrane proteins in H. pylori pathogenesis is needed.
In summary, the phylogeny of HP0638 has geographic characters, and its frame status is correlated with cagA, vacA, and iceA genotypes. The CT dinucleotide repeat pattern in the putative signal sequence of HP0638 also has geographic characters and appears relatively stable in individual patients and families over periods of years. Nevertheless, strains obtained from the same host and appearing otherwise identical can vary in frame status. HP0638 frame status but not the number or pattern of CT repeats is correlated with clinical outcome. If confirmed in independent studies, analysis of HP0638 polymorphisms may be useful as an H. pylori typing system for epidemiological purposes.
Acknowledgments
This work was supported in part by grants R01GM63270, R01DK53707, K08DK02381, and R29CA77955 from the National Institutes of Health and by the Medical Research Service of the Department of Veterans Affairs.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459–470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D, Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akopyanz, N. S., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180. [DOI] [PubMed] [Google Scholar]
- 5.Ando, T., G. I. Perez-Perez, K. Kusugami, M. Ohsuga, K. C. Bloch, and M. J. Blaser. 2000. Anti-CagA immunoglobulin G responses correlate with interleukin-8 induction in human gastric mucosal biopsy culture. Clin. Diagn. Lab. Immunol. 7:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Ando, T., R. M. Peek, Y.-C. Lee, U. Krishna, K. Kusugami, and M. J. Blaser. Host cell responses to genotypically similar Helicobacter pylori isolates from United States and Japan. Clin. Diagn. Lab. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 6.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771–17777. [DOI] [PubMed] [Google Scholar]
- 7.Atherton, J. C., P. M. Sharp, T. L. Cover, G. Gonzalez-Valencia, R. M. Peek, S. A. Thompson, C. J. Hawkey, M. J. Blaser. 1999. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr. Microbiol. 39:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser, M. J. 1999. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J. Infect. Dis. 179:1523–1530. [DOI] [PubMed] [Google Scholar]
- 9.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111–2115. [PubMed] [Google Scholar]
- 10.Bukanov, N. O., and D. E. Berg. 1994. Ordered cosmid library and high-resolution physical-genetic map of Helicobacter pylori strain NCTC11638. Mol. Microbiol. 11:509–523. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, S., A. Fraser, B. Holliss, J. Schmid, and P. W. O’Toole. 1997. Evidence for ethnic tropism of Helicobacter pylori. Infect. Immun. 65:3708–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cover, T. L,. M. K. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566–10573. [PubMed] [Google Scholar]
- 15.Cover, T. L., Y. Glupczynski, A. P. Lage, A. Burette, M. K. Tummuru, G. I. Perez-Perez, and M. J. Blaser. 1995. Serologic detection of infection with cagA+ Helicobacter pylori strains. J. Clin. Microbiol. 33:1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, J. A., and T. L. Cover. 1995. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J. Infect. Dis. 172:290–293. [DOI] [PubMed] [Google Scholar]
- 17.Ito, Y., T. Azuma, S. Ito, H. Miyaji, M. Hirai, Y. Yamazaki, F. Sato, T. Kato, Y. Kohli, and M. Kuriyama. 1997. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J. Clin. Microbiol. 35:1710–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31–43. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers, E. J., D. A. Israel, J. G. Kusters, M. M. Gerrits, J. Weel, A. van Der Ende, R. W. van Der Hulst, H. P. Wirth, J. Hook-Nikanne, S. A. Thompson, and M. J. Blaser. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuipers, E. J., G. I. Pérez-Pérez, S. G. M. Meuwissen, and M. J. Blaser. 1995. Helicobacter pylori and atrophic gastritis; importance of the cagA status. J. Natl. Cancer Inst. 87:1777–1780. [DOI] [PubMed] [Google Scholar]
- 21.Leunk. R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93–99. [DOI] [PubMed] [Google Scholar]
- 22.Mobley, H. L. 1996. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am. J. Med. 100:2S–9S. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497–1500. [DOI] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358. [DOI] [PubMed] [Google Scholar]
- 26.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220–226. [DOI] [PubMed] [Google Scholar]
- 27.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt, W., and R. Haas. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12:307–319. [DOI] [PubMed] [Google Scholar]
- 30.Sharma, S. A., M. K. Tummuru, G. G. Miller, and M. J. Blaser. 1995. Interleukin-8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect. Immun. 63:1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tee, W., J. Lambert, R. Smallwood, M. Schembri, B. C. Ross, and B. Dwyer. 1992. Ribotyping of Helicobacter pylori from clinical specimens. J. Clin. Microbiol. 6:1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telford, J. L., P. Ghiara, M. Dell’Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, and Z. Xiang. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J. Exp. Med. 179:1653–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. [DOI] [PubMed] [Google Scholar]
- 35.Tummuru, M. K., S. A. Sharma, and M. J. Blaser. 1995. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol. Microbiol. 18:867–876. [DOI] [PubMed] [Google Scholar]
- 36.Tummuru, M. K. R., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61:1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ende, A., E. A. J. Rauws, M. Feller, C. J. J. Mulder, G. N. J. Tytgat, and J. Dankert. 1996. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology 111:638–647. [DOI] [PubMed] [Google Scholar]
- 38.van der Ende, A., Z. J. Pan, A. Bart, R. W. van der Hulst, M. Feller, S. D. Xiao, G. N. Tytgat, and J. Dankert. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823–830. [DOI] [PubMed] [Google Scholar]
- 40.van Doorn, L. J., C. Figueiredo, R. Rossau, G. Jannes, M. van Asbroek, J. C. Sousa, F. Carneiro, and W. G. Quint. 1998. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J. Clin. Microbiol. 36:1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Doorn, L. J., C. Figueiredo, R. Sanna, M. J. Blaser, and W. G. Quint. 1999. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J. Clin. Microbiol. 37:2306–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doorn, L. J., Y. Henskens, N. Nouhan, A. Verschuuren, R. Vreede, P. Herbink, G. Ponjee, K. van Krimpen, R. Blankenburg, J. Scherpenisse, and W. Quint. 2000. The efficacy of laboratory diagnosis of Helicobacter pylori infections in gastric biopsy specimens is related to bacterial density and vacA, cagA, and iceA genotypes. J. Clin. Microbiol. 38:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, K. 1995. Preparation of genomic DNA from bacteria, p. 2.4.1–2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 44.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaoka, Y., H. M. Malaty, M. S. Osato, and D. Y. Graham. 2000. Conservation of Helicobacter pylori genotypes in different ethnic groups in Houston, Texas. J. Infect. Dis. 181:2083–2086. [DOI] [PubMed] [Google Scholar]
- 46.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]