Abstract
Mutations in Mycobacterium tuberculosis uvrB result in severe sensitivity to acidified nitrite, a source of nitric oxide (6). In this study, we show that a uvrB mutant is exquisitely sensitive to UV light but not to several sources of reactive oxygen species in vitro. Furthermore, a uvrB mutant was attenuated in mice as judged by an extension of life span. Attenuation in mice was partially reversed by genetic inactivation of nitric oxide synthase 2 (iNOS) and almost completely reversed in mice lacking both iNOS and phagocyte oxidase. Thus, a gene predicted to encode a key element of DNA repair is required for resistance of M. tuberculosis to both reactive nitrogen and reactive oxygen species in mice.
Cellular DNA is subject to damage by exogenous and endogenously generated oxidants and radicals. Pathogens such as Mycobacterium tuberculosis that infect a mammalian host have to deal with phagocytes that produce both reactive oxygen and reactive nitrogen intermediates (ROI and RNI, respectively) that are toxic to bacteria, including M. tuberculosis (5). ROI and RNI can damage lipids, proteins, and nucleic acids (1, 22). Enzymes such as methionine sulfoxide reductases (19, 25) can reverse some oxidative damage of proteins. Oxidized proteins that escape repair may be targeted for degradation, lest they form aggregates that damage the bacterium (6). Numerous pathways operate to repair damaged DNA.
Exposure to M. tuberculosis often results in latent infection in humans rather than sterilizing immunity (10). In wild-type (wt) mice, phagocytes in mice infected with M. tuberculosis express inducible nitric oxide synthase (iNOS) (11, 14) and control M. tuberculosis infection for months or years but fail to eradicate it. In contrast, M. tuberculosis infection progresses rapidly to lethality in mice that lack iNOS (11). These observations suggested that M. tuberculosis, to a relevant degree, resists the damaging effects of iNOS in vivo. We tested this hypothesis by screening for M. tuberculosis mutants that are hypersusceptible to mildly acidified nitrite (a physiological source of nitric oxide [NO]) in vitro (6). Mutations in genes that encode subunits of a presumptive M. tuberculosis proteasome resulted in severe attenuation of M. tuberculosis lethality in mice (5, 6). This phenotype was partially reversed in mice lacking iNOS, supporting the notion that susceptibility to products of iNOS was one of the defects of these M. tuberculosis mutants. Thus, M. tuberculosis must resist products of iNOS to kill wt mice.
Among the other mutants identified in the screen, we found two with mutations in the nucleotide excision repair (NER) gene uvrB. Of all NO-sensitive mutants identified in the screen, uvrB mutants were the most profoundly sensitive to acidified nitrite (6). In earlier studies, a recA mutant of Mycobacterium bovis bacillus Calmette-Guerin (BCG) had no detectable phenotype in a mouse for up to 80 days postinfection (17). A mutation in dnaE2, a homolog of dnaE, attenuated M. tuberculosis in wt C57BL/6 mice but did not affect the bacterial load until late in the infection (2). Thus, although recA and dnaE2 mutants had increased susceptibility to mutagenesis in vitro, they had distinctively different phenotypes in the mouse. This suggested that not all DNA repair pathways in M. tuberculosis contribute equally, if at all, to survival in the host.
In this study, we examined the role of uvrB after exposure of M. tuberculosis to UV light, RNI, and ROI. Furthermore, we determined the contribution of uvrB in a mouse model of infection. Similar to what was observed for the dnaE2 M. tuberculosis mutant, a uvrB mutant did not grow to the same levels as wt M. tuberculosis in a wt mouse. Inactivation of iNOS only partially reduced the attenuation of the uvrB mutant. However, the uvrB mutant was almost fully virulent in mice deficient in both gp91phox and iNOS. This suggested that uvrB is required for M. tuberculosis to resist both RNI and ROI in vivo.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Table 1 lists strains and plasmids. Escherichia coli was grown in Luria Bertani (LB) broth or on LB agar plates (Difco). All M. tuberculosis strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, 0.5% bovine serum albumin, 0.2% dextrose, and 0.085% sodium chloride (7H9-ADN). M. tuberculosis solid medium used Middlebrook 7H11 agar supplemented with Middlebrook enrichment (oleic acid, albumin, dextrose, and catalase [OADC]). For M. tuberculosis, kanamycin and hygromycin were added when needed, each at 50 μg/ml. For E. coli, kanamycin, carbenicillin, and hygromycin were used at 100, 200, and 200 μg/ml, respectively. M. tuberculosis cultures were grown in 75-cm2 vented flasks in humidified incubators with 5% CO2.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype and/or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pMV306 | Hygr; integrates at att site on mycobacterial chromosome | 20 |
| pMV-uvrB | Hygr; same as pMV306 but with M. tuberculosis uvrB | This work |
| Strains | ||
| E. coli DH5α | λF− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK−mK−) supE44 thi-1 gyrA relA1 | Gibco-BRL |
| M. tuberculosis | ||
| H37Rv | wt | American Type Culture Collection |
| MHD18 | H37Rv pMV306 | 6 |
| MHD7 | fbiC::ΦMycoMarT7, codon 314 | 6 |
| MHD9 | uvrB::ΦMycoMarT7, codon 400 | 6 |
| MHD13 | uvrB::ΦMycoMarT7, codon 476 | 6 |
| MHD34 | same as MHD9 with pMV306 | This work |
| MHD35 | same as MHD9 with pMV-uvrB | This work |
| MHD36 | same as MHD13 with pMV306 | This work |
| MHD37 | same as MHD13 with pMV-uvrB | This work |
Plasmid constructions.
Table 2 lists primers (Invitrogen). The complementing plasmid, pMV-uvrB, was constructed by PCR amplification of uvrB. Two primer pairs were designed to amplify slightly overlapping halves of uvrB, each fragment including the unique SacI site that bisects uvrB. In addition, the 5′ fragment included 200 bp of sequence upstream of the start codon. Each amplified fragment was cloned separately into pHG329 and sequenced (Cornell Bio Resource Center). After sequence confirmation of each half, the 5′ region clone was digested with HindIII and SacI, while the 3′ region clone was digested with SacI and XbaI. Both fragments were simultaneously ligated to pMV306 that had been digested with HindIII and XbaI. E. coli was transformed chemically (16), and M. tuberculosis was transformed by electroporation (9).
TABLE 2.
Primers used in this work
| Primer name | Sequence |
|---|---|
| uvrB-HindIII-f1 | CCCAAGCTTGTCTCGTGCTTGGGTGGGTG |
| uvrB-r1 | GAACTCGCCGCCGGTCTG |
| uvrB-f2 | GTGTATCTGTCTGCCACCCC |
| uvrB-XbaI-f2 | GCTCTAGAGCTCACGCCAGCTGCC |
| uvrB-f3 | GCGCTGCTGTCGCGTCG |
Nitrite survival assays.
For NO survival assays, M. tuberculosis from stationary-phase cultures (optical density at 580 nm [OD580] of 1 to 2) were centrifuged in 50-ml conical tubes at 3,310 × g in a Sorvall T21 tabletop centrifuge for 8 min at room temperature. Pellets were resuspended in a half volume of 7H9-ADN acidified with HCl (pH 5.5). The cells were then centrifuged at 130 × g for 8 min. The supernatants were single-cell suspensions, while the pellet contained clumps. The OD580s of the supernatants were measured, and the supernatants were diluted into acidified media to an OD580 of 0.08. One hundred ninety-four microliters of each suspension was placed into 6 wells of a 96-well flat-bottomed plate. Three wells received no treatment (“pH 5.5 only”), and three wells each received 6 μl of 0.1 M sodium nitrite (final NaNO2 concentration, 3 mM). Trays were incubated at 37°C for 6 days. Cultures were diluted and plated for surviving CFU.
UV light exposure.
Single-cell suspensions of M. tuberculosis cultures were made as described for the NO-killing assays but using regular 7H9-ADN. An aliquot (5 to 10 μl) of serially diluted cultures was spotted onto Middlebrook 7H11-OADC and exposed to UV light in a biosafety cabinet, measuring UV intensity with a dosimeter.
Susceptibility to ROI.
M. tuberculosis cultures were grown to stationary phase and diluted to an OD580 of 0.15. An aliquot (150 μl) of culture was spread onto 7H11-OADC agar and allowed to dry for about 10 min. Six-millimeter paper disks (BBL) were placed in the center of the bacterial lawns and spotted with the compound of interest. Duplicate plates were prepared. After 1 week, zones of inhibition were measured. The vehicle controls, dimethyl sulfoxide or ethanol, resulted in no inhibition of M. tuberculosis growth (data not shown).
Bone marrow macrophage isolation and infection.
wt C57BL/6 mice were from Jackson Laboratories. Mice deficient in iNOS or both iNOS and gp91phox on the C57BL/6 background were from our own colony (18) or purchased from Jackson Laboratories. For convenience, mice lacking both phoxgp91 and iNOS are termed “phox/NOS−/−.” Femoral bone marrow cells were seeded into 1-cm-diameter tissue culture wells at 2 × 105/well in 0.5 ml of Dulbecco's minimal Eagle's medium supplemented with 10% fetal bovine serum, 10% L-cell conditioned medium, 10 mM HEPES, 0.6 g/liter l-glutamine, and 1 mM pyruvate, with or without 50 U/ml of pure, recombinant mouse gamma interferon (IFN-γ; Genentech). After 6 to 7 days of differentiation, the medium was replaced with fresh medium with or without 50 U/ml of IFN-γ. To prepare the M. tuberculosis inocula, bacteria from mid-logarithmic-phase cultures (OD580 of 0.4) were centrifuged in 50-ml conical tubes at 3,310 × g for 8 min at room temperature. Pellets were resuspended in phosphate-buffered saline (PBS)-0.05% Tween 80 (PBS-Tween) and then centrifuged at 130 × g for 8 min. The OD580 of the supernatant was measured, and the amount of bacteria per ml was determined. Bacteria were then centrifuged at 3,310 × g for 8 min and resuspended in Dulbecco's minimal Eagle's medium. Macrophages were infected with M. tuberculosis at a multiplicity of infection of 4. Infected macrophages were washed with PBS and lysed with 0.5% Triton X-100 to recover intracellular bacteria at 4, 72, and 144 h. To assess macrophage activation, nitrite production was measured by a Griess assay (7). In the experiments presented, wt macrophages produced 30 to 40 μM nitrite by day 2.
Mouse infections.
To prepare bacteria for aerosol infection, M. tuberculosis from mid-logarithmic-phase cultures (OD580 of 0.4) were centrifuged in 50-ml conical tubes at 3,310 × g for 8 min at room temperature. Pellets were resuspended in a half volume of PBS-Tween and then centrifuged at 130 × g for 8 min. The OD580 of the supernatant was measured, and the suspension was diluted to an OD580 of 0.04 in PBS. CFU (108) were placed into the nebulizer of a Glas-Col inhalation exposure system (Terre Haute, IN). Each mouse received ∼100 to 200 bacilli. This was determined in 3 mice per strain of mouse and per strain of M. tuberculosis 1 day after infection by counting CFU recovered from the lungs. Organs were homogenized in 4 ml of PBS-Tween on day 1 or in 5 ml of PBS-Tween at week 3, 8, or 15. For survival experiments, groups of 4 to 5 mice were infected. Mice were humanely sacrificed when they became cachectic and lethargic.
RESULTS
Complementation of uvrB mutants reverses hypersusceptibility to acidified nitrite; mutants are susceptible to UV light.
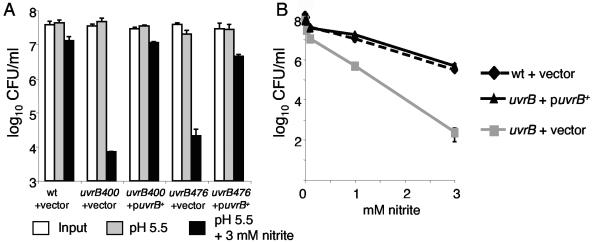
In a previous study, we screened 10,100 ΦMycoMarT7 mutants for M. tuberculosis that were sensitive to acidified nitrite (6). Two uvrB mutants of 12 mutants total were the most sensitive to acidified nitrite. The uvrB stop codon overlaps the putative start codon of Rv1634, which putatively encodes an efflux pump protein (http://genolist.pasteur.fr/TubercuList/). Therefore, it is possible that the two genes are cotranscribed. The ΦMycoMarT7 transposon insertion in both uvrB mutants had the kanamycin gene expressed in the same direction as uvrB and Rv1634. Therefore, it was possible that even if uvrB and Rv1634 were in an operon, the kanamycin resistance promoter allowed for wt Rv1634 expression. To determine whether the nitrite-sensitive phenotype was due to disruption of uvrB or polar effects on Rv1634, we complemented both uvrB mutations in single copy. A plasmid encoding only uvrB was sufficient to fully complement the uvrB mutations for sensitivity to nitrite (Fig. 1A).
FIG. 1.
Complementation of uvrB mutations for nitrite sensitivity. (A) Two independent uvrB::ΦMycoMarT7 mutants were complemented with the integrative plasmid pMV-uvrB. (B) Nitrite dose-response curves of wt, uvrB, and complemented strains. All strains contain either the empty vector pMV306 or pMV-uvrB integrated at the att site of the chromosome. Each datum point represents the average of triplicates from one experiment and is representative of the results from three independent experiments. Error bars represent standard deviations.
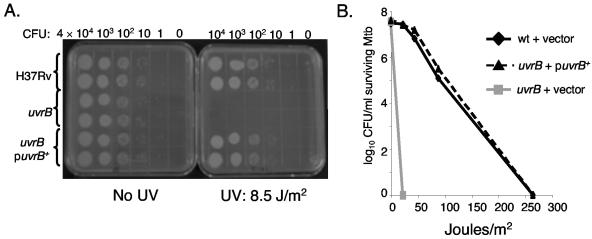
Next we tested the susceptibility of a uvrB mutant to UV light. There were no detectable CFU of the uvrB mutant at the lowest UV dose tested (Fig. 2A). In contrast, there was no loss of viability of either the wt or complemented strains at this UV dose. (Fig. 2B).
FIG. 2.
uvrB mutants are hypersusceptible to UV light. (A) Growth of M. tuberculosis strains after exposure to 8.5 mJ/m2 of UV light. (B) UV dose-response curve. All strains were transformed by either plasmid pMV306 or pMV-uvrB. Each datum point represents the average of duplicates from one experiment and is representative of the results from three independent experiments.
uvrB is not required for protection against several sources of ROI in vitro.
We have previously shown that a uvrB mutant is not hypersusceptible to 5 mM hydrogen peroxide (6). We went on to test the susceptibility of a uvrB mutant to cumene hydroperoxide, an organic peroxide, and plumbagin, a generator of intracellular superoxide. The uvrB mutant was no more sensitive than wt or complemented M. tuberculosis to these chemicals in a disk diffusion assay (Table 3).
TABLE 3.
Susceptibility to ROI
| Strain | Zone of inhibition (cm)awith:
|
||
|---|---|---|---|
| Cumene hydroperoxideb | 5 mM Plumbaginc | 50 mM Plumbaginc | |
| wt + vector | 3.1 | 2.0 | 4.2 |
| uvrB mutant + vector | 3.1 | 2.0 | 4.2 |
| uvrB mutant + pMV-uvrB | 3.0 | 2.0 | 4.0 |
Each value represents the average of the results from duplicate plates from one experiment and is representative of the results from two independent experiments.
0.2% in dimethyl sulfoxide.
The concentrations represent the starting concentrations of the compound placed onto 6-mm filter disks on the bacterial lawns.
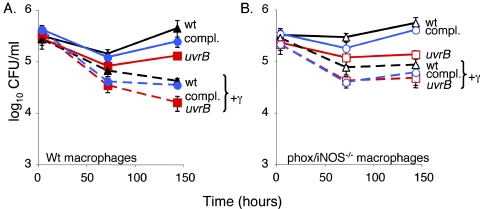
A uvrB mutant has slightly decreased survival in bone marrow-derived macrophages.
During an infection, M. tuberculosis primarily resides in macrophages in the lungs. Activated macrophages produce several antimicrobial molecules, including RNI and ROI (15). A uvrB mutant had a reproducible but subtle growth defect in both wt and phox/iNOS−/− bone marrow-derived macrophages (Fig. 3). By 6 days, there were two- to fourfold more wt or complemented M. tuberculosis CFU than uvrB mutant M. tuberculosis CFU. Replacing uvrB in single copy complemented the defect. Activation of the macrophages with IFN-γ did not enhance the growth restriction. Therefore, additional factors independent of exogenous IFN-γ help control the growth of M. tuberculosis in macrophages in vitro.
FIG. 3.
A uvrB mutant has a slight growth defect in wt and phox/iNOS−/− bone marrow-derived macrophages. Macrophages were infected with M. tuberculosis at a multiplicity of infection of 4. Each datum point represents the average of triplicates ± standard deviation from one of two representative experiments. Differences between wt and uvrB strains in unactivated wt or phox/iNOS−/− macrophages are statistically significant according to Student's unpaired t test (P < 0.05). compl., complemented; γ, IFN-γ.
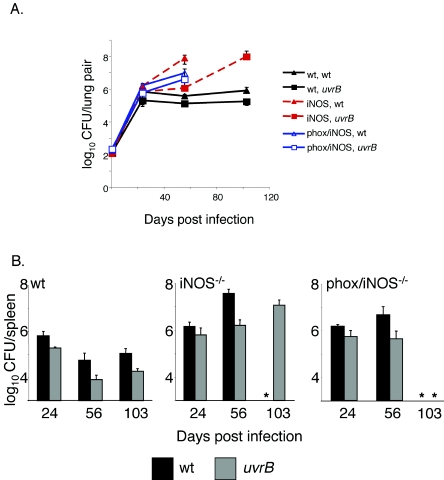
A uvrB mutant is attenuated in wt and iNOS−/− mice, but not in phox/iNOS−/− mice.
To test the survival of a uvrB mutant in mice, we infected groups of wt and iNOS−/− mice by aerosol. During the first 3 weeks of infection, M. tuberculosis grows exponentially, after which time adaptive immunity slows the growth of the bacteria. Induction of iNOS is one of the key events that controls M. tuberculosis infections (11). The uvrB mutant grew exponentially during the first 3 weeks of infection in all mouse strains but did not grow as well as the wt M. tuberculosis strain in the lungs and spleen after 3 weeks (Fig. 4A and B). Moreover, attenuation of the uvrB mutant was evident in the lungs and spleens of iNOS−/− mice. Mice lacking iNOS quickly succumbed to infection with wt M. tuberculosis (Fig. 5). However, iNOS−/− mice infected with the uvrB mutant lived considerably longer. Although the uvrB mutant was attenuated in the iNOS−/− mice, it grew better than it did in wt mice (Fig. 4A). These results suggested that the uvrB mutant was susceptible not only to iNOS but also to other immune defenses.
FIG. 4.
A uvrB mutant does not grow as well as wt M. tuberculosis in either wt or iNOS−/− mice but returns to virulence in phox/iNOS-deficient mice. (A) CFU from the lungs of wt, iNOS, or phox/iNOS-deficient mice. The difference between wt and uvrB M. tuberculosis in wt mice at day 56 was statistically significant based on Student's unpaired t test (P = 0.006). Values between wt and uvrB M. tuberculosis in phox/iNOS−/− mice were not considered statistically significant. (B) CFU from spleens of the same mice represented in panel A. Each datum point represents the average of the results from 3 to 4 mice. Error bars represent ±1 standard deviation. *, no mice survived to this time point.
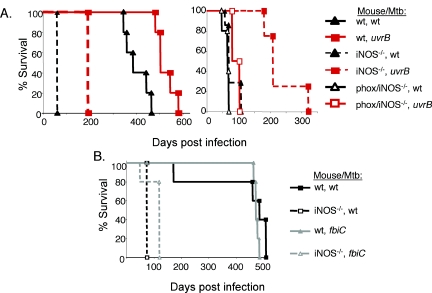
FIG. 5.
A uvrB mutant is attenuated in wt and iNOS−/− mice but returns to virulence in phox/iNOS-deficient mice. (A) The first experiment shows that the uvrB mutant is attenuated in both wt and iNOS−/− mice (left panel). The uvrB mutant is attenuated in iNOS−/− mice but is almost as virulent as wt M. tuberculosis in phox/iNOS−/− mice (right panel). Groups of 4 to 5 mice were infected for each survival curve. wt mice for the second experiment were all still alive at the time the manuscript was submitted. (B) An fbiC mutant is not attenuated in mice. Groups of 5 wt or iNOS−/− mice were infected with an aerosol of an fbiC mutant M. tuberculosis strain as described for the uvrB mutants.
Although the uvrB mutant did not appear to be more susceptible than wt M. tuberculosis to several ROI tested individually in vitro, it was possible that the uvrB mutant was sensitive to a mixture of ROI or to products of phox we did not test, such as ozone (26). We infected wt, iNOS−/−, and phox/iNOS−/− mice by aerosol and examined CFU in the lungs and spleens 3, 8, and 15 weeks after infection. Mice lacking both iNOS and gp91phox−/− were as susceptible to wt M. tuberculosis as iNOS−/− mice (Fig. 5A), consistent with published results (10). However, mice deficient in both gp91phox and iNOS were far more susceptible to the uvrB mutant than iNOS−/− mice (Fig. 4B and 5A).
In addition to the uvrB mutants, two fbiC mutants were identified in our screen for RNI-sensitive mutants (6). Unlike the uvrB mutant, an fbiC mutant did not appear to produce a different course of infection from wt M. tuberculosis in either wt or iNOS−/− mice (Fig. 5B). This suggests that although fbiC is essential for resistance to RNI in vitro, it is dispensable for in vivo exposure to RNI.
DISCUSSION
We identified two independent transposon mutations in the uvrB gene of M. tuberculosis that sensitized the bacteria to acidified nitrite (6). We have confirmed in the present work that the same mutations sensitize the bacteria to UV light and that complementation restores wt resistance both to nitrite and to UV. The new biologic insights in this report are as follows. First, a uvrB mutant was markedly attenuated in both wt and iNOS−/− mice and genetic inactivation of phox allowed for near wt growth of a uvrB mutant in iNOS-deficient mice. These data suggest that both iNOS and phox are important for controlling M. tuberculosis infections in mice. Second, although we did not obtain biochemical evidence for increased DNA damage in uvrB mutant M. tuberculosis in mice, the genetic evidence presented here strongly implicates DNA as one of the first specific molecular targets to be identified in M. tuberculosis whose damage by the host suppresses the virulence of the bacterium. More specifically, these findings suggest that DNA is a critical target in M. tuberculosis for both iNOS and phox. Finally, our studies suggest that the Uvr system in M. tuberculosis is essential for the organism to cause premature death in immunocompetent mice, raising the possibility of targeting UvrB for experimental chemotherapy.
Phagocyte oxidase has not previously been shown to be important for the control of M. tuberculosis infections either alone or in conjunction with deficiency of iNOS (4, 10). However, judging by the life span of the mice, it is apparent that phox plays a potential role in protecting the host that is normally fended off by UvrB, as revealed when uvrB is disrupted.
It is notable that other genes (e.g., uvrA, C, and D) involved in the repair of UV-induced damage were not identified in our screen (6). Other gene products may be able to substitute for their functions. Alternatively, the original screen may not have been saturating. Thus, it remains possible that other uvr mutants would have the same phenotype as the uvrB mutants.
Although NER and other forms of DNA repair have been intensely studied in bacteria (2, 13, 17, 21, 24), the role of NER for bacterial survival in a mammalian host has apparently not been reported. The function of UvrB is to excise damaged nucleotides from DNA (23). Expression of several uvr genes, including uvrB, was increased in M. tuberculosis within human macrophages (8). A mutation in Salmonella enterica serovar Typhimurium uvrB resulted in enhanced mutagenesis by NO donors, which was thought likely to be attributable to deamination of cytosine nucleotides and resultant transition mutations (12). A recombination-defective S. enterica serovar Typhimurium recBC mutant that was attenuated in wt mice (3) was as virulent as wt bacteria in mice deficient in phox or in both phox and iNOS (18). In contrast, a recA mutant of M. bovis BCG was not attenuated in mice more than the parental strain (17), although the fact that the parental strain is already attenuated may have masked a recA phenotype. We did not identify recA, dnaE2, or other components of NER other than uvrB in our screen (6). As mentioned earlier, there are several possible reasons for this, including that the mutagenesis was not saturating or that these genes are individually dispensable for resistance to nitrite in vitro.
Acknowledgments
We thank William Holloman for invaluable advice on DNA repair assays and Jon Nezezon for mouse husbandry. Animal work was approved by and performed under the guidelines of the Institutional Animal Care and Use Committee.
This work was funded by NIH grants HL61241 and AI055548 to C.F.N. and T32 AI07621 to K.H.D.
Editor: V. J. DiRita
REFERENCES
- 1.Alvarez, B., and R. Radi. 2003. Peroxynitrite reactivity with amino acids and proteins. Amino Acids 25:295-311. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933-936. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, A. M., B. H. Segal, A. A. Frank, S. M. Holland, and I. M. Orme. 2000. Transient loss of resistance to pulmonary tuberculosis in p47phox−/− mice. Infect. Immun. 68:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin, K., G. Lin, Z. Chen, H. Li, and C. Nathan. 2005. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 55:561-571. [DOI] [PubMed] [Google Scholar]
- 6.Darwin, K. H., S. Ehrt, N. Weich, J.-C. Gutierrez-Ramos, and C. F. Nathan. 2003. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302:1963-1966. [DOI] [PubMed] [Google Scholar]
- 7.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatfull, G. F., and J. W. R. Jacobs. 2000. Molecular genetics of Mycobacteria. ASM Press, Washington, D.C.
- 10.Hisert, K. B., M. A. Kirksey, J. E. Gomez, A. O. Sousa, J. S. Cox, W. R. Jacobs, Jr., C. F. Nathan, and J. D. McKinney. 2004. Identification of Mycobacterium tuberculosis counterimmune (cim) mutants in immunodeficient mice by differential screening. Infect. Immun. 72:5315-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacMicking, J. D., R. J. North, R. LaCourse, J. S. Mudgett, S. K. Shah, and C. F. Nathan. 1997. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA 94:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maragos, C. M., A. W. Andrews, L. K. Keefer, and R. K. Elespuru. 1993. Mutagenicity of glyceryl trinitrate (nitroglycerin) in Salmonella typhimurium. Mutat. Res. 298:187-195. [DOI] [PubMed] [Google Scholar]
- 13.Merino, D., H. Reglier-Poupet, P. Berche, and A. Charbit. 2002. A hypermutator phenotype attenuates the virulence of Listeria monocytogenes in a mouse model. Mol. Microbiol. 44:877-887. [DOI] [PubMed] [Google Scholar]
- 14.Mogues, T., M. E. Goodrich, L. Ryan, R. LaCourse, and R. J. North. 2001. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J. Exp. Med. 193:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nathan, C. F., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Sander, P., K. G. Papavinasasundaram, T. Dick, E. Stavropoulos, K. Ellrott, B. Springer, M. J. Colston, and E. C. Bottger. 2001. Mycobacterium bovis BCG recA deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. Infect. Immun. 69:3562-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiloh, M. U., J. D. MacMicking, S. Nicholson, J. E. Brause, S. Potter, M. Marino, F. Fang, M. Dinauer, and C. Nathan. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29-38. [DOI] [PubMed] [Google Scholar]
- 19.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 21.Suvarnapunya, A. E., H. A. Lagasse, and M. A. Stein. 2003. The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 48:549-559. [DOI] [PubMed] [Google Scholar]
- 22.Szabo, C. 2003. Multiple pathways of peroxynitrite cytotoxicity. Toxicol. Lett. 140-141:105-112. [DOI] [PubMed] [Google Scholar]
- 23.Theis, K., M. Skorvaga, M. Machius, N. Nakagawa, B. Van Houten, and C. Kisker. 2000. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat. Res. 460:277-300. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, S. A., R. L. Latch, and J. M. Blaser. 1998. Molecular characterization of the Helicobacter pylori uvrB gene. Gene 209:113-122. [DOI] [PubMed] [Google Scholar]
- 25.Weissbach, H., F. Etienne, T. Hoshi, S. H. Heinemann, W. T. Lowther, B. Matthews, G. St. John, C. Nathan, and N. Brot. 2002. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 397:172-178. [DOI] [PubMed] [Google Scholar]
- 26.Wentworth, P., Jr., J. E. McDunn, A. D. Wentworth, C. Takeuchi, J. Nieva, T. Jones, C. Bautista, J. M. Ruedi, A. Gutierrez, K. D. Janda, B. M. Babior, A. Eschenmoser, and R. A. Lerner. 2002. Evidence for antibody-catalyzed ozone formation in bacterial killing and inflammation. Science 298:2195-2199. [DOI] [PubMed] [Google Scholar]