Abstract
Nucleic acid diagnostics is dominated by fluorescence-based assays that use complex and expensive enzyme-based target or signal-amplification procedures1–6. Many clinical diagnostic applications will require simpler, inexpensive assays that can be done in a screening mode. We have developed a ‘spot-and-read’ colorimetric detection method for identifying nucleic acid sequences based on the distance-dependent optical properties of gold nanoparticles. In this assay, nucleic acid targets are recognized by DNA-modified gold probes, which undergo a color change that is visually detectable when the solutions are spotted onto an illuminated glass waveguide. This scatter-based method enables detection of zeptomole quantities of nucleic acid targets without target or signal amplification when coupled to an improved hybridization method that facilitates probe-target binding in a homogeneous format. In comparison to a previously reported absorbance-based method7, this method increases detection sensitivity by over four orders of magnitude. We have applied this method to the rapid detection of mecA in methicillin-resistant Staphylococcus aureus genomic DNA samples.
A colorimetric detection method for nucleic acids based on the distance-dependent optical properties of DNA-modified gold nanoparticles (DNA-GNP) was previously reported7–9. Because the absorption frequency of the surface plasmon band of metal nanoparticles depends on interparticle distance as well as aggregate size, nanoparticle aggregation mediated by DNA hybridization resulted in a red-shift of the nanoparticles’ surface plasmon band and a visual change of solution color from red to purple or blue. This visible color change was observed by spotting a small aliquot (e.g., 1 μl) of the hybridization solution onto a reversed-phase thin layer chromatography (TLC) plate7. More recently this strategy was extended to the detection of proteins10, carbohydrates11 and metal ions12,13 using functionalized gold nanoparticles. Because of the unique hybridization characteristics of DNA-GNP probes, such as sharp melting transitions and higher melting temperatures, this method achieved a remarkable sequence specificity that allowed discrimination of single-base mismatches, deletions or insertions14.
Although the simplicity of spotting the sample followed by visual readout is attractive for diagnostic applications, the method’s utility is limited by the relatively low limit of detection of 10 fmol target7. The two main factors that contribute to the low sensitivity are the inability to detect nanoparticles at lower concentrations and the requirement of a larger aggregate to achieve a detectable colorimetric shift. Experimental data and optical modeling demonstrated that a large number (e.g., hundreds to thousands) of 15-nm diameter gold particles are needed to provide a measurable red-shift in the surface plasmon band9,15. Experimentally, this necessitates a molar excess of target over nanoparticles to promote formation of large aggregates9. Efforts to increase sensitivity have focused on using 50- to 100-nm diameter gold particles, which absorb more light than the 15-nm diameter particles. However, pre- and post-test (aggregation) colors were not easily distinguishable by a visual readout after spotting16.
We sought to improve the sensitivity of the spot test by developing a method that monitors scattered light rather than reflected light from 40- to 50-nm diameter gold particles (Fig. 1). Given the aggregation-induced red-shift in the surface plasmon frequency mentioned above, we reasoned that the color of scattered light should change concomitantly and provide a more sensitive means of detecting nanoparticle hybridization complexes when spotted onto a waveguide17,18. Here we demonstrate that this approach is feasible and provides much higher sensitivity (greater than four orders of magnitude) in nucleic acid detection than the previously reported absorbance-based spot test. Moreover, this method enables the detection of aggregates in the presence of a substantial excess of nonaggregated particles, which is critical to drive the hybridization in the presence of low target concentrations. In addition, we describe assay modifications that facilitate a simple homogeneous hybridization and detection protocol for rapid visual colorimetric detection of specific gene sequences in total bacterial DNA derived from <105 cells.
Figure 1.
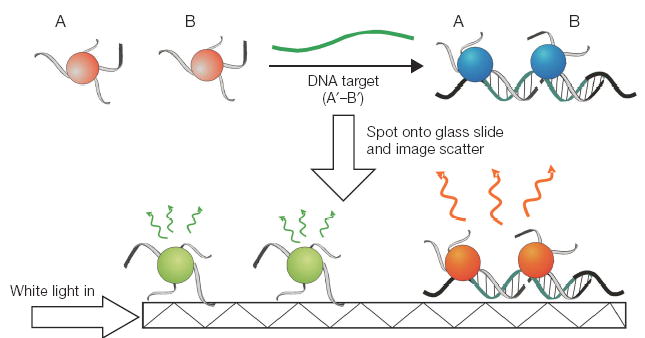
Colorimetric detection of nucleic acid sequences. Step 1: DNA-GNP probes (A and B) are hybridized to a DNA target in solution. Step 2: The samples are spotted onto a glass slide which is illuminated with white light in the plane of the slide. The evanescent induced scatter from the gold nanoparticles is visually observed. Individual 40- to 50-nm diameter gold probes scatter green light, whereas complexed probes scatter yellow to orange light because of a plasmon band red-shift.
Like other colorimetric assays, which use two types of DNA-GNP probes that bind to adjacent regions on a nucleic acid target7, our assay uses two oligonucleotide sequences (A and B) complementary to a target sequence. We chose as a target the bacterial mecA gene, which confers methicillin resistance. Oligos A and B were attached to 50-nm diameter gold particles (GNP50) according to a previously reported procedure, resulting in a high oligo density16. The DNA-GNP50 probes (A and B) exhibit a characteristic plasmon absorption band with a λmax of 528 nm. The probes (1:1 ratio of A and B at 80 pM) were tested by hybridization to a complementary target (A′–B′) or a noncomplementary control (A–B) sequence at 33 nM. Consistent with previous results at a high target/probe ratio, a red-to-purple colorimetric transition was observed visually in the presence of complementary target solution because of DNA-mediated probe aggregation, whereas the negative control solution remained red. This correlates with a strong plasmon band red-shift in the UV-visible spectrum (Fig. 2a), and a visual red-to-blue transition when a 1-μl aliquot is spotted onto a reversed-phase plate (Fig. 2b).
Figure 2.
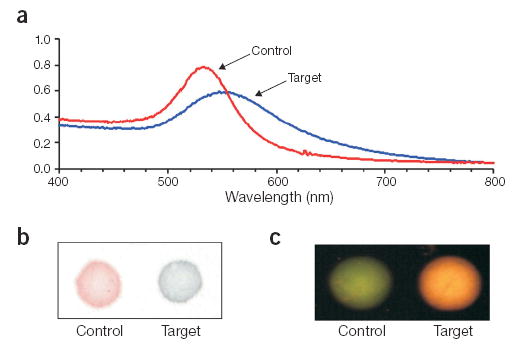
Comparison of detection methodologies for monitoring DNA-GNP binding to target DNA. (a) UV-visible spectrum of target and control samples. (b) 1-μl spot of each sample on a reversed-phase plate. (c) Evanescent light–induced scatter from each sample (1 μl) spotted onto an illuminated glass slide.
For detection of nanoparticle scatter, 1-μl aliquots of each sample were spotted onto a glass microscope slide and excited by planar illumination of the glass slide with white light. This generates an evanescent wave that extends a few hundred nanometers from the slide surface and couples light into nanoparticles that are within the penetration depth of the evanescent field (Fig. 1)17,19. The negative control solution showed green scatter, whereas the complementary target solution showed more intense scattering of orange light (Fig. 2c). This indicates that the DNA-mediated formation of gold probe complexes changes the color and intensity of scattered light.
A titration experiment with a synthetic mecA target (A′–B′) was done to determine assay sensitivity (Fig. 3a). The scattered light from four separate reactions containing probe (1:1 ratio of A and B, 5 × 106 total particles/μl) and decreasing amounts of complementary target DNA (2 × 108 – 2 × 105 copies/μl) was compared to that of a negative control sample (no target) after a 2-h hybridization. Whereas the light scattered from the negative control appeared green, the light scattered from samples containing target ranged from yellow at the lower target concentrations to orange above 2 × 107 copies/μl. Thus, no more than 2 × 105 target molecules (333 zmol) are required under these assay conditions for visual detection of the color change. This sensitivity is roughly four orders of magnitude higher than the previously reported 10-fmol detection limit (3 × 109 copies/μl in a 2-μl reaction) achieved by visual analysis of reflected light from aliquots spotted onto a TLC plate7.
Figure 3.
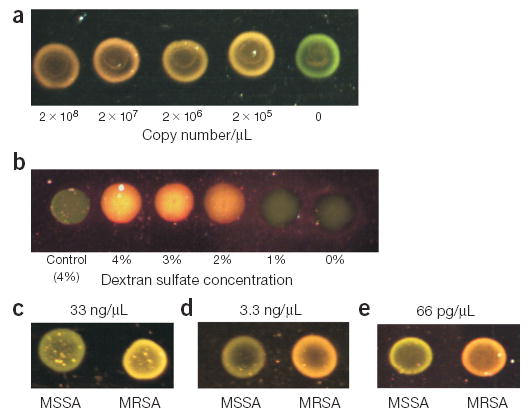
mecA gene detection with DNA-GNPs. (a) High sensitivity detection of a mecA gene sequence using 50-nm gold probes. A 1-μl aliquot of each sample was spotted and dried onto a glass slide, and an image of the evanescent-induced scatter was captured with a color CMOS detector. The color of each spot was clearly detectable with the naked eye. (b) Detection of a mecA PCR product (1.3 × 109 copies/μl) with 40-nm probes (A and B) at various concentrations of dextran sulfate. A no-target control was used for comparison. Color CMOS image of a 1-μl sample aliquot spotted onto a glass slide is shown. (c) Colorimetric detection of MRSA genomic DNA samples isolated from bacterial cells. MSSA genomic DNA served as a negative control. The color of each 1-μl spot was clearly detectable with the naked eye (color CMOS image shown). Two 40-nm probes (A and B) were used to detect a final concentration of 33-ng/μl of genomic DNA. (d) Same experiment as 3c using four 40-nm probes (A, B, C and D) with 3.3 ng/μl of genomic DNA. (e) Same experiment as 3c using four 50-nm probes (A, B, C and D) with 66 pg/μl of genomic DNA.
We attribute this substantial increase in sensitivity to three main factors. First, monitoring scattered light enables detection of lower nanoparticle concentrations as compared to monitoring of absorbed or reflected light20–22. The samples from this experiment could not be detected visually when a 1-μl aliquot was spotted onto a reversed-phase plate because the total probe concentration was only 8 pM. Second, assuming every target is complexed with two particles, there is an approximate 24-fold excess of nonaggregated particles at the lowest target concentration when a color change is still observable. This is critical for the assay because it enables target hybridization to be driven with an excess of probe, and also for imaging because nanoparticle concentrations that are visually detectable by Mie scatter may be used to detect lower concentrations of target. The 50-nm diameter particles were expected to promote a larger colorimetric shift than the 15-nm diameter particles because of increased gold volume15, but this surprising result also indicates that the probe-target complexes must exhibit a large increase in scatter intensity compared to individual probes (also see Fig. 4c).
Figure 4.
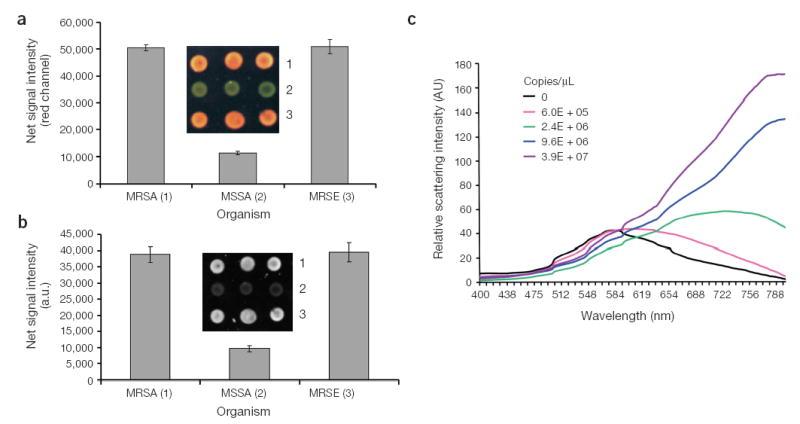
Colorimetric analysis using optical instrumentation. Methicillin-resistant S. aureus (MRSA) and S. epidermidis (MRSE), and methicillin-sensitive S. aureus (MSSA) genomic DNA samples were tested using 40-nm gold probes (A and B). (a) The net signal intensity from the red channel of the color CMOS image (inset) is plotted for each sample. Three replicates were done for each sample. The error bar represents the standard deviation in signal intensity. (b) The glass slide is excited with a red LED, and the image is captured with a monochrome photosensor. The inset shows the captured image in this detection configuration. (c) Analysis of target samples (A′–B′) hybridized to DNA-GNP50 probes using a diode array detector.
Third, the use of dextran sulfate enhances hybridization kinetics23, permitting rapid probe-target hybridization even at low (picomolar) concentrations of probe. To further illustrate this principle, we tested the effect of dextran sulfate in the hybridization mixture as an accelerant of hybridization kinetics. As shown in Figure 3b, gold probe samples (DNA-GNP40 A and B, 20 pM total probe) containing more than 2% dextran sulfate exhibited a color change from green to orange when hybridized for 15 min to a 100-fold molar excess of a 281 base pair mecA PCR fragment, whereas the same test samples containing <1% dextran sulfate did not change color. A no-target control containing 4% dextran sulfate also remained green, demonstrating that the color change was hybridization specific. Because target solutions with less than 1% dextran sulfate remained green even after hours of hybridization, the dextran sulfate is clearly critical for this homogeneous assay format.
To test the feasibility of directly detecting unamplified DNA sequences, we focused on detection of mecA in clinical samples of methicillin-resistant Staphylococcus aureus (MRSA)24. In addition to high sensitivity, this also requires high selectivity because the probes must hybridize specifically to the target sequence in the presence of complex DNA. We tested colorimetric detection of mecA in a sample containing total genomic DNA isolated from MRSA, and total genomic DNA from a methicillin-sensitive S. aureus (MSSA) strain as a negative control. The bacterial DNAs (33 ng/μl) were fragmented by sonication before a 15-min hybridization to 40-nm diameter DNA-GNPs (DNA-GNP40 A and B) at a ratio of 1.6 targets/probe. Aliquots (1 μl) of the control and target samples were spotted onto the glass slide, and the scatter colors were compared (Fig. 3c). A visually detectable color change from green to yellow was observed for only the MRSA sample, demonstrating that the test can be done directly with genomic DNA and with very short hybridization times.
Because the colorimetric shift is dependent on the number of particles within the aggregate structure and the distance between the particles9, we reasoned that increasing the number of gold probes per target may provide higher sensitivity by enhancing the plasmon band red-shift. Two additional mecA probes (C and D) were designed to bind in close proximity to the existing probes (A and B) to test this hypothesis. The mecA-positive and mecA-negative genomic DNA samples were hybridized to a mixture of four DNA-GNP40 probes (A, B, C and D) for 2 h and spotted onto the glass slide. Even with tenfold less genomic DNA in the assay (3.3 ng/μl) and an excess of nanoparticle probe (probe/target ratio of 5), a green to orange colorimetric shift was observed for the MRSA sample, whereas the MSSA sample remained green (Fig. 3d).
A further increase in sensitivity was achieved by using larger 50-nm diameter probes in a similar assay. The combination of four DNA-GNP50 probes (A, B, C and D) generated a visually detectable color change from greenish yellow to orange in the presence of as little as 66 pg/μl of MRSA total genomic DNA (Fig. 3e). This is equivalent to approximately 20,000 copies (33 zmol) of target in the 1-μl volume analyzed on the glass slide, which represents an order of magnitude increase in sensitivity as compared to the same analysis with only two DNA-GNP50 probes (Fig. 3a). Equally important is the increase in the detectable probe/target ratio (~230-fold), which suggests that a four-probe complex enhances both the colorimetric shift and scatter intensity when compared to a two-probe complex.
In addition to visual analysis, the colorimetric signals generated by this assay can be quantified by analyzing signal intensity in the red channel of a red-green-blue sensor such as a color CCD camera. Colorimetric analysis of MRSA and MSSA genomic DNA samples demonstrates that the observed colorimetric changes are quantifiable by this imaging method, and the signals generated by this assay are reproducible (Fig. 4a). Alternatively, a specific excitation frequency may be used to selectively illuminate probe-target complexes, which exhibit a red-shifted surface plasmon band, requiring only a monochrome sensor for signal quantification. Analysis of the same samples using red LED excitation demonstrates that this detection configuration also provides reproducible signal quantification (Fig. 4b). Finally, the relative scattering intensity of probe/target complexes can be monitored using a diode array detector (Fig. 4c). As the target concentration increases, the scattering intensity increases in the wavelength region of 600–800 nm. Therefore, the amount of target in a sample may be quantified by measuring the scattering intensity in this wavelength region at a given probe concentration.
We have described a simple, rapid and sensitive method for detecting DNA targets through hybridization to gold nanoparticle probes in a homogeneous assay format. The resulting nanoparticle complexes are detected by Mie scatter with greatly enhanced detection sensitivity (~four orders of magnitude) compared to a previously reported absorbance-based spot test, enabling colorimetric detection of zeptomole quantities of DNA targets. This sensitivity is possible in a homogeneous format because aggregate formation is detectable even when only a very small fraction of the nanoparticle probes is involved in the hybridization, suggesting a large change in both color and intensity of light scattered from the complexes.
Addition of dextran sulfate to the hybridization mixture facilitates hybridization kinetics, thereby enabling probe-target complexes to form rapidly (<2 h) even at picomolar probe and femtomolar target concentrations. Previously, detection of synthetic oligonucleotides was achieved only after a snap-freeze step to promote sufficient probe aggregation (that is, probe-target hybridization)7. However, snap-freezing is not amenable to automation and promotes mismatch formation at the lower temperature. Our hybridization method overcomes these limitations, enabling rapid detection of specific DNA sequences in more complex PCR or genomic DNA targets. As few as 20,000 target molecules could be detected specifically using short oligonucleotide probes in the presence of total bacterial DNA without the need for enzymatic target or signal amplification. Notably, this homogeneous nanoparticle-based colorimetric assay is approximately four orders of magnitude more sensitive than a more complex enzyme-based solid-phase colorimetric assay that detected 500 ng of mecA25, as well as a recently reported fluorescence-based method which uses nanometer-scale conformational changes in surface-bound polystyrene beads to detect nanomolar concentrations of nucleic acid target26. Combined with visual readout that eliminates complex detection instrumentation, this assay system provides opportunities for the development of low cost, robust and rapid molecular diagnostics that can be done at the site of care. Adaptation to single-nucleotide polymorphism detection in human genomic DNA and detection of other biomolecules such as proteins are underway. We also expect that individual probe complexes should become detectable in this homogeneous format with the appropriate use of imaging instrumentation17, possibly allowing for the efficient detection of single target molecules.
METHODS
General experimental methods
Nanopure water (>18.0 MΩ) from a Millipore Milli-Q gradient water polisher was used for all reported experiments. Oligonucleotide synthesis reagents were purchased from Glen Research or Applied Biosystems. Oligonucleotide synthesis was done using phosphoramidite chemistry on an Applied Biosystems Expedite 8909 DNA synthesizer. All genomic DNA samples were purchased from American Type Culture Collection. Aqueous solutions of 40- or 51.9-nm diameter gold particles were purchased from Ted Pella (mean diameter reported by manufacturer). Modified oligonucleotides were prepared and loaded onto the gold nanoparticles according to literature procedures16. The final nanoparticle concentration was adjusted to 250 pM (50-nm particle) or 750 pM (40-nm particle) in Tris buffer after measuring the absorption at 520 nm (ɛ520 (50) = 1.6 × 1010 M−1cm−1, ɛ520 (40) = 6.7 × 109 M−1cm−1).
All hybridization experiments were done in 0.5-μl centrifuge tubes. A hybridization buffer containing 20% formamide, 16% dextran sulfate and 3.75 mM MgCl2 was used as indicated. UV-visible measurements were done on an Agilent 8453 spectrophotometer. An Epson Expression 1600 flatbed scanner at 300 d.p.i. resolution was used to capture color images of spotted nanoparticle samples on reversed-phase TLC plates (Alltech Associates). A 1-μl sample was spotted onto poly-l-lysine slides (Cel Associates) for scatter detection unless noted otherwise.
Probe and target sequences
The following probe sequences were used in the described experiments:
A: 5′-A15-PEG- ATGGCATGAGTAACGAAGAATA-3′
B: 5′-A15-PEG- TTCCAGATTACAACTTCACCA-3′
C: 5′-A15-PEG- AAAGAACCTCTGCTCAACAAG-3′
D: 5′-A15-PEG- GCACTTGTAAGCACACCTTCAT-3′
The following synthetic target and control sequences were used in the described experiments:
A′–B′ (Target): 5′-TGGTGAAGTTGTAATCTGGAACTTGTTGAGCAGAGGTTCTTTTTTATCTTGGGTTAATTTATTATATTCTTCGTTACTCATGCCAT-3′
A–B (Control): 5′-ATGGCATGAGTAACGAAGAATATAATAAATTAACCCAAGATAAAAAAGAACCTCTGCTCAACAAGTTCCAGATTACAACTTCACCA-3′
It should be noted that probe sequence D is not within the synthetic target sequence shown above but is located within the mecA sequence (not shown). This probe sequence was only used in mecA testing within genomic DNA samples.
Target detection with nanoparticle probes
A target sequence (A′–B′) or control sequence A–B (5 μl, 100 nM) was combined with 6 μl of DNA-GNP50 probe (sequences A and B at a 1:1 ratio, 200 pM total probe), and 4 μl of hybridization buffer. The solutions were heated to 95 °C for 30 s and incubated in a water bath at 37 °C for 10 min. UV-visible measurements of the target and control samples were recorded using a 5-μl sample aliquot (5-μl cuvette, 5-mm path length from Hellma cells). For the absorbance-based spot test, 1 μl of each sample was spotted onto a reversed-phase TLC plate and allowed to dry for 30 min at 22–24 °C. For the Rayleigh scatter-based spot test, a 1-μl aliquot of each sample was pipetted onto a poly-l-lysine slide and imaged wet. For imaging, the glass slide is illuminated with white light in the plane of the slide using a planar fiber optic illuminator (Edmund Industrial Optics). The evanescent induced light scatter was imaged visually or captured with a color CMOS detector. Imaging was done with this illumination/detection method unless otherwise noted.
Target sensitivity with nanoparticle probes
Various target dilutions (5 μl, A′–B′) were mixed with 6 μl of DNA-GNP50 probe (sequences A and B at a 1:1 ratio, 20 pM total probe), and 4 μl of hybridization buffer. A solution containing hybridization buffer and both probes without target was used as a negative control. The solutions were heated at 95 °C for 30 s, followed by incubation at 22–24 °C for 2 h. A 1-μl aliquot of the solution was spotted, and the slide was placed in a dessicator before imaging until the samples were dried (Fig. 3a).
Dextran sulfate comparison
A ~6-nM mecA 281 base pair PCR fragment (5 μl, fragment length and approximate concentration determined with an Agilent Bioanalyzer) was mixed with 6 μl of DNA-GNP40 probe (sequences A and B at a 1:1 ratio, 50 pM total probe), and 4 μl of hybridization buffer. The solutions were heated to 95 °C for 30 s and incubated in a water bath at 40 °C for 15 min. A 1-μl aliquot of each sample was spotted and imaged wet (Fig. 3b).
Genomic DNA detection
Methicillin-resistant S. aureus and Staphylococcus epidermidis (mecA+, ATCC ID Nos. 700699D and 35984D, respectively) and MSSA (mecA-, ATCC ID No. 35556D) genomic DNA samples isolated from cultured bacterial cells were resuspended in 20 mM Tris to a final concentration of 1 μg/μl. The samples were sonicated on ice using three consecutive 10-s, 3-watt pulses and then diluted with Tris buffer to the stated final concentration. Analysis with an Agilent Bioanalyzer indicate average DNA fragment sizes of approximately 500–1,000 base pairs after sonication.
For two-probe detection, a 100 ng/μl solution of MRSA or MSSA (2 μl) was combined with 2.4 μl of DNA-GNP40 probe (sequences A and B at a 1:1 ratio, 25 pM total probe), and 1.6 μl of hybridization buffer. The entire mixture was heated to 95 °C for 1.5 min, followed by incubation at 37 °C for 15 min. One μl of each sample was spotted onto a slide at 43 °C and allowed to dry for 10 min before imaging (Fig. 3c). This same test was done for 1 h using three replicates of each genomic DNA sample (MRSA, MSSA and MRSE), and the net signal intensity in the red channel of the color CMOS sensor was quantified for each spot using Genepix software from Axon instruments (Fig. 4a). In Figure 4b, signal quantification was achieved using the Verigene ID detection system designed at Nanosphere which illuminates the glass slide with a red LED (λex = 630-nm central wavelength) and captures an image of the entire glass slide using a monochrome photosensor. The net signal intensity from each spot was quantified using Genepix software.
For detection with four probes, 5 μl of MRSA or MSSA (10 ng/μl) was combined with 6 μl of DNA-GNP40 probes (sequences A, B, C and D at a 1:1:1:1 ratio, 20 pM total probe), and 4 μl of a hybridization buffer. The sample was heated to 95 °C for 30 s, followed by incubation at 37 °C for 2 h. One μl of each sample was spotted onto a slide at 43 °C and allowed to dry for 10 min before imaging (Fig. 3d). The same assay was done with four DNA-GNP50 probes in place of DNA-GNP40 probes, using 5 μl of a 200-pg/μl sample of MRSA or MSSA (Fig. 3e).
Spectral analysis of samples
Assays were done by hybridizing probes B and C to target A′–B′ using the same experimental conditions described in Fig. 3a. The resulting samples were then spotted and dried onto poly-l-lysine slides and placed in an evanescent illuminator. The spectra were collected using a USB 2000 photodiode spectrometer from Ocean Optics configured for operation in the 350–1,000-nm range (Fig. 4c). To establish spectral correction factors accounting for nuances in the spectrophotometer and the light source, all samples were normalized to a filtered solution of Ludox TM-50.
Acknowledgments
We thank the National Institutes of Health (2 R44 CA85008-02) for support of this work. We also thank Susan Hagenow for assistance in sequence design, and the manufacturing and engineering groups at Nanosphere for technical support.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests (see the Nature Biotechnology website for details).
References
- 1.Kirk BW, Feinsod M, Favis R, Kliman RM, Barany F. Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res. 2002;30:3295–3311. doi: 10.1093/nar/gkf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whelen AC, Persing DH. The role of nucleic acid amplification and detection in the clinical microbiology laboratory. Annu Rev Microbiol. 1996;50:349–373. doi: 10.1146/annurev.micro.50.1.349. [DOI] [PubMed] [Google Scholar]
- 3.Bernard PS, Wittwer CT. Real-time PCR technology for cancer diagnostics. Clin Chem. 2002;48:1178–1185. [PubMed] [Google Scholar]
- 4.Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 5.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall JG, et al. Sensitive detection of DNA polymorphisms by the serial invasive cleavage reaction. Proc. Natl. Acad. Sci. USA. 2000;97:8272–8277. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 8.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 9.Storhoff JJ, et al. What Controls the Optical Properties of DNA-Linked Gold Nanoparticle Assemblies? J. Am. Chem. Soc. 2000;122:4640–4650. [Google Scholar]
- 10.Thanh NTK, Rosenzweig Z. Development of an aggregation-based immunoassay for anti- protein A using gold nanoparticles. Anal Chem. 2002;74:1624–1628. doi: 10.1021/ac011127p. [DOI] [PubMed] [Google Scholar]
- 11.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. Quantitative and reversible lectin-induced association of gold nanoparticles modified with alpha-lactosyl-omega-mercapto- poly(ethylene glycol) J Am Chem Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 12.Kim YJ, Johnson RC, Hupp JT. Gold nanoparticle-based sensing of “spectroscopically silent” heavy metal ions. Nano Lett. 2001;1:165–167. [Google Scholar]
- 13.Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 14.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 15.Lazarides AA, Schatz GC. DNA-Linked Metal Nanosphere Materials: Structural Basis for the Optical Properties. J Phys Chem B. 2000;104:460–467. [Google Scholar]
- 16.Reynolds RA, Mirkin CA, Letsinger RL. Homogeneous, nanoparticle-based quantitative colorimetric detection of oligonucleotides. J Am Chem Soc. 2000;122:3795–3796. [Google Scholar]
- 17.Sonnischen C, et al. Spectroscopy of single metallic nanoparticles using total internal reflection spectroscopy. Appl. Phys. Lett. 2000;77:2949–2951. [Google Scholar]
- 18.Jin RC, et al. Photoinduced conversion of silver nanospheres to nanoprisms. Science. 2001;294:1901–1903. doi: 10.1126/science.1066541. [DOI] [PubMed] [Google Scholar]
- 19.Stimpson DI, et al. Real-Time Detection of DNA Hybridization and Melting on Oligonucleotide Arrays by Using Optical-Wave Guides. Proc. Natl. Acad. Sci. USA. 1995;92:6379–6383. doi: 10.1073/pnas.92.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications - I. Theory. Anal Biochem. 1998;262:137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 21.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications - II. Experimental characterization. Anal Biochem. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 22.Taton TA, Lu G, Mirkin CA. Two-color labeling of oligonucleotide arrays via size-selective scattering of nanoparticle probes. J Am Chem Soc. 2001;123:5164–5165. doi: 10.1021/ja0102639. [DOI] [PubMed] [Google Scholar]
- 23.Wetmur JG. Acceleration of DNA renaturation rates. Biopolymers. 1975;14:2517–2524. doi: 10.1002/bip.1981.360200711. [DOI] [PubMed] [Google Scholar]
- 24.Chambers HF. Methicillin Resistance in Staphylococci: Molecular and Biochemical Basis and Clinical Implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenison R, Yang S, Haeberli A, Polisky B. Interference-based detection of nucleic acid targets on optically coated silicon. Nat Biotechnol. 2001;19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 26.Singh-Zocchi M, Dixit S, Ivanov V, Zocchi G. Single-molecule detection of DNA hybridization. Proc Natl Acad Sci USA. 2003;100:7605–7610. doi: 10.1073/pnas.1337215100. [DOI] [PMC free article] [PubMed] [Google Scholar]


