Abstract
Regulated interactions between microtubules (MTs) and the cell cortex control MT dynamics and position the mitotic spindle. In eukaryotic cells, the adenomatous polyposis coli/Kar9p and dynein/dynactin pathways are involved in guiding MT plus ends and MT sliding along the cortex, respectively. Here we identify Bud14p as a novel cortical activator of the dynein/dynactin complex in budding yeast. Bud14p accumulates at sites of polarized growth and the mother-bud neck during cytokinesis. The localization to bud and shmoo tips requires an intact actin cytoskeleton and the kelch-domain-containing proteins Kel1p and Kel2p. While cells lacking Bud14p function fail to stabilize the pre-anaphase spindle at the mother-bud neck, overexpression of Bud14p is toxic and leads to elongated astral MTs and increased dynein-dependent sliding along the cell cortex. Bud14p physically interacts with the type-I phosphatase Glc7p, and localizes Glc7p to the bud cortex. Importantly, the formation of Bud14p–Glc7p complexes is necessary to regulate MT dynamics at the cortex. Taken together, our results suggest that Bud14p functions as a regulatory subunit of the Glc7p type-I phosphatase to stabilize MT interactions specifically at sites of polarized growth.
Keywords: Bud14p, dynein, Glc7p, microtubule regulation, spindle orientation
Introduction
Microtubules (MTs) are essential structures that organize the cytoplasm and assemble and position the mitotic spindle. Spindle orientation is critical for accurate chromosomal segregation in eukaryotic cells, and represents a major strategy to generate cell diversity by asymmetric cell division during development of metazoans (Pearson and Bloom, 2004). In Saccharomyces cerevisiae, orientation of the mitotic spindle along the mother-bud axis is achieved by long astral MTs emanating from the spindle pole bodies (SPBs) and interacting with the bud cortex (Kusch et al, 2003).
Several components, including the adenomatous polyposis coli (APC)-like protein Kar9p and the kinesin Kar3p and its accessory factor Cik1p, localize to SPBs and/or MT plus ends, and pull the spindle to the bud neck, thereby providing directional cues for spindle orientation (Liakopoulos et al, 2003). Kar3p is required for MT attachment to sites of polarized growth during plus-end depolymerization (Maddox et al, 2003). In a second step, the mitotic spindle elongates through the neck via dynein-dependent sliding of cytoplasmic MTs along the bud cortex (Carminati and Stearns, 1997; Adames and Cooper, 2000). In budding yeast, the two pathways are genetically redundant such that they can partially substitute for each other, and only cells lacking the function of both processes are inviable because nuclear division occurs in the mother cell.
For MTs to slide along the cortex, dynein needs to be anchored at the cortex and walk along MTs towards the SPB. Cytoplasmic dynein and its accessory dynactin complex localize to the plus end of MTs by a mechanism involving Lis1p/Pac1p (Lee et al, 2003) and Bik1p (Sheeman et al, 2003). MT sliding requires the cortical protein Num1p, which interacts with the dynein intermediate chain Pac11p, as well as the formin Bni1p (Heil-Chapdelaine et al, 2000; Farkasovsky and Kuntzel, 2001). The interaction between MT plus ends and cortical Num1p may help to unload dynein and Pac1p from the MT tip to the cortex and/or activate the dynein motor or enhance motor processivity at the cortex (Lee et al, 2005). Dynein has been found at the cell cortex in higher eukaryotes (Busson et al, 1998; Gönczy et al, 1999), and the mechanism by which cortically anchored dynein pulls on astral MTs to move the nucleus or to reorient the spindle is likely to be conserved (Gundersen et al, 2004). However, the components involved in MT capture or stabilization at the cortex remain elusive.
The interaction of MTs with cellular structures is dynamic and evidence from several organisms suggest that type I phosphatases (PP1) regulate the interaction of MTs with segregating chromosomes during anaphase (Axton et al, 1990; Fernandez et al, 1992). In budding yeast, the catalytic subunit of the PP1, Glc7p, regulates the MT-binding activity of kinetochores, possibly through dephosphorylation of the CBF3 subunit Ndc10p (Sassoon et al, 1999). Besides the regulation of MT–kinetochore interactions, Glc7p is involved in many other biological processes, including glucose repression, glycogen accumulation, meiosis, and cytokinesis (Stark, 1996). Consistent with these diverse functions, Glc7p localizes to several cellular compartments and structures, including the cytoplasm, nucleus, spindle pole bodies, and sites of polarized growth (Bloecher and Tatchell, 2000). Glc7p interacts via a conserved binding motif with a set of regulatory subunits, which are thought to recruit the phosphatase to specific compartments and regulate substrate specificity (Bollen and Stalmans, 1992; Egloff et al, 1997).
We are interested in understanding how cortical proteins regulate actin and MT function. Interestingly, Bud14p is localized at the bud cortex (Ni and Snyder, 2001) and is found in a synthetic-genome array (SGA) screen with the MT-binding proteins Kar3p and Bim1p (Tong et al, 2004). In this study, we investigated a potential role for Bud14p in stabilizing MT–cortex interactions. Our results suggest that Bud14p functions as a regulatory subunit of Glc7p to modulate MT–cortex interactions during mitosis.
Results
Bud14p exhibits genetic interactions with several components involved in MT function
To investigate the function of Bud14p, we screened the Euroscarf k.o. collection for genes that are synthetic lethal with BUD14 (Tong et al, 2001), and confirmed the interactions by tetrad analysis (Figure 1A; data not shown). Interestingly, several genes that showed a synthetic interaction with BUD14 are known to affect MT function, including KAR3, CIK1, and PAC10 (Figure 1A; see also Tong et al, 2004). Kar3p and Cik1p physically interact and exhibit a minus-end-directed motor activity important for orientation of the mitotic spindle by coupling MTs with the cell cortex. Weak synthetic growth defects were also detected with KAR9 and BIM1 (Figure 1A), while no interaction was observed between BUD14 and DHC1. Whereas bud14Δ, bim1Δ, dhc1Δ, and bud14Δ dhc1Δ cells all divided with similar kinetics, the doubling time of bud14Δ bim1Δ cells increased by approximately 60 min as compared to the respective single mutants when grown at 30°C (data not shown). In addition to these synthetic interactions, Bud14p physically binds to components localized at the bud cortex, such as the GAP Bem2p, the kelch-domain protein Kel2p, the protein kinase Kin2p, and the PP1 Glc7p (Figure 1A) (Balakrishnan et al, 2005). Taken together, the Bud14p functional network suggests a possible link between the cell cortex and MT dynamics.
Figure 1.
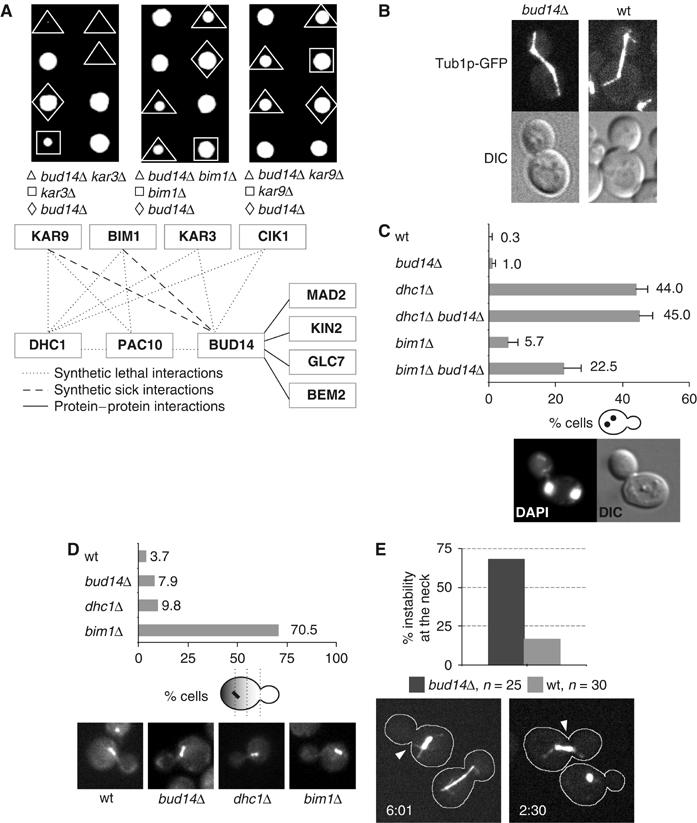
Bud14p is required to stabilize the pre-anaphase spindle at the mother-bud neck. (A) A subset of the synthetic-lethal and protein–protein interaction network identified by SGA analysis, two-hybrid and co-immunoprecipitation assays is shown (source: Saccharomyces Genome Database (SGD) (Balakrishnan et al, 2005)). Tetrad analysis confirmed the synthetic-lethal interaction between BUD14 (yMK22) and KAR3 (Y05556) and the synthetic-sick interactions between BUD14 and BIM1 (Y00147) or KAR9 (Y01023). (B) Live images of the mitotic spindle in wt (yBL100) and bud14Δ cells expressing Tub1p-GFP (yMK13). (C) The accumulation of binucleated mother cells at 16°C was determined by DAPI staining in wt (yBL100), bud14Δ (yMK13), bim1Δ (yMK93), dhc1Δ (yMK94), bud14Δ bim1Δ (yMK141), and bud14Δ dhc1Δ (yMK146) cells, and plotted as percentage (%) of the total number of cells with standard deviations (n=300–400 in each case). (D) Percentages (%) of cells expressing Tub1p-GFP that mis-localize the pre-anaphase spindle at the mother-bud neck. Cells were grown to exponential phase at 16°C, fixed in 4% formaldehyde/0.2% glutaraldehyde, and analyzed microscopically (wt (yBL100): n=107, dhc1Δ (yMK94): n=41, bim1Δ (yMK93): n=61, and bud14Δ (yMK13): n=164). (E) The stability of the mitotic spindle at the bud neck in wt (yBL100: n=30, light grey) and bud14Δ (yMK13: n=25, dark grey) cells expressing Tub1p-GFP was quantified by counting the number of cells where the proximal spindle pole body is displaced from the mother-bud neck by at least half the length of the pre-anaphase spindle. Shown are stills of time-lapse movies of 7-min intervals at the indicated time points (in min) (Supplementary Movies M1 and M2).
bud14Δ cells exhibit a defect in maintaining the position of the pre-anaphase spindle at the bud neck
Consistent with a role of Bud14p in the assembly and/or functioning of the MT cytoskeleton, we occasionally (7.9%; n=400) observed that bud14Δ cells initiated anaphase before nuclear migration to the bud neck was completed (data not shown). However, no significant defects in spindle or astral MT properties were apparent when analyzing time-lapse movies of wild-type (wt) and bud14Δ cells expressing Tub1p-GFP (Figure 1B). To investigate whether Bud14p may play a role in nuclear positioning and/or spindle orientation, we determined by DAPI staining whether nuclear division in bud14Δ occurs in mother cells. As shown in Figure 1C, bud14Δ and bim1Δ single mutants showed only a mild increase of binucleated cells compared to wt, while, as expected, over 40% (n=300) of the dhc1Δ cells underwent nuclear division in the mother cell. However, over 20% (n=400) of the bud14Δ bim1Δ double-mutant cells accumulated binucleated mother cells. In contrast, no further increase of binucleated cells was detected in bud14Δ dhc1Δ double mutants, suggesting that Bud14p and Dhc1p may function in a redundant pathway to Bim1p to regulate spindle orientation during mitosis. To corroborate these results, we analyzed cells expressing Tub1p-GFP to determine the position and stability of the mitotic spindle. As shown in Figure 1D, bud14Δ (8%; n=164) and dhc1Δ (10%; n=41) single mutants exhibited a mild defect in nuclear positioning compared to wt cells (3%; n=107), while, as expected, over 70% (n=61) of bim1Δ cells showed mis-localized nuclei (Adames and Cooper, 2000). Interestingly however, in time-lapse movies, we observed that the position of the pre-anaphase spindle in proximity of the mother-bud neck was unstable in bud14Δ cells (Figure 1E and Supplementary Movies M1 and M2). In contrast to wt cells, the proximal spindle pole body notably moves away from the mother-bud neck in 68% (n=25) of the bud14Δ cells at the G2/M phase of the cell cycle, as analyzed by time-lapse microscopy (wt: 17%; n=30). Thus, while Bud14p was dispensable for the initial positioning of the pre-anaphase spindle, it was required to maintain this position at the mother-bud neck.
Overexpression of Bud14p is toxic and promotes the formation of long astral MTs
As shown in Figure 2A, wt cells overexpressing Bud14p from the GAL1,10 promoter were unable to form colonies. Synchronous release of α-factor-arrested G1 cells revealed that overexpression of Bud14p did not interfere with bud emergence or DNA replication, but the cells uniformly arrested with a large budded morphology and a 2N-DNA content (Figure 2B). Analysis of spinning-disc confocal stacks of live images of Tub1p-GFP cells overexpressing Bud14p revealed that the length of astral MTs emanating from either one of the two spindle poles was greatly increased (Figure 2C) and MTs often reached around the entire bud cortex. In contrast, no significant changes in actin structures were observed (data not shown). Strikingly, the mitotic spindle was mispositioned and often pulled into the bud when spindle dynamics were analyzed by Tub1p-GFP time-lapse microscopy of G2/M cells overexpressing Bud14p (Figure 2D and E and Supplementary Movies M3 and M4). In several movies, the mitotic spindle was pulled to the bud cortex and then rapidly dragged back to the mother cortex (for example, Supplementary Movie M3), indicating that excessive forces emanating from the bud and sometimes also the mother cell cortex are exerted on the premitotic spindle. Together, these observations suggest that increased Bud14p levels result in strengthened MT–cortex interactions preferentially at the bud cortex.
Figure 2.

Bud14p overexpression arrests cells in mitosis with long MTs and the pre-anaphase spindle displaced into the bud. (A) Five-fold serial dilutions of wt (K699) cells harboring an empty control plasmid (eV; pRS415) or a plasmid expressing Bud14p from the inducible GAL1,10 promoter (pMK5) were spotted on selective media containing glucose (GAL-promoter off; upper panel) or galactose (GAL-promoter on; lower panel). The plates were photographed after 3 days at 25°C. (B) wt (K699) cells harboring an empty control plasmid (eV; pRS415) or a plasmid expressing Bud14p from the inducible GAL1,10 promoter (pMK5) were arrested in G1 with α-factor (time 0), and released into media containing 2% galactose. Cell cycle progression was monitored at the times indicated (in h) by FACS analysis. The morphology and budding index was determined microscopically 6 h after release (n=200). (C) The length of astral MTs (in μm) emanating from the proximal and the distal SPB was measured as schematically indicated in maximal projections of 13–19 spinning-disc microscopy z-layers, each separated by 0.2 μm, using ImageJ software (http://www.nih.gov/). Comparison of wt cells (yBL100) overexpressing Bud14p (pMK5) for 6 h and cells carrying a control plasmid (eV; pRS415). (D, E) Analysis of spindle dynamics in wt cells (yBL100) overexpressing Bud14p (pMK5) or carrying a control plasmid (eV; pRS415). Shown in panel D are stills of spinning-disc time-lapse movies of 7-min intervals (Supplementary time-lapse Movies M3 and M4) at the time points indicated (in min). The displacement of the pre-anaphase spindle into the bud was quantified from live epifluorescent images in panel (E) at the indicated times (in h) after galactose induction. Shown is the percentage of Bud14p-overexpressing (dark gray; n=300) and control cells (light gray; n=200) that positioned the pre-anaphase spindle in the bud.
Bud14p accumulates at sites of polarized growth, and this localization is dependent on an intact actin cytoskeleton and the cortical proteins Kel1p and Kel2p
To analyze the subcellular localization of Bud14p, we expressed functional YFP-Bud14p from the constitutive ADH1-promoter in wt or bud14Δ cells. Consistent with previously published results (Ni and Snyder, 2001), YFP-Bud14p accumulated at bud tips and the mother-bud neck at later stages of the cell cycle (Figure 3A), and was also found at the tip of mating projections (+αF). Immunoblotting of α-factor-synchronized cultures showed that endogenous Bud14p levels remained constant throughout the cell cycle (Figure 3B). Costaining of CFP-Tub1p and YFP-Bud14p revealed that the accumulation of Bud14p at the mother-bud neck occurred after completion of anaphase and spindle disassembly, implying that Bud14p at bud tips may be critical with respect to spindle orientation and function.
Figure 3.

The localization of Bud14p to sites of polarized growth requires an intact actin cytoskeleton and the cortical kelch-domain-containing proteins Kel1p and Kel2p. (A) The localization of YFP-Bud14p (pMK113) expressed from the ADH1 promoter (upper panel) and CFP-Tub1p (middle panel) was determined by epifluorescence microscopy in wt (yMK78) cells. (B) The level of Bud14p-myc expressed from its endogenous genomic locus (yMK164) was analyzed by immunoblotting after cell cycle synchronization by α-factor arrest/release. Cell cycle progression was monitored at the times indicated (in min) by immunoblotting for Clb2p (middle panels). Actin served as a loading control (lower panels). An untagged wt strain (K699) controls for the specificity of Bud14p-myc detection. (C) The localization of YFP-Bud14p (pMK113) and CFP-Tub1p was determined in wt (yMK78) cells in the presence (+) or absence (DMSO) of the actin depolymerization drug LAT-A, or the MT-depolymerization drug Nocodazole. Arrows mark the localization of YFP-Bud14p at the bud cortex and bud neck, respectively. (D, E) The localization of GFP-Bud14p (pMK60) expressed from its endogenous promoter was determined in wt (BY4741), kel1Δ (YO2852), kel2Δ (YO6996) and kel1Δ kel2Δ (yMK91) cells. Arrows mark GFP-Bud14p at bud tips, the mother-bud neck, and shmoo tips in cells exposed to α-factor (panels D and E). Asterisks denote loss of GFP-Bud14p at bud or shmoo tips in kel1Δ and kel1Δ kel2Δ cells, while crosses indicate loss of GFP-Bud14p staining at the mother-bud neck. Percentages (%) of cells that have lost polarized GFP-Bud14p localization at shmoo tips (n=200) are shown in (E).
Interestingly, we observed that the bud-tip and neck localization of YFP-Bud14p was reduced after disassembly of the actin cytoskeleton with Latrunculin-A (LAT-A) (Figure 3C). Quantification revealed that 72% of LAT-A-treated cells had no detectable YFP-Bud14p at the tip and 82% no longer showed YFP-Bud14p at the neck (n=200), while YFP-Bud14p localization was largely unaffected after disassembly of the MT network by Nocodazole treatment (Figure 3C). Taken together, these results suggest that Bud14p requires an actin-dependent mechanism to accumulate at sites of polarized growth.
High-throughput analysis suggests that Bud14p interacts with the kelch-domain-containing proteins Kel1p and Kel2p (Ho et al, 2002), both of which localize to sites of polarized growth (Philips and Herskowitz, 1998). Interestingly, the localization of GFP-Bud14p to bud sites, shmoo tips, and the mother-bud neck region was greatly reduced in kel1Δ and kel1Δ kel2Δ cells, while no effect was observed in kel2Δ cells (Figure 3D and E). Quantification revealed that 48% of kel1Δ and 71% of kel1Δ kel2Δ cells (n=200) exposed to α-factor had no detectable GFP-Bud14p at the tips of mating projections (Figure 3E). Western blot analysis showed that GFP-Bud14p was expressed at comparable levels (data not shown), implying that the localization defect was not due to instability of GFP-Bud14p. Together, these results suggest that Kel1p and Kel2p are important for Bud14p localization, and may function as anchors for Bud14p at sites of polarized growth.
Bud14p physically interacts with the PP1 Glc7p through a conserved binding motif
Besides an amino-terminal SH3 domain (amino acids (aa) 262–318), Bud14p contains a conserved motif that is a predicted binding site for the PP1 Glc7p (aa 375–379; Figure 4A). To determine whether these motifs are functionally important, we performed two-hybrid (Table I) and coimmunoprecipitation experiments with wt and Bud14p mutants (Figure 4B). As shown in Table I, Bud14p fused to the activation domain (AD) strongly interacted with Glc7p fused to the DNA-binding domain (DBD). A carboxy-terminal fragment encompassing the putative Glc7p-binding site was sufficient for this interaction, although the SH3 domain also significantly contributed to efficient binding. Supporting this analysis, GST-tagged Bud14p was able to coimmunoprecipitate with HA-tagged Glc7p (Figure 4B) (Lenssen et al, 2005). Importantly, specific point mutations in the Glc7p-interaction motif of Bud14p (V377A and F379A; Figure 4A) strongly diminished its ability to interact with Glc7p both by two-hybrid (Table I) and coimmunoprecipitation assays (Figure 4B), implying that Glc7p indeed interacts with Bud14p through this conserved domain. The mutant protein is still able to function in two-hybrid assays, as Bem2p interacted with both wt and Bud14p mutants V377A and F379A. To corroborate these results, we analyzed several Glc7p mutant proteins for their ability to interact with Bud14p. Interestingly, Glc7p-133 was defective for binding to Bud14p, as shown by two-hybrid assay (Table I) (Lenssen et al, 2005), but still efficiently interacted with the Glc7p regulatory subunit Urip/Bud27p (data not shown) (Venturi et al, 2000). Glc7p-133 contains four mutations changing aa R186A, R187A, R190A, and Q298K, which are predicted to be on the surface of Glc7p (Baker et al, 1997). Together, these results suggest that Bud14p may function as a regulatory subunit of Glc7p at the bud cortex.
Figure 4.

Bud14p binds Glc7p through a conserved motif and is required to localize Glc7p to bud and shmoo tips. (A) The SH3 domain (light gray; aa 262–318) and the conserved Glc7p-binding motif (dark gray; aa 375–379) of Bud14p are schematically indicated (Glc7p-binding motif: dark gray: identical, light gray: strong similarity; NCBI-accession identities: Candida glabrata: XP_447443, Kluyveromyces lactis: XP_45238, Eremothecium gossypii: AQNP_985077, S. pombe: NP_595240). The arrowhead and asterisk highlight the mutated aa in the Glc7p-binding motif that abolish the interaction with Bud14p. (B) HA2-Glc7p (pIP607) was immunoprecipitated from extracts prepared from cells (KT1961) expressing as indicated GST-Bud14p (pCDV476), GST-Bud14p-V377A (pLC742), or GST-Bud14p-F379A (pLC743). The input extracts (upper two panels) or the immunoprecipitates were analyzed by immunoblotting with GST (lower two panels) or HA antibodies (upper panel). (C, D) The localization of GFP-Glc7p (pMK146) expressed from the ADH1 promoter was analyzed by spinning-disc confocal microscopy in wt (K699) (C: lower rows, D: right panels) or bud14Δ (Y00413) cells (C: upper rows, D: left panel). Shown are maximal projections of z-stacks from live images, 13–19 layers, 0.2 μm apart. Cells in panel D were treated with α-factor for 2 h and the loss of shmoo tip localization of GFP-Glc7p was quantified in bud14Δ (n=19) and wt (n=15) cells. Arrowheads mark GFP-Glc7p localization at sites of polarized growth, while asterisks denote loss of GFP-Glc7p localization.
Table 1.
Bud14p and Glc7p specifically interact in yeast two-hybrid assays
| (A) | ||||
|---|---|---|---|---|
| |
β-Galactosidase activity (Miller Units) |
|
||
| AD fusion (pJG4-5) | DBD fusion (pEG202) | |||
| GLC7 | BEM2 | — | ||
| BUD14 | 1361±50 | 576±65 | 8±3 | |
| BUD14[V377A] | 116±20 | 515±34 | ND | |
| BUD14[F379A] | 50±12 | 474±30 | ND | |
| BUD14[1–221] | 5±2 | 101±17 | ND | |
| BUD14[l–340] | 6±5 | 30±5 | ND | |
| BUD14[221–709] | 1538±100 | 1±0 | ND | |
| BUD14[340–709] | 959±35 | 1±0 | ND | |
| BUD14[ΔSH3ΔC-term] | 26±3 | 4±3 | ND | |
| — | 3±0 | 1±0 | 7±0 | |
| (B) | ||||
| |
β-Galactosidase activity (Miller Units) |
|||
| AD fusion (pJG4-5) | DBD fusion (pEG202) | |||
| |
GLC7 |
glc7-133 |
glc7-132 |
glc7-129 |
| BUD14 | 1361±50 | 56±3 | 1623±20 | 876±25 |
| (A and B) Wild type Bud14p (pMK122), Bud14p-V377A (pMK123), and Bud14p-F379A (pMK124) and various truncated Bud14 proteins (pKP555, pKP558, pKP559, pKP554 and pMK117) were tested for their interaction with Glc7p (pCDV471) and Bem2p (pMK71) by two-hybrid analysis as activation (AD)- or DNA-binding-domain (DBD) fusions, as described in Materials and methods. β-Galactosidase activity was measured in liquid assays for at least three independent transformants of each strain, and listed as average Miller Units with standard deviations. The numbers in brackets indicate amino acids of full-length Bud14p starting from the amino-terminal methionine. In panel B, full-length Bud14p (pMK122) was tested for its ability to bind wild-type Glc7p (pCDV471) and the Glc7p point mutants Glc7p-133 (pIP760), Glc7p-132 (pMK136), and Glc7p-129 (pMK135). β-Galactosidase activities were measured as described above. | ||||
| ND: not determined. | ||||
Bud14p is required to localize Glc7p to the bud cortex and shmoo tips
As regulatory subunits are thought to target Glc7p to specific cellular regions (Bloecher and Tatchell, 2000), we compared the localization of GFP-Glc7p expressed from the ADH promoter in wt and bud14Δ cells. In bud14Δ cells, the nuclear and mother-bud neck pools of Glc7p were unaffected (Figure 4C), and Glc7p was still observed on spindle pole bodies during mitosis. In contrast, GFP-Glc7p was no longer detected at the bud cortex (panel C), and also failed to accumulate at shmoo tips in bud14Δ cells arrested with α-factor (panel D). We quantified this defect and found that GFP-Glc7p was lost in all (n=19) of the bud14Δ cells looked at, while only 13% (n=15) of wt cells had no detectable amounts of Glc7p-GFP at shmoo tips. We conclude that Bud14p is required to localize Glc7p at sites of polarized growth, including the bud cortex and shmoo tips. In contrast, GFP-Glc7p remained visible at the mother-bud neck region in bud14Δ cells (Figure 4C), consistent with previous reports that have identified Bni4p as the limiting determinant of Glc7p localization at the mother-bud neck (Kozubowski et al, 2003).
The Bud14p–Glc7p complex may regulate spindle positioning at the bud cortex
We next compared the subcellular localization of wt Bud14p with Bud14p-V377A, Bud14p-F379A, and Bud14p-ΔSH3, using YFP fusions expressed in bud14Δ cells harboring CFP-Tub1p. While an intact SH3 domain was necessary for the accumulation of Bud14p at the cortex, Bud14p-V377A and Bud14p-F379A efficiently accumulated at sites of polarized growth and the mother-bud neck region (Figure 5A). Likewise, wt YFP-Bud14p was found at bud tips and the mother-bud neck in glc7-133 cells (data not shown), confirming that binding of Bud14p to Glc7p is not required for its localization to sites of polarized growth.
Figure 5.

Interaction of Bud14p with Glc7p is functionally important in vivo. (A) The localization of wt YFP-Bud14p (pMK113) and the indicated YFP-Bud14p mutants (pMK114: Bud14p-V377A; pMK115: Bud14p-F379A; pMK168: Bud14p-[1-705, ΔSH3]) expressed from the ADH1 promoter was analyzed in bud14Δ cells harboring CFP-Tub1p (yMK71). (B) Five-fold serial dilutions of an equal number of wt (KT1112) cells transformed with an empty control plasmid (eV; pRS415) or plasmids allowing, as indicated, overexpression of Bud14p (pMK5 and pMK150), Bud14p-V377A (pMK151), Bud14p-F379A (pMK152), or Bud14p-ΔSH3 (pMK156) from the inducible GAL1,10 promoter were spotted on media containing glucose (GAL-promoter off) or galactose (GAL-promoter on). The plates were photographed after 3 days at 25°C. (C) The accumulation of binucleated mother cells at 16°C was determined by DAPI staining in bud14Δ bim1Δ cells (yMK141) transformed with an empty control vector (eV; pRS416) or, as indicated, plasmids expressing wt Bud14p (pMK125), Bud14p-V377A (pMK128), or Bud14p-F379A (pMK131) from the ADH1 promoter. (D) Four-fold serial dilutions of an equal number of wt (KT1112), glc7-133 (KT1636), glc7-133 bud14Δ (yMK173), and glc7-133 bim1Δ (yMK180) transformed with an empty plasmid (eV; pRS416) or plasmids expressing Bud14p (pMK125) or the Glc7p-interacting protein Uri1p (pBL49) from the ADH1 promoter were spotted on plates and photographed after 3 days at 37°C. (E) The accumulation of binucleated mother cells was determined in DAPI-stained wt (KT1112), bud14Δ (yMK163), bim1Δ (yMK172), bim1Δ bud14Δ (yMK180), glc7-133 (KT1636), glc7-133 bud14Δ (yMK173), and glc7-133 bim1Δ (yMK167) cells grown to exponential phase at 16°C, and blotted as percentage of the total number of cells with standard deviations (n=500 in each case).
To address the functional importance of the Bud14p–Glc7p complex, we first compared lethality induced by overexpression of wt and Bud14p mutants unable to interact with Glc7p. As shown in Figure 5B, overexpression of wt Bud14p, but not Bud14p-V377A or Bud14p-F379A, from the inducible GAL1,10 promoter was toxic. Likewise, the toxicity of overexpression of wt Bud14p was much reduced in isogenic glc7-133 cells (data not shown). Simultaneous overexpression of Glc7p and Bud14p did not rescue the toxicity associated with Bud14p overexpression (data not shown), indicating that high levels of Bud14p did not simply titrate Glc7p away from an essential site, but rather functions by a gain-of-function mechanism. Finally, cells overexpressing Bud14p-ΔSH3 were viable, suggesting that the interaction of Bud14p and Glc7p at the bud cortex is functionally important.
Consistent with this conclusion, DAPI staining revealed that neither Bud14p-V377A nor Bud14p-F379A were able to restore the nuclear positioning defect of bud14Δ bim1Δ cells, and over 29% accumulated as binucleate mother cells (Figure 5C; n⩾500). Note that, in this experimental setup, the number of binucleated bud14Δ bim1Δ mother cells was higher compared to Figure 1C, because the cells were grown in synthetic media to select for the plasmids. Moreover, we found that like bud14Δ bim1Δ cells, glc7-133 bim1Δ cells were synthetic-sick (Figure 5D), and an increased number of nuclei divided in mother cells (Figure 5E; n=500). Finally, growth of glc7-133 bim1Δ cells was significantly improved by mild overexpression of Bud14p from the ADH1 promoter, but not Urip (Figure 5D), providing further genetic evidence that the interaction of Bud14p and Glc7p is functionally important.
The Bud14p–Glc7p complex functions as a cortical activator of the dynein/dynactin complex
To test whether Bud14p may regulate the dynein/dynactin complex, we analyzed Bud14p-induced MT sliding in cells deleted for dynein heavy chain (Dhc1p) or its regulator Num1p. Although Bud14p levels were comparable (Figure 6C), the accumulation of the pre-anaphase spindle in the bud was abolished in dhc1Δ and num1Δ cells overexpressing Bud14p for 6 h, while it persisted in bim1Δ cells (Figure 6A and B and Supplementary Movies M5 and M6). While the MT-sliding activity along the cortex was suppressed in dhc1Δ and num1Δ cells, astral MTs were still significantly longer compared to vector controls (Figure 6A and Supplementary Movies M5 and M6). Moreover, Bud14p-induced MT sliding was dependent on its ability to interact with Glc7p as overexpression of Bud14p mutants V377A and F379A did not displace the spindle into the bud (data not shown), implying that the Bud14p–Glc7p complex may increase dynein function.
Figure 6.
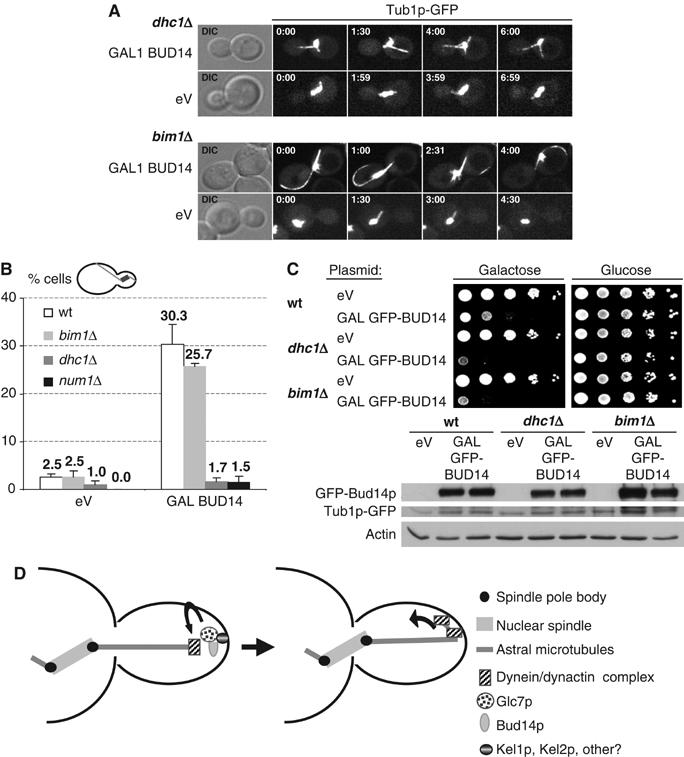
Bud14p-induced excessive cortical sliding of MTs, but not lethality, is dependent on Dhc1p and Num1p. (A, B) Spindle and astral MTs were visualized by spinning-disc confocal and epifluorescent microscopy of Tub1p-GFP in wt (yBL100), dhc1Δ (yMK94), num1Δ (yMK214), or bim1Δ (yMK93) cells that expressed either an empty control vector (eV; pRS415) or Bud14p (pMK5) from the inducible GAL1,10 promoter for 6 h. Shown in panel A are stills of maximal projections of the time-lapse Supplementary Movies M5 and M6 and empty vector control movies at the time points indicated (in min; movie window: 7 min). (B) The number of cells displacing the pre-anaphase spindle into the bud was plotted as percentage of the total number of cells overexpressing Bud14p (pMK5) or an empty control vector (eV; pRS415) for 6 h as counted from epifluorescent live images (eV: n=200, BUD14: n=300). (C) Five-fold serial dilutions of wt (yBL100), dhc1Δ (yMK94), and bim1Δ (yMK93) cells harboring an empty control plasmid (eV; pRS414) or a plasmid expressing GFP-BUD14 from the inducible GAL1,10 promoter (pMK18) were spotted on media containing glucose (GAL-promoter off) or galactose (GAL-promoter on). The plates were photographed after 3 days at 25°C. GFP-Bud14p levels were analyzed by immunoblotting with GFP antibodies of yeast extracts after 6 h induction with 2% galactose. Actin and tubulin control for equal loading of the Western blot. (D) A speculative model depicting the role of the Bud14p–Glc7p complex in the regulation of MT cortex interactions. The budding yeast dynein–dynactin complex is recruited to astral MT plus ends by Bik1p and Pac1p. Bud14p localizes Glc7p to the cortex by interacting with Kel1p and Kel2p, and this complex may directly or indirectly promote dynein unloading or stimulate cortical dynein activity, to induce sliding movement of astral MTs along the bud cortex.
However, while deletion of DHC1 or NUM1 was able to rescue the spindle positioning defects associated with Bud14p overexpression, overexpression of Bud14p was still toxic and the cells arrested at the metaphase–anaphase transition (Figure 6C and data not shown). Surprisingly, this mitotic arrest was independent of the Mad2p spindle-assembly/attachment checkpoint pathway (data not shown), implying that, besides the dynein–dynactin pathway, the Bud14p–Glc7p complex must have additional targets involved in the control of mitosis.
Discussion
In this study, we identified Bud14p as a novel regulatory subunit of the PP1 Glc7p involved in the regulation of MT interactions at the bud cortex. Since Bud14p is conserved in other systems (Martin et al, 2005), it is possible that Bud14p-like activities also function to activate the dynein/dynactin pathway in higher eukaryotes.
The Bud14p/Glc7p complex may regulate MT dynamics at sites of polarized growth
Our results suggest that Bud14p is a novel regulatory subunit of Glc7p, which is specifically involved in the regulation of MT attachment at sites of polarized growth. Glc7p has previously been implicated in the regulation of MT–kinetochore interactions during mitosis (Sassoon et al, 1999). Phosphorylation of the essential kinetochore protein Ndc10p is thought to prevent MT binding to kinetochores, and, as a result, glc7-10 cells arrest at the metaphase–anaphase transition due to the activation of the Mad2p-mitotic checkpoint pathway (Sassoon et al, 1999). Capture of spindle MTs at kinetochores also requires the MT tip binding proteins Stu2p, Bim1p, and Bik1p (Tanaka et al, 2005), which may be regulated by Glc7p. The adaptor that targets Glc7p to kinetochores is not known, but is unlikely to be Bud14p because bud14Δ cells are viable and we were unable to detect Bud14p at kinetochores by spinning-disc confocal microscopy. However, these results suggest that stable interactions of MTs with cellular structures may generally be regulated by phosphorylation.
The Bud14p–Glc7p complex functions as a cortical activator of the dynein complex
We propose that the Bud14p–Glc7p complex may function as a specific activator of the dynein complex at the bud cortex. It is unlikely that dynein activation triggered by Bud14p overexpression results indirectly from the increased length of astral MTs, as, for example, kip3Δ cells with long MTs do not promote cortical sliding and pulling of the spindle into the bud (Yeh et al, 2000). At least three distinct processes are required to regulate dynein function in vivo. First, dynein must be loaded onto MTs most likely via SPBs, from where it is transported to the plus end by a mechanism that involves the Kip2, Bik1, and Lis1/Pac1 proteins (Lee et al, 2003; Carvalho et al, 2004). Second, dynein unloading is triggered presumably by Num1p when a growing MT contacts the cell cortex (Heil-Chapdelaine et al, 2000). Finally, unloaded cortical dynein must be activated to promote MT sliding along the cortex. Preliminary experiments suggest that neither loss nor overexpression of Bud14p affects the localization or expression level of dynein (S Grava and Y Barral, personal communication), implying that Bud14p may affect the unloading and/or activity of the dynein/dynactin complex. Interestingly, two large scale screens revealed that the bud-tip protein Kel1p physically interacts with Dhc1p, Pac1p (Newman et al, 2000), and Glc7p (Ho et al, 2002). As Kel1p and Kel2p also bind Bud14p (Ho et al, 2002) and are required for its cortical localization, it is tempting to speculate that Bud14p may target Glc7p to sites of polarized growth to regulate the interaction or activity of the dynein complex with the cortical kelch-domain proteins Kel1p or Kel2p.
Interestingly, the Bud14p homolog in Schizosaccharomyces pombe, tea4, was recently identified as a binding partner of the Kelch protein tea1 (Martin et al, 2005). Deletion of tea4 results in cells with an aberrant shape caused by MT defects, suggesting that the role of both Bud14/tea4-like proteins in MT regulation and their binding partners may have been conserved. However, tea4 was found to localize to the cell ends as well as plus ends of MTs (Martin et al, 2005), indicating that in S. pombe cortical localization of tea4 may at least in part be regulated by a MT-dependent mechanism. This difference may reflect the different growth patterns, as, in contrast to budding yeast, MTs determine the site of cell growth in S. pombe.
What is the critical target of the cortical Bud14p–Glc7p phosphatase complex?
At present, the Bud14p–Glc7p substrate involved in dynein regulation is not known. Although dynein and several subunits of the dynein–dynactin complex are phosphorylated (King, 2000; Vaughan et al, 2001), genetic evidence suggests that the Bud14p–Glc7p complex functions upstream or at the level of Num1p. Cortical Num1p has been proposed to help unloading of dynein and Pac1p from MT ends to the bud cortex (Heil-Chapdelaine et al, 2000; Lee et al, 2005), and Bud14p–Glc7p may thus regulate capturing of the dynein complex by Num1p. In S. pombe, tea4 directly binds and possibly activates the formin for3 at growing cell ends, suggesting that tea4 may link the actin and MT cytoskeletons (Martin et al, 2005). However, although the budding yeast formin Bni1p is known to be phosphorylated during the cell cycle (Ubersax et al, 2003), bni1Δ cells are sensitive to increased Bud14p levels (Cullen and Sprague, 2002). Moreover, bni1Δ cells were shown to translocate the pre-anaphase spindle into the bud (Lee et al, 1999), similar to Bud14p overexpression described here. Together, these results indicate that Bud14p rather inhibits than increases formin function at the cortex. Alternatively, it is possible that Glc7p dephosphorylates a substrate associated with the plus end of MTs, which may need to be dephosphorylated at the cortex to stabilize the interaction. For example, the Clip170/Bik1p protein is a plus-end-binding protein that is phosphorylated during mitosis (J Kusch and Y Barral, personal communication). Interestingly, Bud14p was shown to coimmunoprecipitate with the PAR-1/MARK-like protein kinase Kin2p (Ho et al, 2002). PAR-1/MARK kinases regulate MT dynamics at the cortex in several organisms (Drewes et al, 1997), suggesting that the Bud14p–Glc7p complex may counteract phosphorylations by Kin2p.
The Bud14p–Glc7p complex may regulate other cellular functions
While the MT-sliding phenotype along the bud cortex induced by Bud14p overexpression is suppressed by deletion of DHC1 or NUM1, the length of astral MTs remains increased and the cells remain arrested at the metaphase/anaphase transition. Surprisingly, this cell cycle arrest is not caused by activation of the Mad2p-dependent mitotic checkpoint (data not shown), as it has been shown for glc7-10 cells, which arrest in mitosis because of a defect in MT–kinetochore attachments (Sassoon et al, 1999). However, this cell cycle arrest depends on the ability of Bud14p to interact with Glc7p, suggesting that additional Bud14p–Glc7p targets regulate progression through mitosis. Moreover, cells deleted for BUD14 exhibit a budding pattern defect in haploid and diploid cells (Ni and Snyder, 2001), which is unlikely due to its role in MT regulation. The Bud14p–Glc7p complex has been shown to be regulated by the global transcriptional regulatory complex Ccr4–Not during starvation (Lenssen et al, 2005). As both CCR4 and NOT5 affect the bipolar bud site selection pattern (Ni and Snyder, 2001), the role of Bud14p in bud site selection may be linked to its regulation by the Ccr4–Not complex.
Materials and methods
Strain constructions and genetic manipulations
The genotypes of the yeast strains are: W303 (ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15, ura3, ssd1-d2) and S288C (his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), unless noted otherwise (Supplementary Table II). Deletion of BUD14 in yMK22 was obtained by exchanging the KANMX4 cassette in Y00413 for a NATMX4 cassette by homologous recombination of transformed EcoRI-linearized p4339. Other gene deletions were obtained by transformation of PCR-amplified KANMX4 or NATMX4 cassettes using 3′ and 5′ UTR-specific primers. Correct deletions were confirmed by resistance to G418 (KAN) (200 μg/ml) or Nourseothricin (NAT) (100 μg/ml) and PCR. Tagging of BUD14 (yMK164) at the endogenous locus was achieved as described (Longtine et al, 1998).
DNA manipulations and two-hybrid assays
Plasmids are described in Supplementary Table III. Site-directed mutagenesis was performed by PCR and confirmed by sequencing. Two-hybrid assays were performed as described previously (Butty et al, 1998).
Microscopy and morphological examination
Proteins tagged with green- (GFP), cyan- (CFP), or yellow (YFP) fluorescent protein were visualized with a Zeiss Axiovert 200M fluorescence microscope equipped with a spinning-disc head and an argon laser (458, 488, and 514 nm) (Visitec), and an Orca-ER CCD camera (Hamamatsu, Japan). For time-lapse microscopy, z-stacks of 13 spinning-disc confocal images separated by 0.2 μm each were taken at 15 time-points with 30 s intervals and maximally projected.
Cell cycle synchronization, arrest/release experiments, and fluorescent activated cell sorting (FACS) analysis
Exponentially growing cells were arrested in G1 by the addition of 25 μg/ml (bar1-1 strains) or 50 μg/ml α-factor (LIPAL-Biochemicals). After 3 h, cells were released and samples for protein extracts, budding index determination, and FACS analysis were taken at the indicated time points. For FACS analysis, cells were fixed in 70% EtOH, treated with RNaseA (0.4 μg/μl) overnight, and pepsin (5 mg/ml) shaking for 30 min at 37°C. The DNA was stained with propidium iodide (50 μg/ml).
Exponentially growing cells expressing YFP-Bud14p were treated at 25°C with LAT-A (0.2 mM in DMSO) for 30 min or Nocodazole (15 μg/ml in DMSO) for 2 h, and analyzed by epifluorescence microscopy. Actin depolymerization was monitored by staining with rhodamine-labeled phalloidin (Molecular Probes, Inc.).
Co-precipitation experiments, antibodies, and immunoblotting
Western blotting was performed using α-myc 9E11 (ISREC), α-Clb2 (kindly provided by D Kellogg, UCSC), α-actin (Roche), α-GFP (B34/8 and C163/8, ISREC), α-HA11 (IgG1, Covance), and α-GST (Santa Cruz Biotechnology, Inc.) antibodies. Co-precipitation experiments between Glc7p and various Bud14p constructs were carried out in KT1961 co-transformed with YCplac22-GAL-HA2-GLC7 (expressing HA2-Glc7p from the GAL1,10 promoter) and plasmids expressing either GST-Bud14p, GST-Bud14p-V377A, or GST-Bud14p-F379A, under the control of the GAL1,10 promoter. Cells were grown to exponential phase and induced with 4% galactose for 4 h. Harvested cells were broken up with acid-washed glass beads in a cell disruptor (FastPrep FP120), and the extract cleared by centrifugation. HA2-tagged Glc7p was purified with the protein G-agarose Immunoprecipitation kit (Roche) using monoclonal mouse anti-HA antibodies and associated proteins were analyzed by immunoblotting.
Supplementary Material
Supplementary Tables
Supplementary Movies
Supplementary Movie M1
Supplementary Movie M2
Supplementary Movie M3
Supplementary Movie M4
Supplementary Movie M5
Supplementary Movie M6
Acknowledgments
We thank C Boone, D Kellogg, Y Barral, B Luke and K Petrovic for providing plasmids, strains, and antibodies. We are grateful to M Gersbach and C Rupp for expert technical assistance, J Kusch, D Liakopoluos, P Wiget, and the ETHZ Light Microscopy Center (LMC) for their help with microscopy. We thank Y Barral and S Grava, and members of the IBC polarity club for stimulating discussion, and Y Barral, J-C Labbé, and M Gotta for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation, the ETH/Zurich, and the FGCZ.
References
- Adames NR, Cooper JA (2000) Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol 149: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axton JM, Dombradi V, Cohen PT, Glover DM (1990) One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell 63: 33–46 [DOI] [PubMed] [Google Scholar]
- Baker SH, Frederick DL, Bloecher A, Tatchell K (1997) Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Christie KR, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Nash R, Oughtred R, Skrzypek M, Theesfeld CL, Binkley G, Lane C, Schroeder M, Sethuraman A, Dong S, Weng S, Miyasato S, Andrada R, Botstein D, Cherry JM (2005) Saccharomyces Genome Database, http://www.yeastgenome.org/ [Google Scholar]
- Bloecher A, Tatchell K (2000) Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Biol 149: 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M, Stalmans W (1992) The structure, role, and regulation of type 1 protein phosphatases. Crit Rev Biochem Mol Biol 27: 227–281 [DOI] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR (1998) Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol 8: 541–544 [DOI] [PubMed] [Google Scholar]
- Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M (1998) The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282: 1511–1516 [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T (1997) Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol 138: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML Jr, Hoyt MA, Pellman D (2004) Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell 6: 815–829 [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr (2002) The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot Cell 1: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89: 297–308 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16: 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H (2001) Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol 152: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Brautigan DL, Lamb NJ (1992) Protein phosphatase type 1 in mammalian cell mitosis: chromosomal localization and involvement in mitotic exit. J Cell Biol 116: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Pichler S, Kirkham M, Hyman AA (1999) Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol 147: 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Gomes ER, Wen Y (2004) Cortical control of microtubule stability and polarization. Curr Opin Cell Biol 16: 106–112 [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75: 791–803 [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA (2000) The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- King SM (2000) The dynein microtubule motor. Biochim Biophys Acta 1496: 60–75 [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Panek H, Rosenthal A, Bloecher A, DeMarini DJ, Tatchell K (2003) A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol Biol Cell 14: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, Liakopoulos D, Barral Y (2003) Spindle asymmetry: a compass for the cell. Trends Cell Biol 13: 562–569 [DOI] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D (1999) Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol 144: 947–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA (2005) The offloading model for dynein function: differential function of motor subunits. J Cell Biol 168: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA (2003) The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol 160: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E, James N, Pedruzzi I, Dubouloz F, Cameroni E, Bisig R, Maillet L, Werner M, Roosen J, Petrovic K, Winderickx J, Collart MA, De Virgilio C (2005) The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol Cell Biol 25: 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y (2003) Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112: 561–574 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K (2003) The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol 13: 1423–1428 [DOI] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR III, Chang F (2005) Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell 8: 479–491 [DOI] [PubMed] [Google Scholar]
- Newman JR, Wolf E, Kim PS (2000) A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 13203–13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M (2001) A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12: 2147–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Bloom K (2004) Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol 5: 481–492 [DOI] [PubMed] [Google Scholar]
- Philips J, Herskowitz I (1998) Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J Cell Biol 143: 375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA (1999) Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev 13: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D (2003) Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13: 364–372 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MJ (1996) Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12: 1647–1675 [DOI] [PubMed] [Google Scholar]
- Stuart JS, Frederick DL, Varner CM, Tatchell K (1994) The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol 14: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU (2005) Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Leszyk JD, Vaughan KT (2001) Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J Biol Chem 276: 26171–26179 [DOI] [PubMed] [Google Scholar]
- Venturi GM, Bloecher A, Williams-Hart T, Tatchell K (2000) Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics 155: 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K (2000) Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell 11: 3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R (1993) Mxi1, a protein that specifically interacts with Max to bind Myc–Max recognition sites. Cell 72: 223–232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Movies
Supplementary Movie M1
Supplementary Movie M2
Supplementary Movie M3
Supplementary Movie M4
Supplementary Movie M5
Supplementary Movie M6


