Abstract
Cell morphogenesis is of fundamental significance in all eukaryotes for development, differentiation, and cell proliferation. In fission yeast, Drosophila Furry-like Mor2 plays an essential role in cell morphogenesis in concert with the NDR/Tricornered kinase Orb6. Mutations of these genes result in the loss of cell polarity. Here we show that the conserved proteins, MO25-like Pmo25, GC kinase Nak1, Mor2, and Orb6, constitute a morphogenesis network that is important for polarity control and cell separation. Intriguingly, Pmo25 was localized at the mitotic spindle pole bodies (SPBs) and then underwent translocation to the dividing medial region upon cytokinesis. Pmo25 formed a complex with Nak1 and was required for both the localization and kinase activity of Nak1. Pmo25 and Nak1 in turn were essential for Orb6 kinase activity. Further, the Pmo25 localization at the SPBs and the Nak1-Orb6 kinase activities during interphase were under the control of the Cdc7 and Sid1 kinases in the septation initiation network (SIN), suggesting a functional linkage between SIN and the network for cell morphogenesis/separation following cytokinesis.
Keywords: cell morphogenesis, Mor2, MO25, SIN, SPB
Introduction
Cell morphogenesis and the cell cycle are coordinately regulated. The fission yeast Schizosaccharomyces pombe is an ideal system in which to study cell morphogenesis (Snell and Nurse, 1994; Verde et al, 1995; Mata and Nurse, 1997; Hirata et al, 1998; Brunner and Nurse, 2000; Chang and Peter, 2003). Rod-like fission yeast cells identify growing regions precisely at the opposite ends of the cell, depending on the cell cycle stages (Mitchison, 1970). The growth polarity dynamically changes during three stages of the cell cycle (Marks et al, 1986), that is, the initiation of growth upon cell division, NETO (new end take off; Mitchison and Nurse, 1985), and septum formation. Cortical F-actin moves from the new end, which is newly produced by division, to the old end, which existed in the previous cell cycle, and the cell growth is initiated from only the old end. Upon NETO in the G2 phase, F-actin localization shifts from the old end to both ends, thereby changing the growth polarity from monopolar to bipolar. This growth polarity is maintained during the following interphase. At the onset of mitosis, the cell growth ceases, and the cortical F-actin patches at both ends disappear, having been translocated to the medial ring, which corresponds to the following division site. Then, the onset of cytokinesis is triggered by the septation initiation network (SIN), which results in septum formation in the middle of the cell (reviewed in Simanis, 2003; Seshan and Amon, 2004). The SIN comprises GTPase Spg1 and a downstream kinase cascade that includes Cdc7, Sid1, and Sid2.
The correct positioning of the growth zone(s) in fission yeast requires microtubules (MTs), Tea1, the kinesin-like protein Tea2, CLIP170-like protein Tip1, and Mod5 (Mata and Nurse, 1997; Browning et al, 2000; Brunner and Nurse, 2000; Sanith and Sawin, 2003). Mutations in these genes result in branched cell morphology, and this abnormality is attributed to impairment of the spatial information that defines the position of cell growth. Nonetheless, all of these genes are dispensable for cell division, and even in their absence, cell polarity itself is maintained, as cell morphology is still rod-like. mor2+ and orb6+, encoding the conserved Drosophila Furry-like protein and the NDR/Tricornered kinase, respectively, are unique in this context, as they are essential for cell viability, and mutant cells lose their cell polarity completely and become round (Verde et al, 1998; Hirata et al, 2002). Mor2, in concert with Orb6, becomes localized at the polarized growing regions and plays an important role in the restriction of growth zone(s) to which Tip1 is targeted. Further, a mutation in mor2 induces a Wee1-dependent G2 delay, indicating that the mutation activates the mechanism coordinating growth polarity with cell cycle progression (Hirata et al, 2002). Orb6 directly interacts with Mob2, another essential and conserved protein, and this complex is required for coordination of cell polarity with cell cycle progression (Hou et al, 2003). In Drosophila and Caenorhabditis elegans, the Tricornered kinase/SAX-1 and Furry/SAX-2 are important for neuronal polarity control, tiling, branching, and termination, indicating that the Furry/Mor2–Tricornered/Orb6-dependent cell morphogenesis pathway may be conserved in all eukaryotes (Geng et al, 2000; Zallen et al, 2000; Cong et al, 2001; Emoto et al, 2004; Gallegos and Bargmann, 2004). Any molecule(s) functionally interactive with the Furry/Mor2–Tricornered/Orb6 pathway, however, remains elusive.
MO25 was first identified as a gene expressed at the early cleavage stage of mouse embryogenesis (Miyamoto et al, 1993) and was shown to be an evolutionarily conserved protein in all eukaryotes (Nozaki et al, 1996; Karos and Fischer, 1999). In vertebrates, a gene encoding the serine/threonine kinase LKB1 is mutated in the Peutz–Jeghers cancer syndrome (PJS), and MO25 interacts with STRAD (STE20/PAK-related adaptor pseudokinase), thereby enhancing its ability to bind, activate, and tether the LKB1 in the cytoplasm (reviewed in Baas et al, 2004; Boudeau et al, 2004; Milburn et al, 2004). The mechanism resulting in PJS as a consequence of this mutation is unclear. The budding yeast MO25-like protein Hym1 is involved in a network, RAM, that regulates Ace2 transcriptional activity and polarized growth (Nelson et al, 2003), although its precise role remains elusive.
Here we show that a fission yeast novel morphogenesis network is essential for growth polarity and cell separation. The network consists of the evolutionarily conserved essential proteins Pmo25, GC kinase Nak1, Orb6, and Mor2. Pmo25 forms a complex with Nak1 and alters its location from the mitotic spindle pole body (SPB) to the dividing medial region. Pmo25 and Nak1 are important for the kinase activity of Orb6. Furthermore, our results indicate a linkage between the SIN and the network essential for cell morphogenesis.
Results
Search for a molecule(s) functionally interacting with Mor2
mor2+ was originally identified as a temperature-sensitive (ts) mutant with defects in cell morphology (morphological round; Radcliffe et al, 1998; Hirata et al, 2002). To search for a molecule(s) functionally interacting with Mor2, we screened for other mor mutants and identified mor4-125. mor4 is allelic to orb3 (Snell and Nurse, 1994) and encodes an essential germinal center kinase, designated Nak1 (Huang et al, 2003; Leonhard and Nurse, 2005; hereafter mor4/orb3 is called nak1). In the nak1-125 mutant, the conserved threonine-198 (ACT) is substituted by isoleucine (ATT; mutated nucleotide is underlined) in protein kinase domain IX. The nak1 mutant cells showed defects in growth polarity and cell separation, as well as a temperature-dependent reversible morphological change, as did the mor2 mutant cells (Supplementary Figure 1A). The morphology of the double or triple mutants between nak1 and mor2 or orb6 was similar to that of the single mutants (Supplementary Figure 1B). These results suggest that Nak1 functions in the same pathway as Mor2–Orb6. Nak1 belongs to the STE20/PAK-related kinase family and has homology with STRAD, which forms a complex with the evolutionarily conserved protein MO25. The budding yeast MO25-like protein Hym1 is involved in the RAM pathway that consists of the budding yeast homologs of fission yeast Mor2, Orb6, and Nak1 (Nelson et al, 2003). The fission yeast genome contains one ORF that shows a high homology to MO25 (SPAC1834.06c; Figure 1A). This gene was designated pmo25+ and was characterized in this study.
Figure 1.
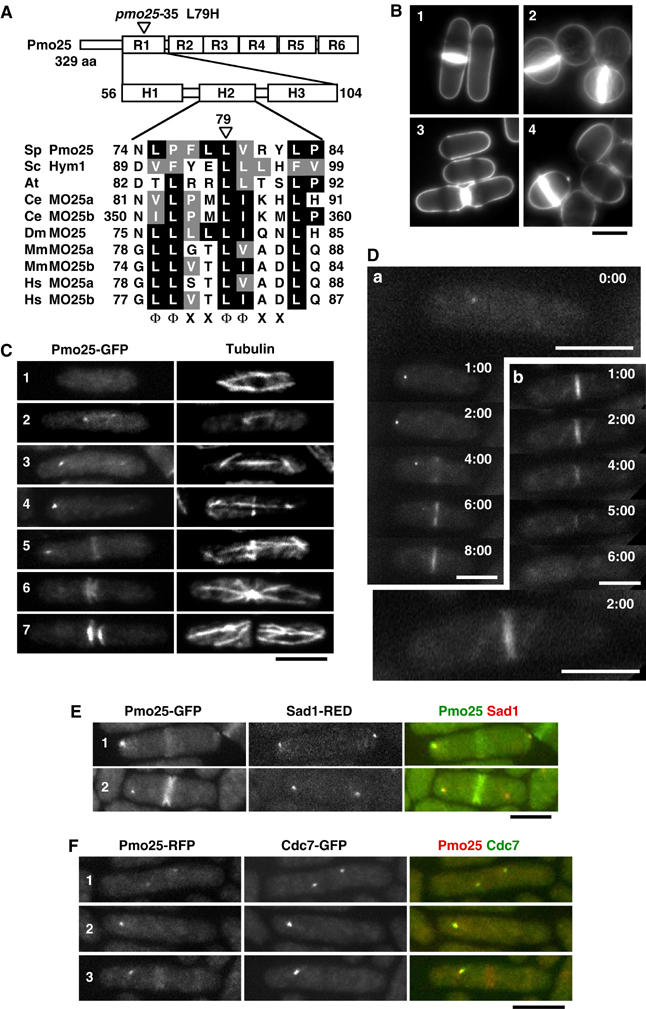
Structure and localization of Pmo25. (A) Pmo25 consists of six repeats (R1–R6) each having three helices (H1–H3). The mutation site of the pmo25-35 allele is located within H2 of R1. The H2 of Pmo25 (NCBI accession no. NP_594685) is aligned with Saccharomyces cerevisiae Sc Hym1 (NP_012732), Arabidopsis thaliana (CAB78730), C. elegans Ce MO25a (CAB16486), Ce MO25b (NP_508691), Drosophila melanogaster Dm MO25 (P91891), Mus musculus Mm MO25a (NP_598542), Mm MO25b (AAH16128), Homo sapiens Hs MO25a (NP_057373), and Hs MO25b (CAC37735). The conserved motif (Φ, hydrophobic; X, any residue) is shown. (B) Calcofluor staining of WT cells (1), pmo25-deleted cells (2), pmo25-deleted cells with pREP41-Pmo25 ((3) in EMM, induced condition; (4) in EMM containing thiamine, repressed condition). (C, D) WT cells having pmo25+:GFP:kanr grown in YES medium at 25°C were stained with anti-tubulin antibody (C). Pmo25 localization at mitosis (a) and cell separation (b) of the living cells was observed by time lapse (minutes) (D). (E, F) WT cells having pmo25+:GFP:kanr and sad1+:dsRED:LEU2 (E) or pmo25+:mRFP:kanr and cdc7+:GFP:kanr (F) grown in YES medium at 25°C were used. Bars indicate 5 μm.
The essential Pmo25 protein is localized at SPB(s) and the septation site
Gene disruption by one-step replacement showed that the pmo25+ gene was essential for cell viability. The nonviable pmo25-deleted spores failed to germinate (19% of the spores) or ceased growth after several divisions (81%). The pmo25-deleted cells that germinated from spores showed an unseptated/septated round morphology (Figure 1B, cell 2), suggesting that Pmo25 is important for cell polarity and cell separation. The pmo25-deleted cells carrying episomally expressed pmo25+ showed a normal rod-shape as wild-type (WT) cells (cell 3) and the ‘loss of plasmid' phenotype was the same as the terminal phenotype of the pmo25-deleted spores after germination (cell 4). These phenotypes of the pmo25-deleted cells are reminiscent of those seen in orb6-, mor2-, and nak1-deleted cells (Verde et al, 1998; Hirata et al, 2002; Huang et al, 2003).
To investigate the cellular localization of Pmo25, we constructed cells expressing Pmo25-GFP from a chromosomal native promoter. The tagged strains grew as well as WT cells, indicating that the tagging did not interfere with the Pmo25 function. Although the level of Pmo25 protein did not noticeably change during the cell cycle (data not shown), its cellular localization showed drastic alterations, as described below. Pmo25 appeared to be localized throughout the cells during interphase (Figure 1C, cell 1), and then accumulated as two separate dots in early mitosis/anaphase A (cells 2 and 3), as one dot during anaphase B and in the medial region of the cell in post-anaphase (cells 4 and 5), and finally on either side of the late septum (cells 6 and 7). In order to visualize the localization dynamics of Pmo25-GFP in a single cell, we performed time-lapse live imaging (Figure 1D). As shown earlier in still images, Pmo25-GFP was localized as two dots (one is very faint) in early mitosis and as one dot during anaphase and in the medial region of the cell in post-anaphase (cell a). The localization of Pmo25 to split rings was observed (cell b, 2 min; large panel). Pmo25 at the new ends of the cell disappeared within 2 min after cell separation, and no specific localization was evident afterward (cell b).
To determine whether the Pmo25-containing dot(s) was the SPB, we created WT cells expressing Pmo25-GFP and the SPB marker Sad1-RFP (Hagan and Yanagida, 1995). Pmo25 was localized at one of the two SPBs and in the medial region in post-anaphase (Figure 1E). It was previously shown that Cdc7, an upstream kinase of SIN, becomes localized at both of the SPBs during early mitosis and then, during anaphase B, associates with only one of the SPBs, to which GTP-bound Spg1 is preferentially localized (Sohrmann et al, 1998). Recently, it was further shown that this Cdc7-bound SPB is the new SPB, in which the SIN is active (Grallert et al, 2004). Given the asymmetrical nature of the SIN pathway, we addressed whether Pmo25 becomes localized at the Cdc7-associated new SPB or not. To this end, we used WT cells expressing Pmo25-RFP and Cdc7-GFP. It turned out that Pmo25 was localized at both SPBs during early mitosis (Figure 1F, cell 1) and then exclusively to the Cdc7-associated new SPB during anaphase B (cells 2 and 3). Thus, Pmo25 and Cdc7 were colocalized during early mitosis and anaphase B.
Physical interactions among Pmo25, Nak1, Mor2, and Orb6
Given the phenotypic similarities of each mutant cell, we examined the physical interaction among Pmo25, Nak1, Orb6, and Mor2 using the S. cerevisiae two-hybrid system (Figure 2A). The WT and mutant proteins (m1/Y558C and m2/G1709D; Hirata et al, 2002) of Mor2 were separated into three fragments, Mor2N, M, and C (Figure 2B). Pmo25 interacted with Nak1 but not with Mor2 or Orb6 (Figure 2A). Orb6 interacted with Mor2N and Mor2C (Figure 2A), but not with Pmo25. These interactions were abolished by the mor2 mutations, m1 and m2, respectively (Figure 2A and B). Nak1 interacted with Mor2N, Orb6, and Pmo25. Interestingly, unlike the case for Orb6 and Mor2N, the interaction between Nak1 and Mor2N was not affected by the mor2-m1 mutation (Figure 2A). This finding may imply that Nak1 and Orb6 interact with Mor2N at distinct interaction sites. The above interactions were also observed in the opposite combinations between BD and AD in the two-hybrid system (data not shown).
Figure 2.
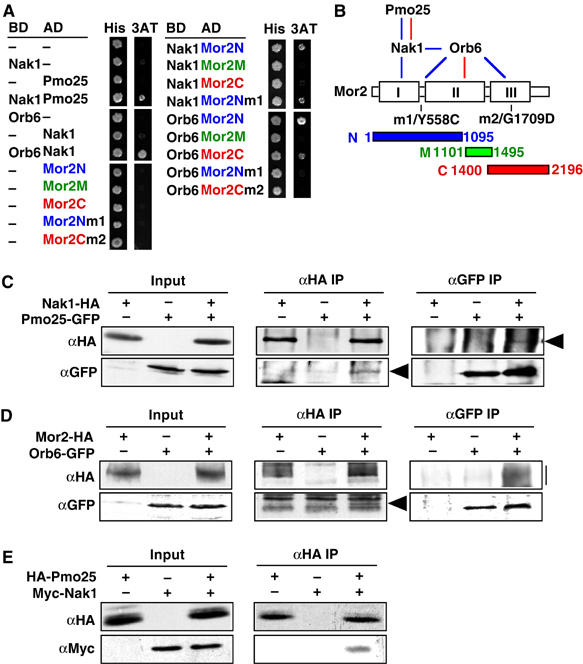
Physical interactions among the proteins. (A) Protein–protein interactions were examined using the yeast two-hybrid system. S. cerevisiae Y190 (104 cells) cells transformed with Gal4-binding domain-fused (BD) and Gal4 activation domain-fused genes (AD) were spotted on SD containing histidine (His) or 3-aminotriazole (3AT). (B) A summary of the physical interactions observed is depicted by blue (two hybrid) and red lines (IP). The boxes in Mor2 indicate the homologous regions among Furry-like proteins (Hirata et al, 2002). (C, D) Extracts of S. pombe cells expressing Nak1-HA and/or Pmo25-GFP (C), or Mor2-HA and/or Orb6-GFP proteins (D) from the chromosomal native promoter were immunoprecipitated with anti-HA or anti-GFP antibodies. Precipitated materials were then immunoblotted using antibodies against HA and GFP. The arrowhead and bar indicate the position of the immunoprecipitated proteins. (E) Pmo25 binds to Nak1 in vitro. Binding of bacterially expressed HA-Pmo25 and Myc-Nak1 proteins was assayed by co-immunoprecipitation. The protein mixture was immunoprecipitated with anti-HA antibody. Precipitated materials were then immunoblotted using antibodies against HA and Myc.
To substantiate the interactions observed in the two-hybrid system, we performed co-immunoprecipitation using S. pombe WT cells expressing Nak1-HA and Pmo25-GFP or Mor2-HA and Orb6-GFP, which were expressed from the chromosomal native promoter. In both cases, anti-HA immunoprecipitates were immunoblotted with anti-HA and anti-GFP antibodies. We found that Pmo25-GFP co-immunoprecipitated with Nak1-HA (Figure 2C), and Orb6-GFP, with Mor2-HA (Figure 2D). The interaction of these proteins was confirmed by reciprocal immunoprecipitation, in which anti-GFP antibodies immunoprecipitated HA-tagged proteins in either case (Figure 2C and D). We also performed co-immunoprecipitation between Nak1-GFP and Orb6-HA, but no physical interaction was detected under the experimental conditions that enabled us to detect interaction between Nak1 and Pmo25 or between Mor2 and Orb6. It is possible that this interaction is transient in S. pombe cells.
To further investigate the interaction between Nak1 and Pmo25, we tested the interactions using bacterially expressed proteins. A bacterially expressed HA-Pmo25 co-immunoprecipitated with Myc-Nak1 in vitro (Figure 2E) but not with GST (data not shown), showing that this interaction between Pmo25 and Nak1 was likely direct. We could not perform this experiment between Mor2 and Orb6, since bacterially expressed Orb6 was unstable. Our results indicate that these proteins form the physical interaction network summarized in Figure 2B. In addition to four proteins, it is likely that Mob2 is involved in this network, since Orb6 directly interacts with Mob2 (Hou et al, 2003).
Pmo25 is required for growth polarity control and cell separation
In order to explore the roles of Pmo25 in cell morphogenesis, we created a ts pmo25 mutant (pmo25-35) by error-prone PCR mutagenesis. The pmo25-35 mutant contained a histidine (CAC) in place of the evolutionarily conserved leucine-79 (CTC) in the region called H2 in R1 (Milburn et al, 2004; see Figure 1A). At 36°C, pmo25-35 mutant cells were, like the pmo25-deleted cells shown earlier (see Figure 1B), round or septated round, supporting further the notion that Pmo25 is important for polarity control and cell separation. Previously, we showed that Mor2 is important for the relocalization of F-actin at the cell end(s) after the shift to 36°C (Hirata et al, 2002). We next examined the distribution of F-actin in the pmo25-35 mutant cells at 36°C. Cortical F-actin in the pmo25 mutant cells was localized as patches at the growing end(s) and the septation site at 25°C as in WT cells (Figure 3A, 0 time). In WT cells, F-actin at the cell end(s) was dispersed within 20 min after the shift. By 2 h after the shift, however, F-actin had been relocalized to the cell end(s). In the pmo25 mutant cells, on the other hand, F-actin was dispersed as in WT cells upon the up shift; however, the relocalization of F-actin to the cell end(s) was not observed, as in mor2 and nak1 mutants. These results indicate that Pmo25 is required for the relocalization of F-actin to the cell end(s).
Figure 3.
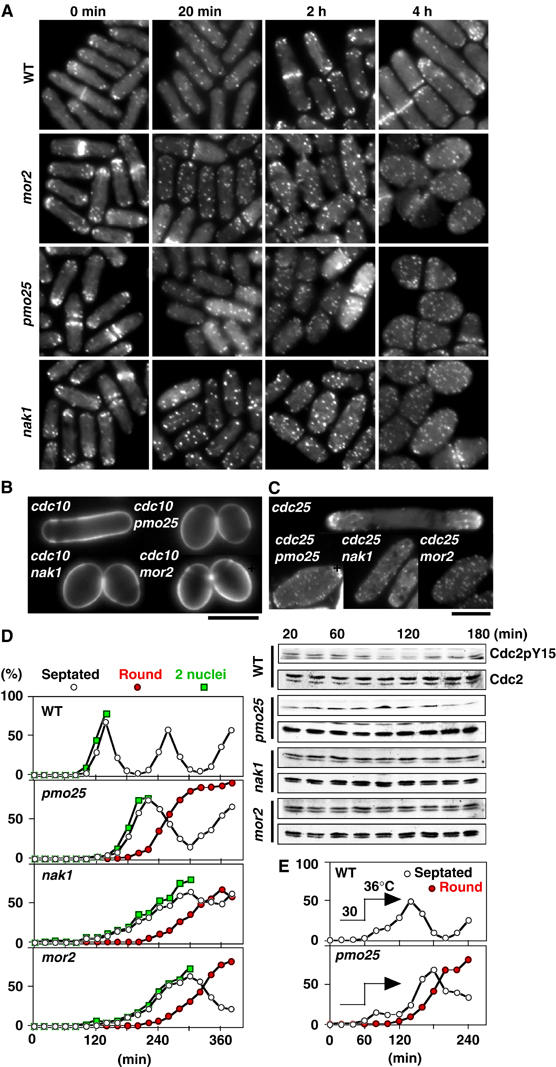
Role of Pmo25 in cell morphogenesis. (A) Cells grown in YES medium at 25°C were transferred to 36°C (time 0), and taken at the indicated times for observation of F-actin localization after being staining with rhodamine-phalloidin. (B, C) Calcofluor (B) and F-actin staining (C) of the double mutants between pmo25 and cdc10 (B) or cdc25 (C). The cells grown at 25°C were shifted to 36°C and incubated for 4 h. (D) Early G2 cells of the mutants collected by elutriation were cultured at 36°C and taken at the indicated times for observation of morphology (left) and for immunoblotting with antibodies specific for Cdc2 phosphorylated on tyrosine-15 (Cdc2pY15) and for PSTAIR (Cdc2; right). (E) Early G2 cells of the mutants collected by elutriation were cultured at 30°C for 1 h and then shifted to 36°C.
To address whether Pmo25 is important for the maintenance of cell polarity, we examined the phenotype of the double mutants between pmo25 and the G1-arrest mutant cdc10 or the G2-arrest mutant cdc25. As shown in Figure 3B and C, the double mutant cells (either cdc10pmo25 or cdc25pmo25) showed abnormal spherical morphology. In the cdc25pmo25 mutant, F-actin was dispersed throughout the cells, although the partial localization of F-actin to the cell end(s) was observed. Likewise, cdc10nak1 and cdc10mor2 also showed round morphology and cdc25nak1 and cdc25mor2 displayed the dispersed F-actin localization. These results indicate that Pmo25, like Mor2 and Nak1, is vital for the maintenance of growth polarity during interphase, both G1/monopolar growth and G2/bipolar growth phases.
We next asked whether the pmo25 mutation induces G2 delay like the mor2 mutation (Hirata et al, 2002). To this end, we prepared synchronous cultures of the mutant grown at the permissive temperature (25°C) via centrifugal elutriation. Small early G2 cells were subsequently shifted to the restrictive temperature (36°C), and the morphological change, nuclear number, and the amount of the tyrosine-phosphorylated form of Cdc2 were then followed afterward (Figure 3D). In WT cells, the level of the phosphorylated Cdc2 was decreased by 100 min upon mitotic entry. In the pmo25 mutant cells, the phosphorylated Cdc2 was maintained until 140 min, indicating that the pmo25 mutation induced G2 delay. Upon 2 h of the shift, the morphology of the pmo25 mutant was still rod-like with the dispersed F-actin localization. After passing through first mitosis, however, the pmo25 mutant cells could not re-establish cell polarity, thereby becoming completely round in shape (Figure 3D), suggesting that Pmo25, like Mor2 and Nak1, is important for polarity establishment following cell division.
To confirm the importance of Pmo25 in polarity establishment following cell division, we elutriated small early G2 cells and cultured these cells at the permissive temperature (30°C) for 1 h, followed by shift to 36°C (Figure 3E). Under this condition, a short G2 delay mutant cells were still observed in the pmo25 mutant upon shift, and after passing through mitosis, the pmo25 mutant cells became completely round with septum (Figure 3E). These results indicate that Pmo25 plays a crucial role in polarity establishment and cell separation following cytokinesis and that its mutant phenotypes are very similar, if not identical, to those of mor2 and nak1.
Pmo25 is involved in the restriction of the growth zone targeted by MT
The MT cytoskeleton is known to regulate cell polarity by determining the region of polarized growth (Mata and Nurse, 1997). In order to examine MT integrity in the pmo25 mutant, we immunostained the MT cytoskeleton in this mutant. At the permissive temperature (25°C), a normal MT cytoskeleton was observed as in WT cells (Figure 4A). At 36°C, for up to 2 h of incubation, the cell end(s) where MT targets was maintained, but after 6 h no clear MT-targeted regions were observed and MT arrays were no longer parallel, as in the mor2 and nak1 mutants (Figure 4A). This result suggests that Pmo25 is important for the restriction of the growth zone targeted by MT.
Figure 4.
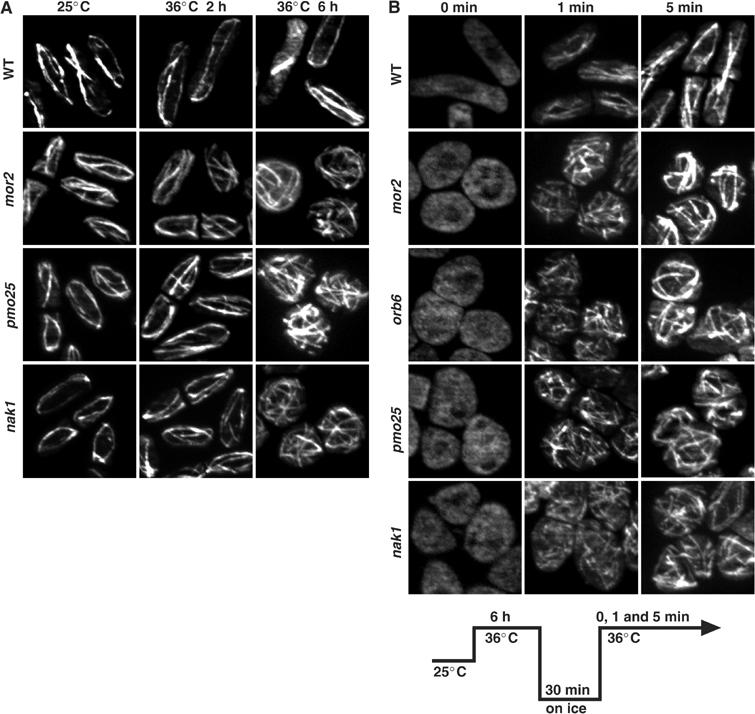
Role of Pmo25 in the organization of the MT cytoskeleton. (A) Cells grown in YES medium at 25°C were transferred to 36°C, fixed, and immunostained with anti-tubulin antibodies at the times indicated. (B) Cells grown at 25°C were transferred to 36°C for 6 h, incubated on ice for 30 min, rewarmed to 36°C, and collected at the indicated times.
To investigate further whether Pmo25 is involved in the polymerization of MTs, we examined the nucleation of MTs disrupted by cold shock. The pmo25 mutant cells grown at 25°C were shifted up to 36°C for 6 h, incubated on ice for 30 min, and then returned to 36°C (Figure 4B, lower panel). Upon cold shock, no MTs were observed (Figure 4B, 0 min). The disrupted MTs were nucleated within 5 min after the return to 36°C, as in WT cells (Figure 4B). These results indicate that Pmo25 is not required for the nucleation of MTs per se.
Pmo25 and Nak1 are important for the cellular localization of Mor2
To address whether Pmo25 and Nak1 are required for the Mor2 localization, we examined the Mor2 localization on these mutant backgrounds. The GFP-Mor2 expression levels in these mutants were indistinguishable from those in WT cells (data not shown). As shown previously (Hirata et al, 2002), in the orb6 mutant, no localization of Mor2 at any polarized growing regions was observed (Figure 5A and B). In the pmo25 and nak1 mutants, the Mor2 localization at the cell cortex in the single round cells (maintenance of polarity) and at the septation site in the septated round cells (septation) was lost in approximately 50% of the cells (Figure 5A and B). Further, no clear localization of Mor2 at the new cell end (establishment of polarity) after cytokinesis was observed in these mutants (Figure 5A and B). These results suggest that Pmo25 and Nak1 are important for the localization of Mor2 to the polarized growing regions, cell end(s) and septum, and support that this network is required for the maintenance and the establishment of cell polarity.
Figure 5.
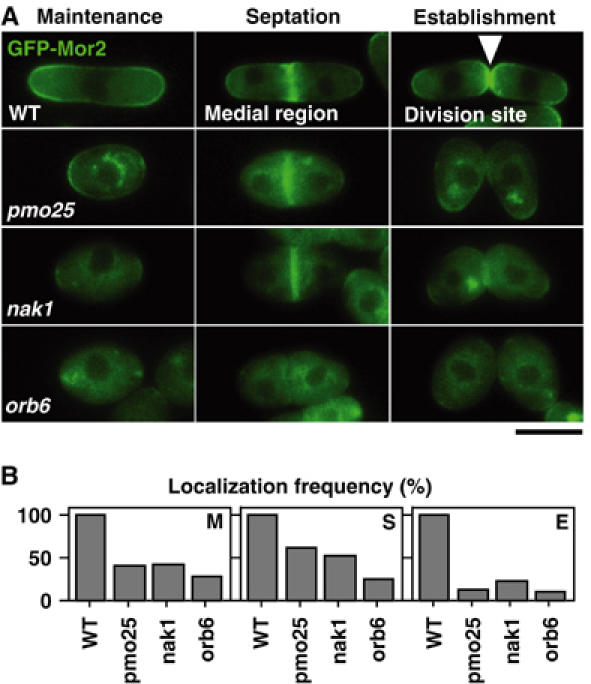
Role of Pmo25 in Mor2 localization. (A, B) Cells having kanr:nmt1:GFP:mor2+ grown in YES medium (repressed condition) at 25°C were shifted to 36°C and incubated for 4 h. Localization of GFP-Mor2 to the cell cortex in interphase (maintenance, M), medial region (septation, S), and cell division site after cytokinesis (establishment, E) is shown in panel A. Quantification of the localization frequency in the cells is shown in panel B.
Pmo25 and Nak1 act upstream of Orb6
We examined whether the Pmo25 localization required the normal functioning of Mor2, Orb6, and Nak1. On nak1, orb6, and mor2 mutant backgrounds, Pmo25 was still clearly localized at one of the SPBs during anaphase B (Figure 6A, marked with arrowheads), and also in the medial region, albeit weakly (Figure 6A). These results suggest that the localization of Pmo25 at SPB(s) and in the medial region does not require the functioning of these proteins.
Figure 6.
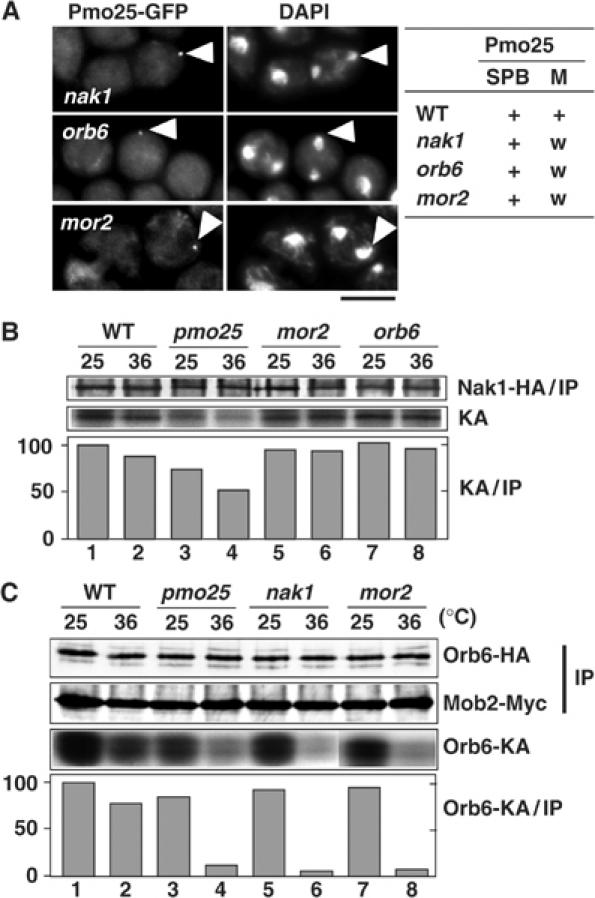
Hierarchical position of Pmo25 in the morphogenesis network. (A) Mutants having pmo25+:GFP:kanr grown in YES medium at 25°C were shifted to 36°C, incubated for 4 h, and fixed. A summary of the Pmo25 localization at the SPB or medial region (M) is shown at the right. w, weaker than in WT. (B) Cells expressing Nak1-HA from the chromosomal native promoter grown in YES medium at 25°C were shifted to 36°C and incubated for 90 min. Cell extracts were then prepared and immunoprecipitated with anti-HA antibody. Half of the immunoprecipitates were immunoblotted with anti-HA (Nak1-HA/IP) and the other half were assayed for autokinase activity. The kinase activities (KA) were quantified with an Intelligent Quantifier (Bio Image) and normalized to the amount of Nak1-HA (IP). (C) The cells expressing Mob2-Myc and Orb6-HA from the chromosomal native promoter grown at 25°C were shifted to 36°C and incubated for 90 min. Cell extracts were prepared and immunoprecipitated with anti-Myc antibody. Half of the immunoprecipitates were immunoblotted with anti-Myc (Mob2-IP) and anti-HA (Orb6-HA) and the other half were assayed for phosphorylation of MBP (by Orb6-KA). The kinase activities (Orb6-KA) were quantified as in panel B.
We next addressed whether the Nak1 kinase activity required the functioning of Pmo25, Mor2, and Orb6. The Nak1 kinase activity can be measured by autophosphorylation of Nak1 (Huang et al, 2003). Our experimental conditions detected the autophosphorylation of Nak1 in the ts nak1 mutant at 25°C but not at 36°C (data not shown). The Nak1 kinase activity in the pmo25 mutant at 36°C was decreased by 50%, compared with that in the WT cells (Figure 6B, lanes 1–4). In clear contrast, in the mor2 and orb6 mutants, the kinase activity was maintained (Figure 6B, lanes 5–8). These results suggest that the Nak1 kinase activity requires Pmo25 but not Mor2 and Orb6 and, furthermore, that Nak1 and Pmo25 act upstream of Orb6.
To further substantiate this relationship, we measured Orb6 kinase activity in the pmo25, nak1, and mor2 mutants. It has been shown that Orb6 specifically co-immunoprecipitates with Mob2 and that its kinase activity can be detected in Mob2 immunoprecipitates (IP, Mob2-Myc; Hou et al, 2003; Wiley et al, 2003). We found that the Orb6 kinase activity in the pmo25 and nak1 mutants was decreased by 90% at 36°C (Figure 6C, lanes 3–6) compared with that in WT cells (lanes 1 and 2). Note that this kinase activity was diminished in the immunoprecipitates prepared from the orb6 mutant (data not shown). This result is consistent with the data presented earlier, showing that Pmo25 and Nak1 act upstream of Orb6. In addition, Orb6 kinase activity was substantially impaired in the mor2 mutant (Figure 6C, lanes 7 and 8). This result supports our previous proposal that the functions of Mor2 and Orb6 are interdependent (Hirata et al, 2002).
Pmo25 and Nakl are colocalized at the SPB and septum
Next, we examined whether Pmo25 is involved in Nak1 localization. As previously shown (Leonhard and Nurse, 2005), Nak1-GFP expressed from a chromosomal native promoter in WT cells was found throughout the cell and in the cell end(s) during interphase (Figure 7A, cell 1), in the SPB(s) in early mitosis (cells 2 and 3, pointed by arrows), in one SPB in anaphase B (cell 4), and in the medial region and double ends upon division (cells 5 and 6). At 36°C in WT cells (Figure 7B, upper panel), the localization of Nak1 to SPB (cell 1, pointed by arrow) and double ends (cell 2) was observed. In the pmo25 mutant cells (Figure 7B, lower panel), on the other hand, no specific localization of Nak1 was observed in anaphase B (cell 1) and cell division (cell 2). We examined the colocalization of Pmo25 and Nak1 using WT cells expressing Pmo25-RFP and Nak1-GFP. Pmo25 and Nak1 were colocalized at one of the two SPBs during mitosis (Figure 7C, cells 1 and 2) and in the medial region in post-anaphase (cells 3 and 4). Nak1 at the septation site overlapped partially with that of Pmo25 there, suggesting that some of the Nak1 form a complex with Pmo25 in the medial region and that Nak1 interacts with Pmo25 on the SPB(s).
Figure 7.
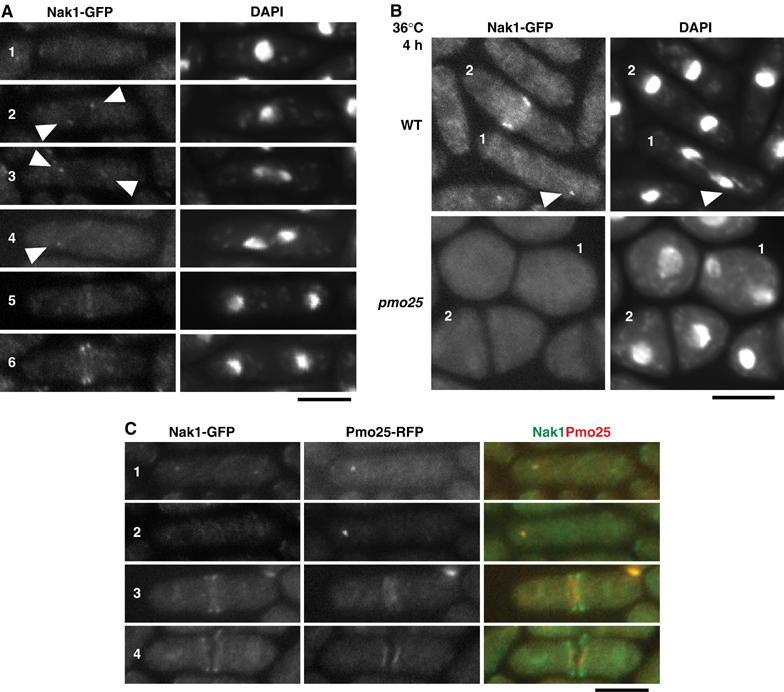
Colocalization of Pmo25 and Nak1. (A) WT cells having nak1+:GFP:kanr were grown in YES medium at 25°C. (B) WT and pmo25 mutant cells having nak1+:GFP:kanr grown at 25°C were shifted to 36°C for 4 h. (C) WT cells having nak1+:GFP:kanr and pmo25+:mRFP:kanr grown at 25°C were used.
Pmo25 localization at the SPB is under the control of the SIN
The localization patterns presented earlier (see Figure 1F) indicate that Pmo25 and Cdc7 were colocalized at the mitotic SPB(s). Accordingly, we addressed the possibility of a functional interaction between the SIN and Pmo25. For this purpose, first we examined the hierarchy of the localization between SIN and Pmo25. As shown in Figure 8A, in the pmo25 mutant, Cdc7 was still found at the SPB. In sharp contrast, on a cdc7-24 or sid1-239 mutant background, no Pmo25 was detected at the mitotic SPB(s) (Figure 8B and C). In the sid2-250 mutant cells, on the other hand, Pmo25 was still capable of being localized to the mitotic SPB(s) (Figure 8B and C). Previously, the cdc7-i10 mutant, which is supposed to activate the SIN without stable association with the SPB at the permissive temperature of 25°C, was identified (Lu et al, 2002). In the cdc7-i10 mutant cells at 25°C, the localization of Pmo25 to mitotic SPB was observed, although the signal was weaker than that in WT cells (Figure 8B and C). This result suggests that the SPB localization of Pmo25 requires the Cdc7 kinase activity rather than the stable association of Cdc7 to the SPB. We next examined Pmo25 localization in the cdc16-116 mutant. Cdc16 is a GAP for Spg1-GTPase, and in the cdc16 mutant, Spg1 is locked in a GTP-bound form, thereby making the SIN constitutively active (Simanis, 2003). As shown in Figure 8B (top two rows) and Figure 8C, Pmo25 was found in the SPB(s) in this mutant. Finally, we examined the localization of Pmo25 in the cps1/drc1 mutant, which is defective in β-glucan synthase. It is known that in addition to having cytokinesis defects, this mutant activates the cytokinesis checkpoint, which requires the SIN (Goff et al, 1999; Liu et al, 2000). As shown in Figure 8D, in 80% of the cps1/drc1 mutant cells, the SPB localization of Pmo25 was observed. Taken together, these results indicate that Pmo25 targeting to the SPB(s) is under the control of the SIN.
Figure 8.
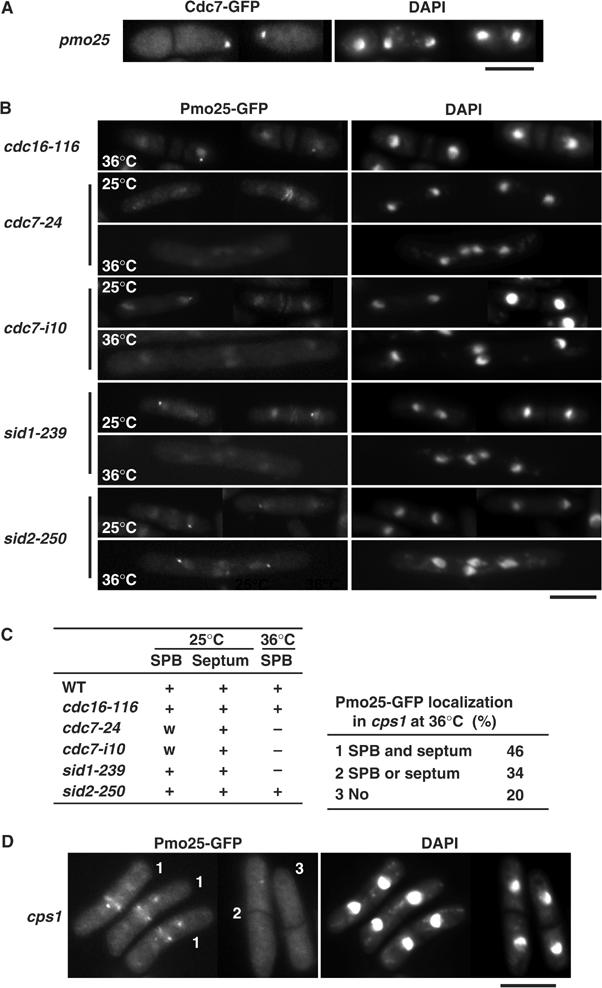
Localization of Pmo25 in SIN mutants. (A) pmo25 mutants having cdc7+:GFP:kanr grown in YES medium at 25°C were shifted to 36°C, incubated for 4 h, and fixed. (B, C) SIN mutants having pmo25+:GFP:kanr grown at 25°C were shifted to 36°C, incubated for 4 h, and fixed (B). A summary of the Pmo25 localization is shown in panel C. w, weaker than in WT. (D) The cps1 mutants having pmo25+:GFP:kanr grown at 25°C were shifted to 36°C, incubated for 4 h, and fixed.
Cdc7 and Sid1 regulate the Nak1 and Orb6 kinase activities in interphase
Pmo25 was found to be important for the Nak1 and Orb6 kinase activities (Figure 6B and C). The SPB localization of Pmo25 required the functioning of Cdc7 and Sid1, but not that of Sid2 (Figure 8B and C). If the SPB localization of Pmo25 was essential for regulation of Nak1 and Orb6 kinases by Pmo25, Cdc7 and Sid1 would be important for the Nak1 and Orb6 kinase activities. Given this assumption, we measured the Nak1 and Orb6 kinase activities in synchronous cultures of WT and SIN mutant cells during the cell cycle. Small early G2 cells grown at the permissive temperature (25°C) were collected and subsequently shifted to the restrictive temperature (36°C), and the Nak1 and Orb6 kinase assays were performed afterward. As previously described (Leonhard and Nurse, 2005), in WT cells, the Nak1 kinase activity did not change during the cell cycle (Figure 9). In clear contrast, in the cdc7 and sid1 mutant, while the kinase activity was decreased after the first mitosis, no clear increase was observed in the following interphase (Figure 9). In sid2 mutant cells, the constitutive Nak1 activity was observed as in WT cells. These results indicate that Cdc7 and Sid1, but not Sid2, are important for upregulation of the Nak1 kinase activity during the early interphase, which corresponds to the period of cell morphogenesis/separation following cytokinesis.
Figure 9.
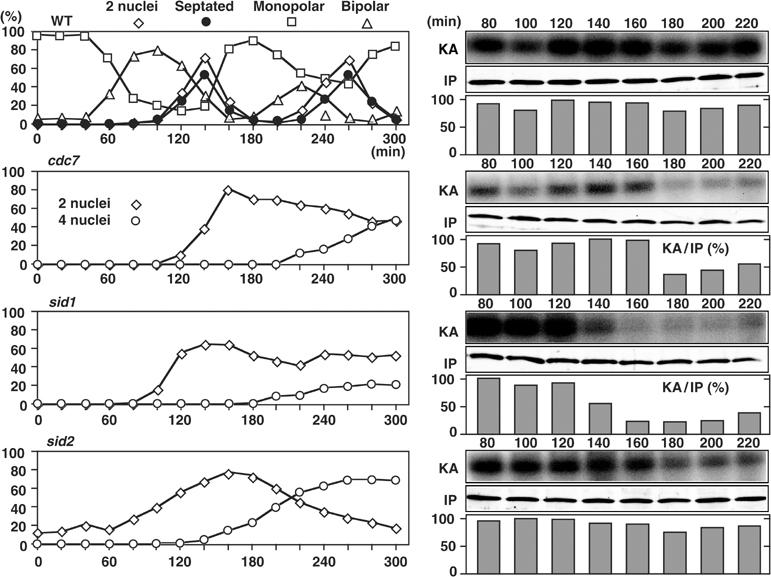
Nak1 kinase activity during the cell cycle in SIN mutants. Early G2 cells of the mutants expressing Nak1-HA from the chromosomal native promoter were collected by elutriation, cultured in YPD medium at 36°C, and taken at the indicated times for the Nak1 kinase assay.
As for Orb6 kinase activities, in WT cells, cell cycle-dependent oscillation of the Orb6 kinase activity was observed (Figure 10). The kinase activity was decreased by 120 min upon mitotic entry, and following cell division it was increased again during the interphase (200 min) and maintained in the later interphase (240 min). In the cdc7 and sid1 mutants, on the other hand, although the kinase activity was decreased upon mitotic entry as in the WT cells, no clear increase was observed in the following late interphase (Figure 10). In sid2 mutant cells, the oscillation of the Orb6 kinase activity was observed as in WT cells. These results indicate that Cdc7 and Sid1, but not Sid2, are important for the regulation of the Nak1 and Orb6 kinase activities in interphase.
Figure 10.
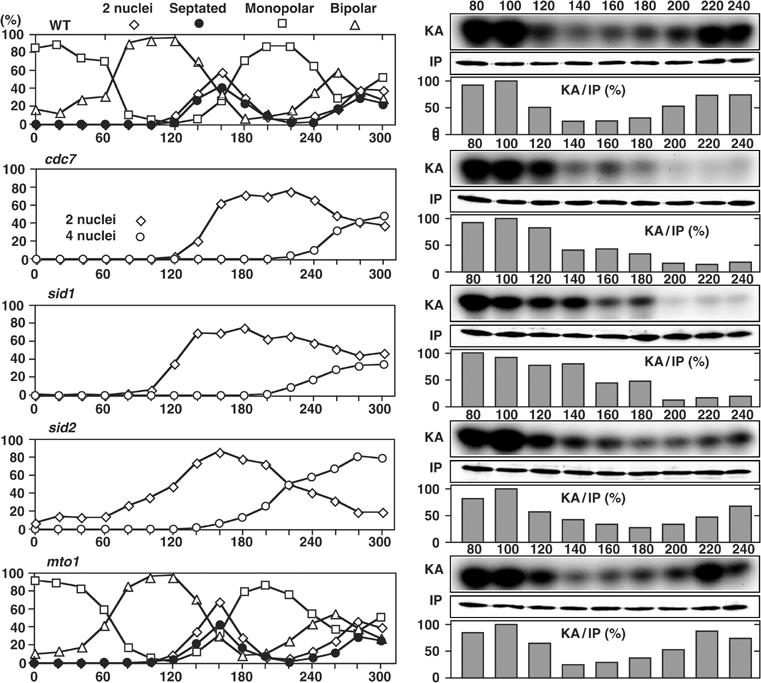
Orb6 kinase activity during the cell cycle in the SIN mutants. Early G2 cells of the mutants expressing Mob2-Myc from the chromosomal native promoter were collected by elutriation, cultured in YPD medium at 36°C, and taken at the indicated times for the Orb6 kinase assay.
It was reported that SIN mutants, such as ts cdc7, are defective in not only septation but also formation of equatorial MTOC (EMTOC; Heitz et al, 2001). To examine whether the defect in reactivation of Orb6 kinase in the cdc7 mutant is caused by the defect in the EMTOC formation, we examined the kinase activity in the mto1-deleted mutant, in which the EMTOC formation is specifically impaired without perturbation of septation (Sawin et al, 2004; Venkatram et al, 2004). In the mto1-deleted mutant cells, normal oscillation of the Orb6 kinase activity was observed as in WT cells (Figure 10). This result indicates that reactivation of Orb6 kinase after mitosis does not require the EMTOC formation. Thus the SIN, in particular its role in septation, regulates both Nak1 and Orb6 kinase activities during interphase.
Discussion
The present study was initiated by screening mor mutants to identify genes functionally related to the Mor2/Furry-Orb6/Ndr/Tricornerd kinase. We identified the GC kinase Nak1/Orb3 as Mor4, and then focused on the evolutionarily conserved MO25-like Pmo25. Our findings may be summarized as follows: (1) Pmo25, Nak1, Orb6, and Mor2 function in concert and play a critical role in both cell polarity and cell separation; (2) Pmo25 becomes localized at the SPB(s) specifically during mitosis and is then translocated to the septation site upon mitotic exit; (3) Pmo25 forms a complex with Nak1 and acts upstream of Orb6; and (4) the mitotic SPB localization of Pmo25 and the kinase activities of Nak1 and Orb6 during interphase are under the control of the SIN.
Network for cell morphogenesis
We demonstrated that Pmo25, Nak1, Orb6, and Mor2 form a physically interacting network important for both cell polarity and cell separation. Mob2 would be involved in this network, since in addition to its physical interaction with Orb6, the phenotypes of the mob2-deleted cells appear indistinguishable from those of the pmo25-, nak1-, orb6-, and mor2-deleted cells (Hou et al, 2003). Our results further indicate that this network is vital not only for the establishment of cell polarity following cytokinesis, but also for the maintenance of cell polarity during interphase. It was shown earlier that the budding yeast homologs of fission yeast Pmo25, Nak1, Mor2, Orb6, and Mob2 are involved in a signaling network, RAM, which regulates polarized growth (Nelson et al, 2003). Thus, morphogenesis network in fission yeast appears to be homologous to budding yeast RAM. However, in budding yeast, the SPB localization of any of the RAM components has not been shown and therefore, the linkage between SPB and this network in fission yeast is novel.
Role of Pmo25 in cell morphogenesis
Pmo25 forms a complex with Nak1 and is important for the kinase activities of Nak1 and Orb6, indicating that the function of Pmo25 in cell morphogenesis is performed through the regulation of these two kinases. We showed that the network is important for the localization of F-actin at the cell end(s) but not in the medial region, suggesting that the mechanism determining cell polarity at the septation site is different from that acting in interphase. How these two kinases regulate the growth polarity is the next subject to be addressed. In budding yeast, RAM controls cell separation through regulation of the Ace2 transcription factor, which activates the expression of the genes for septum degradation (Colman-Lerner et al, 2001; Doolin et al, 2001; Nelson et al, 2003). Interestingly, it was recently shown that fission yeast Ace2 also regulates the expression of the endo-β-1,3-glucanase gene eng1+ (Martin-Cuadrado et al, 2003). It is conceivable that Ace2 also functions downstream of Pmo25 and Nak1.
It should be noted that the period of G2 delay of the pmo25 mutant was shorter than that of the mor2 or nak1 mutant and that, unlike these two mutants, the pmo25 mutant cells proceeded to a second cell cycle without complete cell separation (see septated cells of pmo25 in Figure 3D). It is possible that Pmo25 plays an additional cell cycle-related role that is not shared by Mor2 or Nak1. However, these phenotypic differences might be simply caused by residual Pmo25 action in the ts mutant, since the terminal phenotype of the pmo25-deleted cells was indistinguishable from that of the mor2- and nak1-deleted cells. Further analysis is necessary to clarify this point.
SIN regulates the morphogenesis network
SIN components Cdc7 and Sid1, but not Sid2, were required for both the SPB localization of Pmo25 and the kinase activities of Nak1 and Orb6 in interphase. These results suggest that Cdc7–Sid1 plays a role in the activation of Nak1–Orb6 kinases, which is mediated through Pmo25. We envision that Cdc7–Sid1, on the one hand, stimulates septum formation in a Sid2-dependent manner and, on the other hand, regulates cell morphogenesis/separation following cytokinesis, which require the morphogenesis network. It is worth pointing out that, unlike Cdc7 and Sid1, Sid2 becomes localized at not only the mitotic SPBs but also the medial ring upon cell division (Sparks et al, 1999). This localization difference between Cdc7–Sid1 and Sid2 may indicate that SIN signaling cascades from the SPB bifurcate, one acting through Sid2 and the other via Pmo25–Nak1–Orb6. It is of note that independent work performed by another group has detected kinase activity in Pmo25-immunoprecipitates even in the ts nak1 mutant (Damian Brunner, personal communication). As presented in this study, Nak1 kinase activity was substantially reduced in ts pmo25 mutants. Given the fact that vertebrate MO25 binds to both STRAD and LKB1, we think that Pmo25 may be associated with some protein kinase(s) other than Nak1, such as the kinase(s) components of SIN.
The results indicated that Pmo25 plays a connecting role between SIN and the morphogenesis network. Pmo25, Nak1, Mor2, Orb6, and Mob2 are all evolutionarily conserved essential proteins. In addition, at least some of the SIN components have also been conserved through evolution (Simanis, 2003; Seshan and Amon, 2004). Homologous budding yeast RAM also regulates cell morphogenesis. However, in this organism, a functional linkage between RAM and MEN (SIN equivalent) has not been established. In Drosophila and C. elegans, the NDR/Tricornered kinase/SAX-1–Furry/SAX-2 signaling pathway is important for the control of neuronal polarity (Emoto et al, 2004; Gallegos and Bargmann, 2004). In vertebrates, MO25 is involved in PJS and polarity control of single intestinal epithelial cells (Baas et al, 2004). It would be of great interest to know whether the functional interaction between the SIN and MO25 has been evolutionarily conserved.
The morphogenesis network regulates cell polarity (establishment and maintenance) and cell separation. The mutants in the SIN but not those in the morphogenesis network show polarized growth, suggesting that SIN regulates cell separation but not polarity control. Indeed, in the cdc7 mutant, Mor2 was still localized at both cell ends (data not shown), indicating that the SIN is not essential for the maintenance of polarity. However, it remains possible that SIN regulates polarity establishment following cell separation, since the SIN mutants do not undergo cell separation. Further analysis is necessary to clarify the cellular event(s) regulated by SIN and the network.
Concluding remarks
In summary, we have presented evidence that in fission yeast, a series of evolutionarily conserved proteins, which become localized at the SPB in a cell cycle-dependent manner, play an essential role in cell morphogenesis following cytokinesis. In higher eukaryotes, the centrosome is vital for polarity establishment, and this role is independent of its MT-organizing activity (Piel et al, 2001; Stevenson et al, 2001; Cowan and Hyman, 2004). Our present data should help in understanding of the molecular mechanisms underlying the MT-independent role of the centrosome in cell morphogenesis in higher eukaryotes.
Materials and methods
Strains and general methods
Strains are listed in Supplementary Table S1. The general methods including PCR tagging were used throughout (Moreno et al, 1991; Bähler et al, 1998; Hirata et al, 1998).
Supplementary Material
Supplementary Figure S1
Supplementary Table 1
Supplementary Information
Supplementary Information
Acknowledgments
We thank P Nurse, I Hagan, V Simanis, D McCollum, KL Gould, K Sawin, O Niwa, J Ishiguro, and T Kuno for strains, K Gull for antibodies, and K Mizuta for the two-hybrid system. We are grateful to P Nurse for encouragement throughout this study and to D Brunner for communicating results prior to publication. DH thanks T Miyakawa, I Yamashita, T Yamada, and K Hirata for their support. DH was supported by grants from the Ministry of Education, Science and Culture of Japan. TT was supported by Cancer Research UK. FV was supported by grants from the National Science Foundation and the Sylvester Comprehensive Cancer Center.
References
- Baas AF, Smit L, Clevers H (2004) LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol 14: 312–319 [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Boudeau J, Scott JW, Resta N, Deak M, Kieloch A, Komander D, Hardle DG, Prescott AR, van Aalten DMF, Alessi DR (2004) Analysis of the LKB1–STRAD–MO25 complex. J Cell Sci 117: 6365–6375 [DOI] [PubMed] [Google Scholar]
- Browning H, Hayles J, Mata J, Avelinne L, Nurse P, McIntosh JR (2000) Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol 151: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P (2000) CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102: 695–704 [DOI] [PubMed] [Google Scholar]
- Chang F, Peter M (2003) Yeasts make their mark. Nat Cell Biol 5: 294–299 [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R (2001) Yeast Cbk1 and Mob2 activates daughter-specific genetic programs to induce asymmetric cell fates. Cell 14: 739–750 [DOI] [PubMed] [Google Scholar]
- Cong J, Geng W, He B, Liu J, Charlton J, Adler PN (2001) The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development 128: 2793–2802 [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA (2004) Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature 431: 92–96 [DOI] [PubMed] [Google Scholar]
- Doolin MT, Johnson AL, Johnston LH, Butler G (2001) Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol 40: 422–432 [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan Y-N (2004) Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119: 245–256 [DOI] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI (2004) Mechanosensory neurite termination and tiling depend on SAX-2 and SAX-1 kinase. Neuron 44: 239–249 [DOI] [PubMed] [Google Scholar]
- Geng W, He B, Wang M, Adler PN (2000) The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156: 1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff XL, Woollard A, Simanis V (1999) Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol Gen Genet 262: 163–172 [DOI] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM (2004) Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129: 1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz MJ, Petersen J, Valovin S, Hagan IM (2001) MTOC formation during mitotic exit in fission yeast. J Cell Sci 114: 4521–4532 [DOI] [PubMed] [Google Scholar]
- Hirata D, Kishimoto N, Suda M, Sogabe Y, Nakagawa S, Yoshida Y, Sakai K, Mizunuma M, Miyakawa T, Ishiguro J, Toda T (2002) Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G2 delay. EMBO J 18: 4863–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T (1998) Essential role of tubulin-folding cofactor D in microtubules assembly and its association with microtubules in fission yeast. EMBO J 17: 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M-C, Wiley DJ, Verde F, McCollum D (2003) Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci 116: 125–135 [DOI] [PubMed] [Google Scholar]
- Huang TY, Markley NA, Young D (2003) Nak1, an essential germinal center (GC) kinase regulates cell morphology and growth in Schizosaccharomyces pombe. J Biol Chem 278: 991–997 [DOI] [PubMed] [Google Scholar]
- Karos M, Fischer R (1999) Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol Gen Genet 260: 510–521 [DOI] [PubMed] [Google Scholar]
- Leonhard K, Nurse P (2005) Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J Cell Sci 118: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Balasubramanian MK (2000) A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci 113: 1223–1230 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sugiura R, Yada T, Cheng H, Sio SO, Shuntoh H, Kuno T (2002) Calcineurin is implicated in the regulation of the septation initiation network in fission yeast. Genes Cells 7: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan I, Hyams JS (1986) Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci 5: 229–241 [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Duenas E, Sipiczki M, Vazquez de Aldana CR, del Rey F (2003) The endo-β-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J Cell Sci 116: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P (1997) tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89: 939–949 [DOI] [PubMed] [Google Scholar]
- Milburn CC, Boudeau J, Deak M, Alessi DR, Aalten DMFv (2004) Crystal structure of MO25α in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat Struct Mol Biol 11: 193–200 [DOI] [PubMed] [Google Scholar]
- Mitchison JM (1970) Physiological and cytological methods for Schizosaccharomyces pombe. In Methods in Cell Physiology, Prescott DM (ed) Vol. 4, pp. 131–165. New York, NY: Academic Press [Google Scholar]
- Mitchison JM, Nurse P (1985) Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci 75: 357–376 [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Matsushiro A, Nozaki M (1993) Molecular cloning of a novel mRNA sequence expressed in cleavage stage mouse embryo. Mol Reprod Dev 34: 1–7 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nelson B, Kurischoko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LDB, Hughes TR, Boone C, Luca FC (2003) RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell 14: 3782–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M, Onishi Y, Togashi S, Miyamoto H (1996) Molecular characterization of the Drosophila Mo25 gene, which is conserved among Drosophila, mouse and yeast. DNA Cell Biol 15: 505–509 [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science 291: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T (1998) Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell 9: 1757–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanith HA, Sawin KE (2003) Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423: 647–651 [DOI] [PubMed] [Google Scholar]
- Sawin KE, Lourenco PCC, Sanith HA (2004) Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol 14: 763–775 [DOI] [PubMed] [Google Scholar]
- Seshan A, Amon A (2004) Linked for life: temporal and spatial coordination of late mitotic events. Curr Opin Cell Biol 16: 41–48 [DOI] [PubMed] [Google Scholar]
- Simanis V (2003) Events at the end of mitosis in the budding and fission yeasts. J Cell Sci 116: 4263–4275 [DOI] [PubMed] [Google Scholar]
- Snell V, Nurse P (1994) Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J 13: 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V (1998) Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev 12: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D (1999) Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol 146: 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson VA, Kramer J, Kuhn J, Theurkauf WE (2001) Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat Cell Biol 3: 68–75 [DOI] [PubMed] [Google Scholar]
- Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ, Gould KL (2004) Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol Biol Cell 15: 2287–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P (1995) Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol 131: 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Wiley DJ, Nurse P (1998) Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA 95: 7526–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DJ, Marcus S, D'Urso G, Verde F (2003) Control of cell polarity in fission yeast by association of Orb6p kinase with the highly conserved protein Methyltransferase Skb1p. J Biol Chem 278: 25256–25263 [DOI] [PubMed] [Google Scholar]
- Zallen JA, Peckol EL, Tobin DM, Bargmann CI (2000) Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/Warts serine/threonine kinase family. Mol Biol Cell 11: 3177–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Table 1
Supplementary Information
Supplementary Information


