Abstract
Calcineurin phosphatase activity regulates the nuclear localization of the nuclear factor of activated T cells (NFAT) family of transcription factors during immune challenge. Calcineurin inhibitors, such as the cyclosporin A–cyclophilin A and FK506–FKBP12 complexes, regulate this enzymatic activity noncompetitively by binding at a site distinct from the enzyme active site. A family of endogenous protein inhibitors of calcineurin was recently identified and shown to block calcineurin-mediated NFAT nuclear localization and transcriptional activation. One such inhibitor, Down Syndrome Critical Region 1 (DSCR1), functions in T cell activation, cardiac hypertrophy, and angiogenesis. We have identified a small region of DSCR1 that is a potent inhibitor of calcineurin activity in vitro and in vivo.
Keywords: inhibitor, phosphatase, calcipressin
Calcineurin, a member of the Ser/Thr phosphatase family of enzymes, plays critical roles in many biological processes, such as cell proliferation, cardiovascular and skeletal muscle development, and apoptosis (1–3). Calcineurin's most-studied role is during immune challenge, when calcineurin dephosphorylates and promotes the nuclear localization of the nuclear factor of activated T cells (NFAT) family of transcription factors that increase transcription of T cell-activation genes (4).
Calcineurin is a heterodimer composed of CnA (60 kDa) and CnB (19 kDa) subunits (5). The CnA subunit consists of a catalytic domain, a CnB-binding domain, and a C-terminal regulatory region, consisting of a calmodulin-binding and an autoinhibitory domain (6–8). The latter is susceptible to proteolysis and mostly disordered in the 2.1-Å crystal structure of calcineurin, with the exception of an 18-residue segment of the autoinhibitory region that interacts with the active site of the catalytic domain (7). CnB is the Ca2+-binding regulatory subunit of the phosphatase (9).
Calcineurin activity is regulated by two immunosuppressant drugs. Cyclosporin A and FK506 bind to intracellular proteins called immunophilins, and the resultant cyclosporin A–cyclophilin A and FK506–FKBP12 complexes inhibit calcineurin activity toward large synthetic peptides with inhibitor dissociation constants (Kis) = 270 nM (10) and 6 nM (11), respectively. However, the immunosuppressant complexes induce a 2- to 3-fold increase in activity toward small-molecule substrates, such as p-nitrophenylphosphate (pNPP) (12). Crystal structures of calcineurin in complex with immunosuppressant immunophilins reveal a partially exposed catalytic site in which the active site residue Arg 122 is at least 10 Å from any of the immunophilin and immunosuppressant components (13–15), consistent with the observed noncompetitive mode of inhibition by the drugs and the residual activity of calcineurin toward small substrates (10, 12).
A family of endogenous protein inhibitors of calcineurin has recently been identified (16–18). Human calcipressin 1, or Down Syndrome Critical Region 1 (DSCR1), is encoded by a segment of chromosome 21 that is duplicated in Down Syndrome patients (19). DSCR1 has been shown to inhibit calcineurin-mediated NFAT nuclear translocation and transcriptional activation (18–20). The biological roles of DSCR1 include protection against calcium-mediated oxidative stress (20, 21), cardiac hypertrophy (22, 23), VEGF-mediated signaling during angiogenesis (24, 25), and the formation of aggresomes in Alzheimer's disease (26).
Our preliminary analyses of DSCR1 revealed that it inhibits calcineurin activity toward pNPP, possibly functioning as a calcineurin inhibitor that contacts calcineurin at or near the active site. Crystallization was attempted to determine the structure of the DSCR1–calcineurin complex and reveal the physical basis for enzyme inhibition. Because no crystals were obtained by using full-length proteins, we have generated recombinant fragments of DSCR1 and discovered a region that interacts tightly with calcineurin and competitively inhibits phosphatase activity in vitro and in vivo. Here, we describe a characterization of this inhibitory fragment that provides insights into the endogenous regulation of calcineurin activity and future design of agents targeted to inhibit calcineurin by direct interactions with the enzyme active site.
Materials and Methods
Limited Proteolysis and N-Terminal Sequencing. Partial proteolysis of DSCR1 (25 μg) or CNα–DSCR1 complex (100 μg) purified from overexpressing strains of Escherichia coli (see Supporting Methods, which is published as supporting information on the PNAS web site) was performed in a 15-μl volume. The proteins were incubated for 30 min on ice in reaction buffer (20 mM Tris, pH 8/100 mM NaCl/1 mM DTT), the indicated amounts of trypsin, papain, and subtilisin A were added, and the reaction was incubated for 10 min at room temperature. Proteolytic digestion was stopped with 2× SDS loading buffer (100 mM Tris, pH 6.8/1.4 M 2-mercaptoethanol/4% SDS/0.2% bromophenol blue/20% glycerol), and the samples were heated for 5 min at 95°C, separated on 20% SDS/PAGE, and stained with Coomassie blue R-250 in methanol/acetic acid/water (50/10/40 vol/vol). For N-terminal sequencing, the proteins were transferred onto poly(vinylidene difluoride) membranes (Bio-Rad) in Towbin buffer (25 mM Tris, pH 8.5/192 mM glycine/20% methanol), briefly stained with Coomassie blue R-250 in 40% methanol, and destained with 50% methanol. The protein bands of interest were excised and analyzed by N-terminal sequencing at the Molecular Biology Core Facility, Dana–Farber Cancer Institute (Boston).
Interaction of GST-DSCR1 Fragments with Calcineurin. Cell pellets from 500 ml of E. coli culture expressing GST-DSCR1 fragments (see Supporting Methods for the constructs) were resuspended in 20 ml of lysis buffer (50 mM Tris, pH 8/300 mM NaCl/10% glycerol/2 mM 2-mercaptoethanol/0.01% Nonidet P-40/0.5 mM EDTA/1 mM PMSF/1 mM benzamidine). Glutathione-Sepharose beads (0.5 ml) (Amersham Pharmacia) that had been equilibrated with lysis buffer were added to the soluble extract, and the mixtures were incubated for 4 h at 4°C, with gentle mixing. The resin containing bound GST-DSCR1 fragments was washed three times with 10 ml of lysis buffer. Cultures expressing CNα, CNΔ372, and CNΔ347 from bicistronic constructs (see Supporting Methods) were resuspended and lysed in the above lysis buffer. The amount of soluble protein in the extracts was estimated by electrophoresis of samples along with known concentrations of BSA on SDS/PAGE. Resin containing the GST-DSCR1 fragments was incubated with an equal amount of CNα, CNΔ372, or CNΔ347 lysates for2hat 4°C, with gentle mixing. After washing the beads three times with 1 ml of lysis buffer, the bound proteins were eluted with 2× SDS loading buffer, analyzed by SDS/PAGE, and stained with Coomassie blue.
DSCR1 Binding to Calcineurin. Purified DSCR1-ex7 was reacted with tetramethylrhodamine-5 maleimide (Molecular Probes) in buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, and 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) at a 7:1 fluor-to-protein ratio. The reaction was incubated overnight at 4°C. A NAP-5 column (Amersham Pharmacia) was used to separate labeled DSCR1-ex7 from the unreacted fluor. We calculated the extent of labeling to be 70%. Fluorescence anisotropic emission spectra were collected on a fluorospectrophotometer (Photon Technology International, Lawrenceville, NJ). The total intensity (S) and the anisotropy (R) were calculated according to the equations: S = IVV + 2GIVH and R = (IVV – GIVH)/S, where I indicates the fluorescence intensity observed with the excitation polarizer in the V (vertical) orientation and the emission polarizer in the H (horizontal) orientation. G, the emission polarization bias factor, was calculated by using horizontally polarized excitation (IHV/IHH) values. Titration experiments were performed by adding purified CNα or CNΔ347 to solutions containing a fixed amount (50 nM) of fluorescently labeled DSCR1-ex7 in buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, and 100 μg/ml BSA. Competition experiments were performed by titrating increasing amounts of unlabeled full-length DSCR1 into solutions containing fixed concentrations of fluorescence-labeled DSCR1-ex7 (50 nM) and calcineurin (50 nM). By using the program sigmaplot 8.0, the data were fitted to a nonlinear regression equation for saturation binding: f = Co + (Cf – Co) × 1/KD × [S]/(1/KD × [S] + 1), where Co is initial anisotropy, Cf is final anisotropy, [S] is calcineurin concentration, and KD is the dissociation constant for the calcineurin–DSCR1 complexes. All protein concentrations were determined by using the Bradford assay and extinction coefficients for full-length DSCR1, DSCR1-ex7, CNα, and CNΔ347, with a correction factor obtained from the UV absorbance and calculated extinction coefficients, which are 15,010, 2,680, 51,320, and 41,430 cm–1·M–1, respectively.
Calcineurin Activity Assays. Calcineurin phosphatase activity toward p-nitrophenyl phosphate was assayed by using published methods (27), and inhibition by DSCR1 proteins was measured as described in Supporting Methods.
NFAT Nuclear Localization in Baby Hamster Kidney (BHK) Cells. Full-length DSCR1 and fragments T88 and ex7 were amplified with the forward primers 5′-CCGCTCGAGATGGAGGAGGTGGACCTGCAG-3′, 5′-CCGCTCGAGACCTTACACATAGGAAGCTCACACCTG-3′, and 5′-CCGCTCGAGGGAGAAAAGTATGAATTGCACGCAGCG-3′, respectively, and the reverse primer 5′-AAGGAAAAAAGCGGCCGCTCAGCTGAGGTGGATCGGCGT-3′ (XhoI and NotI restriction sites added are in boldface). DSCR1 fragments ex5/6 and ex6 were amplified with the forward primers 5′-CCGCTCGAGGCCAAATTTGAGTCCCTCTTTAGGACG-3′ and 5′-CCGCTCGAGACCTTACACATAGGAAGCTCACACCTG-3′, respectively, and the reverse primer 5′-AAGGAAAAAGCGGCCGCTCATGGCCCCAGCT TGGAGATGGCATA-3′. The PCR products were digested and ligated into the pMX vector (28), which was modified to include an N-terminal MYC-epitope tag.
GPG-293 cells used for retroviral packaging were maintained in DMEM with 10% FBS, tetracycline (2 mg/ml), puromycin (2 mg/ml), and G418 (0.3 mg/ml), and DSCR1 constructs were transfected into these cells by using TransIT 293 transfection reagent (Mirus, Madison, WI). Retroviral supernatant was collected and used to transduce BHK cells that were stably expressing GFP-NFATc3. The BHK cells were grown and maintained in DMEM supplemented with 10% FBS, 2 mM glutamine, and penicillin and streptomycin at 100 units·ml–1 and 100 μg·ml–1, respectively (GIBCO).
Twenty-four hours after transduction, BHK cells were treated with 500 nM calcium ionophore (ionomycin, Sigma) for 1 h, fixed in 3% formaldehyde for 10 min, and blocked in 3% skim milk in PBS plus 0.1% Triton X-100. The mouse 9E10 anti-MYC and rabbit anti-GFP antibodies (Medical & Biological Laboratories, Nagoya, Japan) were used for detection of MYC-DSCR1 and GFP-NFATc3 fusion proteins, respectively, by immunofluorescence.
Results
Architecture of the DSCR1 Protein. A multiple sequence alignment of human DSCR1 and its orthologues reveals that the calcipressins are well conserved in eukaryotes ranging from yeast to human (see Fig. 6, which is published as supporting information on the PNAS web site). The genes for these proteins comprise seven exons. Exons 1–4 are alternative first exons, whereas exons 5–7 are shared by all isoforms (29, 30). A signature motif FLISPPXSPP of the calcipressins is encoded by residues 105–114 of exon 6. This conserved region resembles the SP repeats found in NFAT proteins, in which the serines are calcineurin substrates for dephosphorylation (31). The motif PXIIXT, encoded by residues 181–186 of exon 7, resembles the PXIXIT motif of NFAT proteins that binds to calcineurin but does not block the catalytic site (32, 33).
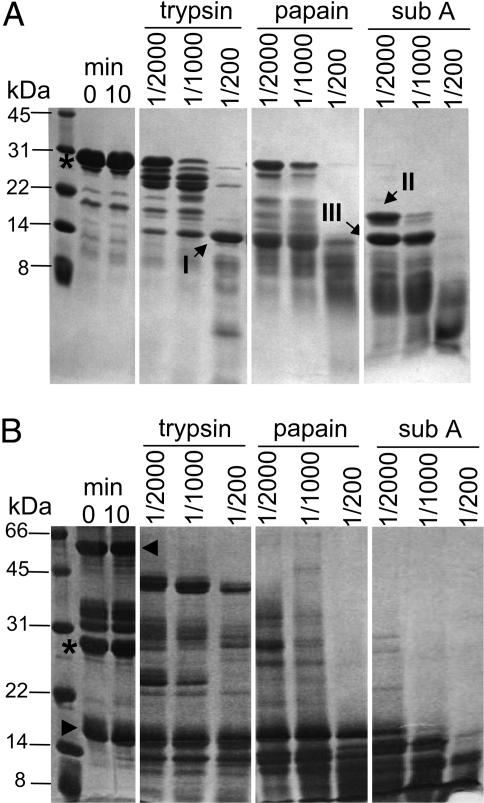
DSCR1 Is Proteolytically Sensitive. Efforts to crystallize the complex of full-length calcineurin–DSCR1 complex failed and, based on secondary structure predictions, we suspected that this was because of unstructured regions of DSCR1. To identify stable domains of DSCR1, limited proteolysis was performed with increasing amounts of trypsin, papain, and subtilisin A. The results indicate that DSCR1 is prone to proteolytic degradation at both the N and C termini (Fig. 1A), suggesting that DSCR1 does not adopt a compact protein fold. Treatment with trypsin or subtilisin A produces somewhat stable fragments of ≈14 kDa and 18 kDa, and similarly sized fragments are obtained in digests with papain. The N terminus of fragment III was identified by peptide sequencing as residue H17 of DSCR1, whereas the N termini of fragments I and II are intact; however, these fragments are further degraded during prolonged incubation with papain and subtilisin A. Importantly, DSCR1 remains sensitive to proteolysis, even in the presence of a binding partner. The pattern of DSCR1 proteolytic fragments changes in complex with CNα, but DSCR1 remains susceptible to proteolysis (Fig. 1B). The A and B subunits of CNα are fairly resistant to proteolysis, although CnA is rapidly converted to a stable polypeptide of ≈43 kDa, corresponding to the catalytic domain (6). Taken together, DSCR1 has a loosely structured protein fold that is not markedly protected from proteolysis in the complex with calcineurin. This result suggests that a limited region of DSCR1 may bind to calcineurin and inhibit its activity.
Fig. 1.
Limited proteolysis of free DSCR1 and DSCR1 in complex with calcineurin. (A) 25 μg of purified DSCR1 (*) was incubated for 10 min with enzyme/protease ratios of 1/2,000, 1/1,000, and 1/200. A reaction without proteases was carried out as a control (left-most panel). Reactions were stopped with SDS loading buffer, and the digested products were analyzed by 20% SDS/PAGE and stained by Coomassie blue. Digested fragments I, II, and III (indicated by arrows) represent fragments lacking N and C termini, indicating that DSCR1 is proteolytically sensitive. (B) An equimolar ratio of DSCR1 (*) and calcineurin (CnA, ◂; CnB, ▸) were incubated together to assemble the complex, subjected to proteolysis, and analyzed as described above. The pattern of DSCR1 proteolytic fragments changes when in a complex but remains susceptible to proteolysis.
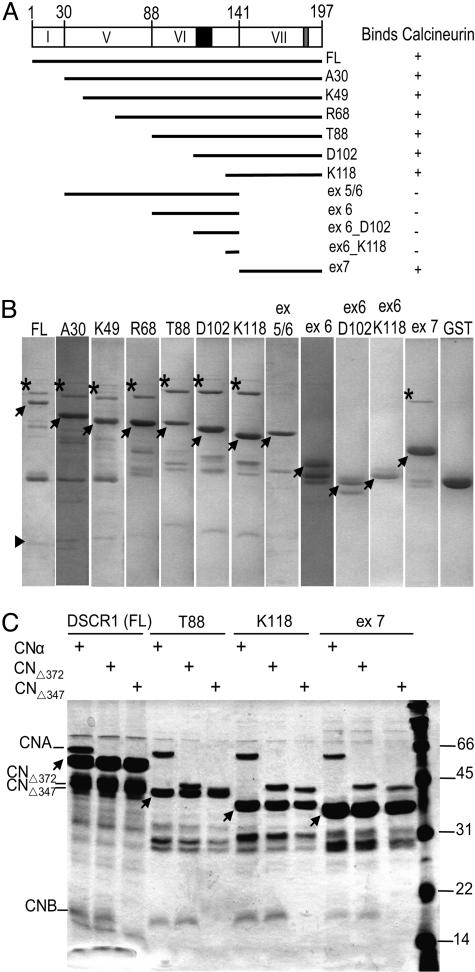
Exon 7 of DSCR1 Is Required to Bind Calcineurin. Because limited proteolysis did not reveal stable protein domains of DSCR1, we relied on the structural organization of the DSCR1 gene to generate protein fragments that might be sufficient for interaction with calcineurin. Fragments corresponding to regions encoded by different exons of the DSCR1 gene (Fig. 2A) were expressed as GST fusion proteins. These fusion proteins were immobilized on glutathione-Sepharose resin and incubated with calcineurin. After washing the resin, the bound proteins were analyzed by SDS/PAGE. Calcineurin is pulled down by full-length DSCR1 (Fig. 2B) but not in the control reaction with GST only, indicating that the presence of the GST tag does not interfere with calcineurin interaction. Surprisingly, DSCR1 fragments lacking the conserved FLISPPXSPP signature motif of exon 6 retain calcineurin-binding activity (Fig. 2 A and B). Furthermore, exon 7 alone binds to calcineurin, whereas DSCR1 fragments lacking exon 7 fail to pull down calcineurin. These results indicate that the region encoded by exon 7 is necessary and sufficient for calcineurin binding.
Fig. 2.
Physical interaction between DSCR1 and calcineurin. (A) Schematic overview depicting full-length DSCR1 and fragments used in our studies. The conserved FLISPPXSPP and PXIIXT motifs are shaded in black and gray, respectively. (B) Glutathione-Sepharose beads containing purified GST-DSCR1 fusion fragments (indicated by the arrows) were incubated with soluble cell lysates containing full-length calcineurin. Bound proteins were eluted from the beads with SDS loading buffer and analyzed by 15% SDS/PAGE. The A (*) and B (▸) subunits of calcineurin are indicated. The B subunit can be observed when more protein is loaded in each lane, as presented in C. Only DSCR1 fragments containing exon 7 pulls down calcineurin. (C) Soluble cell lysates containing full-length calcineurin and truncations of calcineurin were each incubated with the different GST-DSCR1 fusion fragments (indicated by the arrows). The bound proteins were processed as described in Materials and Methods. In lanes depicting CNΔ347 pull-downs, CnB does not interact with DSCR1, although it is expressed, indicating that the catalytic core of calcineurin is sufficient for interaction with DSCR1.
To determine whether the regulatory regions of calcineurin contribute to the interaction with DSCR1, calcineurin fragments lacking both the calmodulin-binding and autoinhibitory domains of CnA (CNΔ372) and additionally lacking the CnB binding domain (CNΔ347) were assayed for DSCR1 binding. We observe that CnB is pulled down with CNΔ372, but not with CNΔ347, when these fragments are incubated with resin containing immobilized DSCR1 fragments (Fig. 2C). The truncated CNΔ347 protein does not interact with the coexpressed CnB subunit in the presence or absence of DSCR1, indicating that CnB associates with the full-length calcineurin in complex with DSCR1 by interacting with CnA and not with DSCR1. These results indicate that the catalytic core of calcineurin alone is sufficient for interaction with DSCR1.
DSCR1-ex7 Adopts a Random-Coil Conformation. The secondary structure of DSCR1-ex7 was examined by CD. The CD spectrum of the exon-7 peptide is characteristic of a random-coil structure (Fig. 7A, which is published as supporting information on the PNAS web site), with minima at 222 and 205 nm. Additionally, we do not observe a cooperative unfolding transition at elevated temperatures (Fig. 7B), and we conclude that DSCR1-ex7 is largely unstructured in solution.
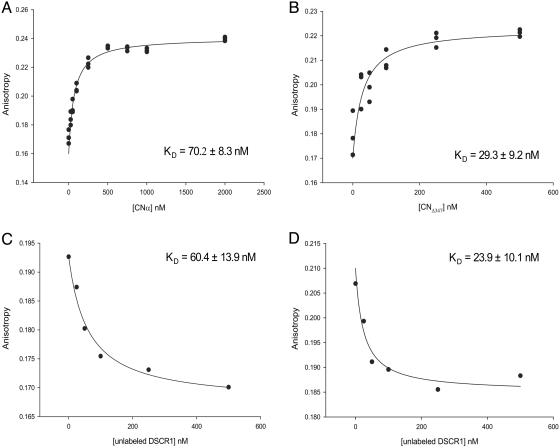
DSCR1-ex7 Binds Calcineurin with High Affinity. Fluorescence anisotropy was used to measure equilibrium binding constants for DSCR1–calcineurin complexes. DSCR1-ex7, which contains a single cysteine at residue 161, was labeled by reacting the peptide with a thiol-reactive fluor. The labeled peptide was titrated with CNα or CNΔ347, and binding was recorded as an increase in anisotropic fluorescent emission upon complex formation. Because our CD spectra of DSCR1-ex7 reveal a random-coil conformation, we expect the fluor to be exposed and unlikely to interfere with binding. This expectation was confirmed by the similar binding constants measured for the labeled peptide and the unlabeled peptide bound in a competition binding experiment (data not shown). These binding studies demonstrate that DSCR1-ex7 binds to full-length calcineurin (Fig. 3A) and to the catalytic core (Fig. 3B) with KD values of 70.2 ± 8.3 and 29.3 ± 9.2 nM, respectively. The apparent weaker binding to full-length calcineurin most likely results from interference by the autoinhibitory domain, which is absent in CNΔ347 and may compete for binding to the active site.
Fig. 3.
Saturable binding of DSCR1 to calcineurin. Fluorescently labeled DSCR1-ex7 peptide (50 nM) was titrated with increasing amounts of CNα (A)orCNΔ347 (B). DSCR1-ex7 binds CNα and CNΔ347, with KD values of 70.2 ± 8.3 nM and 29.3 ± 9.2 nM, respectively. DSCR1-ex7 binding was competed with increasing amounts of unlabeled full-length DSCR1 at a fixed CNα (C)orCNΔ347 (D) concentration of 50 nM. The values of anisotropic fluorescent emission are shown as means (±SD) from three separate experiments. DSCR1 binds CNα and CNΔ347, with KD values of 60.4 ± 13.9 nM and 23.9 ± 10.1 nM, respectively. Apparent binding constants (KD) are shown with the calculated uncertainty of fitting the data to a simple binding equilibrium.
Competition experiments were also performed to determine binding constants of full-length DSCR1 for CNα and CNΔ347. Fluorescently labeled DSCR1-ex7 was titrated with increasing amounts of DSCR1 in the presence of 50 nM calcineurin. The KD values of DSCR1 for CNα and CNΔ347 are 60.4 ± 13.9 nM (Fig. 3C) and 23.9 ± 10.1 nM (Fig. 3D), respectively. Full-length DSCR1 binds to calcineurin with high affinity, comparable to DSCR1-ex7, and the energetically significant contacts of DSCR1 are made to the catalytic core of calcineurin.
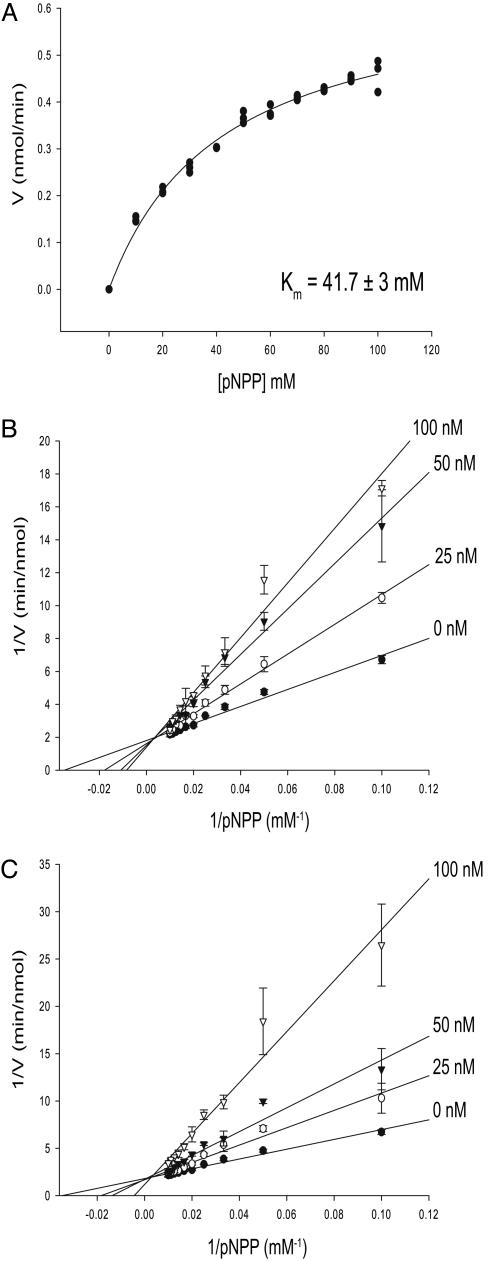
DSCR1 Inhibits Calcineurin Activity Competitively. Kinetic studies were carried out with CNα (see Fig. 8, which is published as supporting information on the PNAS web site) or CNΔ347 (Fig. 4) to investigate the mechanism of inhibition by DSCR1. The chromogenic compound pNPP is a small-molecule substrate that is dephosphorylated by calcineurin, and interference with this substrate would most likely suggest a direct interaction of DSCR1 with the enzyme active site. Hydrolysis of pNPP was measured spectrophotometrically in the presence of increasing concentrations of full-length DSCR1 or DSCR1-ex7. The Km of pNPP for the catalytically active CN347Δ is 41.7 ± 3 mM (Fig. 4A). For each concentration of inhibitor, the concentration of pNPP was varied from 0–100 mM, and reactions were performed in triplicate. The average rates of reactions at varying [pNPP], plotted as double-reciprocal plots, are shown for full-length DSCR1 (Fig. 4B) and DSCR1-ex7 (Fig. 4C). The data are consistent with competitive inhibition, with a Ki = 68.6 ± 3.2 nM for full-length DSCR1 and a Ki = 51.6 ± 2.1 nM for DSCR1-ex7. The competitive mode of inhibition by DSCR1 is suggestive of a direct interaction with the enzyme active site to inhibit phosphatase activity. Furthermore, the DSCR1-ex7 fragment inhibits calcineurin activity with a potency similar to full-length DSCR1, in agreement with the binding studies described above (Fig. 3).
Fig. 4.
Kinetic analysis of the inhibitory mechanism of DSCR1. (A) Activity of the catalytic domain of calcineurin toward pNPP was measured. The Km for pNPP is 41.7 ± 3 mM. CNΔ347 was incubated with full-length DSCR1 (B) and DSCR1-ex7 fragment (C) at different concentrations [0 nM (•), 25 nM (○), 50 nM (▾), and 100 nM (▿)], with pNPP concentration increasing from 10 to 100 mM. Each graph represents the means (±SD) from triplicate reactions. The Ki values for DSCR1 (68.6 ± 3.2 nM) and DSCR1-ex7 (51.6 ± 2.1 nM) are similar.
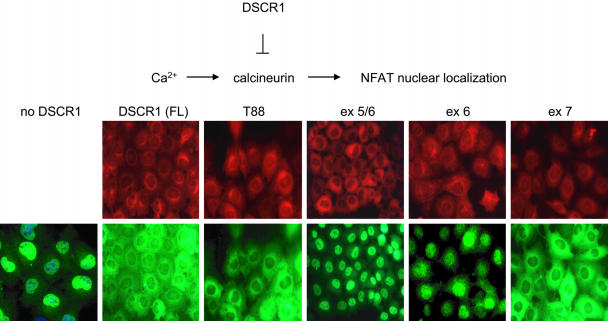
DSCR1-ex7 Is Necessary and Sufficient to Inhibit Calcineurin-Mediated NFAT Nuclear Localization in BHK Cells. Our in vitro studies indicate that the DSCR1-ex7 peptide is a potent inhibitor of calcineurin activity, similar to full-length DSCR1. We next asked whether DSCR1-ex7 is sufficient to block the calcineurin-mediated nuclear localization of NFATc3 in vivo. BHK cells stably expressing GFP-NFATc3 and MYC epitope-tagged DSCR1 fragments (full-length, T88, ex5/6, ex6, and ex7; see Fig. 2A) were treated with a calcium ionophore and fixed with formaldehyde, and the proteins were visualized by immunofluorescence microscopy. In the absence of DSCR1, NFATc3 localizes in the nucleus after calcium release by ionophore (Fig. 5). We observe that full-length DSCR1, DSCR1-T88 (which is encoded by exons 6 and 7), and DSCR1-ex7 all inhibit NFATc3 nuclear localization. In contrast, DSCR1 fragments lacking exon 7, such as DSCR1-ex5/6 and DSCR1-ex6, do not affect the nuclear localization of NFATc3 in response to a calcium stimulus. These results indicate that exon 7 of DSCR1 is necessary and sufficient to inhibit calcineurin activity in vivo.
Fig. 5.
Inhibition of calcineurin-mediated NFATc3 nuclear localization by DSCR1. BHK cells stably expressing GFP-NFATc3 and MYC-DSCR1 fragments were treated with 500 nM ionomycin, a calcium ionophore, and fixed with 3% formaldehyde. Anti-MYC and anti-GFP antibodies were used for detection of DSCR1 (red) and NFATc3 (green), respectively, by immunofluorescence. DSCR1 fragments lacking exon 7 are unable to inhibit calcineurin-mediated nuclear localization of NFATc3.
Discussion
Our results show that DSCR1 is a competitive inhibitor of calcineurin, with a Ki in the low nanomolar range (Fig. 4). This indicates that DSCR1 is a calcineurin inhibitor that is likely to directly contact the enzyme active site, in contrast to cyclosporin A–cyclophilin and FK506–FKBP12 complexes in which essential catalytic residues of calcineurin are at least 10 Å from either the drug or immunophilin component (13–15).
We have identified the region of DSCR1 that is necessary and sufficient for interaction with calcineurin and its inhibition in vitro and in vivo. We show that the C-terminal 57 residues of DSCR1 encoded by exon 7 are required to bind calcineurin (Figs. 2 and 3). This fragment binds with high affinity, and it inhibits calcineurin activity with a potency similar to full-length DSCR1. Our results are consistent with previous studies demonstrating that DSCR1 mutants lacking the C terminus are unable to bind calcineurin and inhibit calcineurin-mediated NFAT nuclear localization (19, 34).
We were surprised that the highly conserved FLISPPXSPP sequence motif is apparently dispensible for calcineurin binding and inhibition. We expected that these conserved residues might function as a competitive inhibitor, mimicking substrates like the NFATs, in which dephosphorylation of phosphoserines within the SPP repeats triggers nuclear localization (35). However, our results are consistent with a previous study showing that mutants of DSCR1 with amino acid substitutions of the serines in this motif are still able to bind and inhibit calcineurin, albeit with some decrease in the extent of inhibition (36). Functional roles of this serine-rich motif of DSCR1 and paralogous proteins remain to be discovered.
A second conserved motif PXIIXT, found in DSCR1, may contribute to interactions with calcineurin. This motif, encoded by exon 7, resembles a consensus PXIXIT sequence of NFAT proteins that binds to calcineurin with low affinity (KD = 10–30 μM) (4). Peptides containing this motif that were selected for binding to the calcineurin catalytic domain failed to inhibit enzymatic activity, suggesting that they bind to a site that is distinct from the active site (32). In contrast, exon 7 of DSCR1 binds tightly to calcineurin (Fig. 3) and in competition with substrates (Fig. 4), implying an intimate interaction with the active site. We find that a further C-terminal truncation of the region spanning the PXIXIT motif (residues 181–191) eliminates the inhibitory activity of DSCR1 (data not shown).
The identification of a DSCR1 peptide fragment that competitively inhibits calcineurin activity could facilitate the development of new inhibitory agents with a different mode of action from the current drugs cyclosporin A and FK506. These approved calcineurin inhibitors prevent graft rejection after organ transplantation (35, 37), but they are neurotoxic and nephrotoxic (38, 39). Thus, there is a need for more selective therapeutic agents targeting calcineurin.
Other partner proteins of DSCR1 have recently been identified. DSCR1 physically interacts with integrin αvβ3 during adhesion of endothelial cells (24) and down-regulates inflammatory marker genes in endothelial cells (25), implying roles in angiogenesis and, possibly, tumor metastasis. DSCR1 is also found to colocalize with ubiquitin in mammalian cells as well as huntintin and ataxin-3, proteins associated with Alzheimer's disease (26). The multifunctionality of DSCR1 suggests that it may regulate other activities in addition to its role in blocking calcineurin activity. Different regions of DSCR1 could regulate these other biological processes.
Supplementary Material
Acknowledgments
We thank Sandy Ryeom (Children's Hospital, Boston) for providing the intial full-length calcineurin and DSCR1 constructs and Antoine van Oijen (Harvard Medical School) for advice on peptide labeling. This work was supported by grants from the National Institutes of Health (NIH) (to T.E. and F.M.) and Phillip Morris (to F.M.) and a NIH Training Grant in the Pharmacological Sciences. T.E. is the Hsien Wu and Daisy Yen Wu Professor of Biological Chemistry and Molecular Pharmacology at Harvard Medical School.
Author contributions: B.C., F.M., and T.E. designed research; B.C. and G.G. performed research; B.C., G.G., F.M., and T.E. analyzed data; and B.C., F.M., and T.E. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BHK, baby hamster kidney; DSCR1, Down Syndrome Critical Region 1; NFAT, nuclear factor of activated T cells.
References
- 1.Kahl, C. R. & Means, A. R. (2003) Endocr. Rev. 24, 719–736. [DOI] [PubMed] [Google Scholar]
- 2.Schulz, R. A. & Yutzey, K. E. (2004) Dev. Biol. 266, 1–16. [DOI] [PubMed] [Google Scholar]
- 3.Groenendyk, J., Lynch, J. & Michalak, M. (2004) Mol. Cells 17, 383–389. [PubMed] [Google Scholar]
- 4.Hogan, P. G., Chen, L., Nardone, J. & Rao, A. (2003) Genes Dev. 17, 2205–2232. [DOI] [PubMed] [Google Scholar]
- 5.Klee, C. B., Draetta, G. F. & Hubbard, M. J. (1988) Adv. Enzymol. Relat. Areas Mol. Biol. 61, 149–200. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard, M. J. & Klee, C. B. (1989) Biochemistry 28, 1868–1874. [DOI] [PubMed] [Google Scholar]
- 7.Kissinger, C. R., Parge, H. E., Knighton, D. R., Lewis, C. T., Pelletier, L. A., Tempczyk, A., Kalish, V. J., Tucker, K. D., Showalter, R. E., Moomaw, E. W., et al. (1995) Nature 378, 641–644. [DOI] [PubMed] [Google Scholar]
- 8.Klee, C. B., Ren, H. & Wang, X. (1998) J. Biol. Chem. 273, 13367–13370. [DOI] [PubMed] [Google Scholar]
- 9.Stemmer, P. M. & Klee, C. B. (1994) Biochemistry 33, 6859–6866. [DOI] [PubMed] [Google Scholar]
- 10.Etzkorn, F. A., Chang, Z. Y., Stolz, L. A. & Walsh, C. T. (1994) Biochemistry 33, 2380–2388. [DOI] [PubMed] [Google Scholar]
- 11.Aldape, R. A., Futer, O., DeCenzo, M. T., Jarrett, B. P., Murcko, M. A. & Livingston, D. J. (1992) J. Biol. Chem. 267, 16029–16032. [PubMed] [Google Scholar]
- 12.Liu, J., Farmer, J. D., Jr., Lane, W. S., Friedman, J., Weissman, I. & Schreiber, S. L. (1991) Cell 66, 807–815. [DOI] [PubMed] [Google Scholar]
- 13.Huai, Q., Kim, H. Y., Liu, Y., Zhao, Y., Mondragon, A., Liu, J. O. & Ke, H. (2002) Proc. Natl. Acad. Sci. USA 99, 12037–12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin, L. & Harrison, S. C. (2002) Proc. Natl. Acad. Sci. USA 99, 13522–13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith, J. P., Kim, J. L., Kim, E. E., Sintchak, M. D., Thomson, J. A., Fitzgibbon, M. J., Fleming, M. A., Caron, P. R., Hsiao, K. & Navia, M. A. (1995) Cell 82, 507–522. [DOI] [PubMed] [Google Scholar]
- 16.Kingsbury, T. J. & Cunningham, K. W. (2000) Genes Dev. 14, 1595–1604. [PMC free article] [PubMed] [Google Scholar]
- 17.Strippoli, P., Lenzi, L., Petrini, M., Carinci, P. & Zannotti, M. (2000) Genomics 64, 252–263. [DOI] [PubMed] [Google Scholar]
- 18.Rothermel, B., Vega, R. B., Yang, J., Wu, H., Bassel-Duby, R. & Williams, R. S. (2000) J. Biol. Chem. 275, 8719–8725. [DOI] [PubMed] [Google Scholar]
- 19.Fuentes, J. J., Genesca, L., Kingsbury, T. J., Cunningham, K. W., Perez-Riba, M., Estivill, X. & de la Luna, S. (2000) Hum. Mol. Genet. 9, 1681–1690. [DOI] [PubMed] [Google Scholar]
- 20.Ermak, G., Harris, C. D. & Davies, K. J. (2002) FASEB J. 16, 814–824. [DOI] [PubMed] [Google Scholar]
- 21.Lin, H. Y., Michtalik, H. J., Zhang, S., Andersen, T. T., Van Riper, D. A., Davies, K. K., Ermak, G., Petti, L. M., Nachod, S., Narayan, A. V., et al. (2003) Free Radical Biol. Med. 35, 528–539. [DOI] [PubMed] [Google Scholar]
- 22.Lange, A. W., Molkentin, J. D. & Yutzey, K. E. (2004) Dev. Biol. 266, 346–360. [DOI] [PubMed] [Google Scholar]
- 23.van Rooij, E., Doevendans, P. A., Crijns, H. J., Heeneman, S., Lips, D. J., van Bilsen, M., Williams, R. S., Olson, E. N., Bassel-Duby, R., Rothermel, B. A. & De Windt, L. J. (2004) Circ. Res. 94, e18–26. [DOI] [PubMed] [Google Scholar]
- 24.Iizuka, M., Abe, M., Shiiba, K., Sasaki, I. & Sato, Y. (2004) J. Vasc. Res. 41, 334–344. [DOI] [PubMed] [Google Scholar]
- 25.Hesser, B. A., Liang, X. H., Camenisch, G., Yang, S., Lewin, D. A., Scheller, R., Ferrara, N. & Gerber, H. P. (2004) Blood 104, 149–158. [DOI] [PubMed] [Google Scholar]
- 26.Ma, H., Xiong, H., Liu, T., Zhang, L., Godzik, A. & Zhang, Z. (2004) J. Neurochem. 88, 1485–1496. [DOI] [PubMed] [Google Scholar]
- 27.Sagoo, J. K., Fruman, D. A., Wesselborg, S., Walsh, C. T. & Bierer, B. E. (1996) Biochem. J. 320, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onishi, M., Kinoshita, S., Morikawa, Y., Shibuya, A., Phillips, J., Lanier, L. L., Gorman, D. M., Nolan, G. P., Miyajima, A. & Kitamura, T. (1996) Exp. Hematol. 24, 324–329. [PubMed] [Google Scholar]
- 29.Fuentes, J. J., Pritchard, M. A. & Estivill, X. (1997) Genomics 44, 358–361. [DOI] [PubMed] [Google Scholar]
- 30.Cao, X., Kambe, F., Miyazaki, T., Sarkar, D., Ohmori, S. & Seo, H. (2002) Biochem. J. 367, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, C., Shaw, K. T., Raghavan, A., Aramburu, J., Garcia-Cozar, F., Perrino, B. A., Hogan, P. G. & Rao, A. (1996) Proc. Natl. Acad. Sci. USA 93, 8907–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G. & Rao, A. (1999) Science 285, 2129–2133. [DOI] [PubMed] [Google Scholar]
- 33.Li, H., Rao, A. & Hogan, P. G. (2004) J. Mol. Biol. 342, 1659–1674. [DOI] [PubMed] [Google Scholar]
- 34.Vega, R. B., Yang, J., Rothermel, B. A., Bassel-Duby, R. & Williams, R. S. (2002) J. Biol. Chem. 277, 30401–30407. [DOI] [PubMed] [Google Scholar]
- 35.Shaw, K. T., Ho, A. M., Raghavan, A., Kim, J., Jain, J., Park, J., Sharma, S., Rao, A. & Hogan, P. G. (1995) Proc. Natl. Acad. Sci. USA 92, 11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genesca, L., Aubareda, A., Fuentes, J. J., Estivill, X., De La Luna, S. & Perez-Riba, M. (2003) Biochem. J. 374, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber, S. L. & Crabtree, G. R. (1992) Immunol. Today 13, 136–142. [DOI] [PubMed] [Google Scholar]
- 38.Sigal, N. H., Dumont, F., Durette, P., Siekierka, J. J., Peterson, L., Rich, D. H., Dunlap, B. E., Staruch, M. J., Melino, M. R., Koprak, S. L., et al. (1991) J. Exp. Med. 173, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumont, F., Staruch, M. J., Koprak, S. L., Siekierka, J. J., Lin, C. S., Harrison, R., Sewell, T., Kindt, V. M., Beattie, T. R., Wyvratt, M. & Sigal, N. H. (1992) J. Exp. Med. 176, 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mondragon, A., Griffith, E. C., Sun, L., Xiong, F., Armstrong, C. & Liu, J. O. (1997) Biochemistry 36, 4934–4942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.