Abstract
Bruton's tyrosine kinase (BTK) deficiency results in a differentiation block at the pre-B cell stage. Likewise, acute lymphoblastic leukemia cells are typically arrested at early stages of B cell development. We therefore investigated BTK function in B cell precursor leukemia cells carrying a BCR-ABL1, E2A-PBX1, MLL-AF4, TEL-AML1, or TEL-PDGFRB gene rearrangement. Although somatic mutations of the BTK gene are rare in B cell precursor leukemia cells, we identified kinase-deficient splice variants of BTK throughout all leukemia subtypes. Unlike infant leukemia cells carrying an MLL-AF4 gene rearrangement, where expression of full-length BTK was detectable in only four of eight primary cases, in leukemia cells harboring other fusion genes full-length BTK was typically coexpressed with kinase-deficient variants. As shown by overexpression experiments, kinase-deficient splice variants can act as a dominant-negative BTK in that they suppress BTK-dependent differentiation and pre-B cell receptor responsiveness of the leukemia cells. On the other hand, induced expression of full-length BTK rendered the leukemia cells particularly sensitive to apoptosis. Comparing BTK expression in surviving or preapoptotic leukemia cells after 10-Gy γ radiation, we observed selective survival of leukemia cells that exhibit expression of dominant-negative BTK forms. These findings indicate that lack of BTK expression or expression of dominant-negative splice variants in B cell precursor leukemia cells can (i) inhibit differentiation beyond the pre-B cell stage and (ii) protect from radiation-induced apoptosis.
Keywords: pre-B cell receptor, differentiation block, apoptosis
Recent findings by us and others (1, 2) suggested a role for the pre-B cell receptor and related signaling molecules as a tumor suppressor to prevent the development or limit the proliferation of leukemic cells. BCR-ABL1+ pre-B lymphoblastic leukemia cells frequently exhibit defective expression of the pre-B cell receptor related signaling molecule SLP65 (1) and acquire independence from pre-B cell receptor-dependent survival signals (2). The analysis of mouse mutants of the pre-B cell receptor-related signaling molecules SLP65 and Bruton's tyrosine kinase (BTK) demonstrated that SLP65 and BTK cooperate to suppress leukemic transformation (3).
Pre-B lymphoblastic leukemia cells typically exhibit a differentiation block at the pre-B cell stage of development (2); likewise, BTK deficiency in humans leading to X-linked agammaglobulinemia results in a breakdown of pre-B cell receptor signals and a differentiation block at the pre-B cell stage (4). To elucidate a possible role for BTK in leukemic transformation of human B cell precursors, we investigated BTK function in pre-B acute lymphoblastic leukemia cells.
Materials and Methods
Patient Samples, Cell Lines, and Cell Purification. Normal CD19+ μ-chain- pro-B cells and CD19+ VpreB+ pre-B cells were sorted from human bone marrow from four healthy donors (purchased from Cambrex, Baltimore) by using immunomagnetic beads against CD19 (Miltenyi Biotech, Bergisch Gladbach, Germany) and cell sorting using antibodies against CD19, VpreB (BD Biosciences, Heidelberg, Germany), and the μ-chain (Jackson ImmunoResearch). Similarly, CD5+ CD19+ B1 cells, IgD+ CD27- naïve B cells, CD19+ CD27+ memory B cells, and CD19+ CD138+ plasma cells were sorted from peripheral blood of four healthy donors by using antibodies against CD5, CD19, CD27, CD138, and IgD (BD Biosciences).
In total, 29 B cell precursor leukemias including 12 cell lines and 17 primary cases were studied. Eleven cases of B cell precursor leukemia with MLL-AF4 gene rearrangement [t (4, 11)(q21;q23)] including eight primary cases (I–VIII, Table 1, which is published as supporting information on the PNAS web site) and three cell lines (BEL1, RS4;11, and SEM) were analyzed. Eleven samples carrying a BCR-ABL1 gene rearrangement [t (9, 22)(q34;q11)] including seven primary cases (IX–XV, Table 1) and four cell lines (BV173, Nalm1, SD1, and SUP-B15) were studied. In addition, three leukemia cell lines carrying an E2A-PBX1 gene rearrangement [t (1, 19)(q23;p13); 697, Kasumi2, and MHH-CALL3], three cases of pre-B lymphoblastic leukemia with TEL-AML1 fusion gene [t (12, 21)(p12;q22)] including two primary cases (XVIII and XIX, Table 1), and the cell line REH and one pre-B lymphoblastic leukemia cell line harboring a TEL-PDGFRB gene rearrangement [Nalm6; t (5, 12)(q33.2;p13.2)] were studied. For all cases, fusion transcripts resulting from oncogenic gene rearrangements were detected by PCR as described (5). Clinical data for all primary cases were described previously (2).
Western Blotting. For the detection of tyrosine-phosphorylated BTK by Western blot, a phosphotyrosine-specific antibody against BTKY223 and EIF4E (Cell Signaling Technology, Beverly, MA; Santa Cruz Biotechnology) were used. Western blot experiments were carried out as described (6).
Inhibitors of BCR-ABL1 and BTK. For inhibition of BCR-ABL1 kinase activity, the antileukemic drug STI571 (Novartis, Basel) was used at a concentration of 10 μmol/liter. For inhibition of BTK, cells were incubated with α-cyano-β-hydroxy-β-methyl-N-(2,5-dibromophenyl) propenamide (LFM-A13; Calbiochem) at a concentration of 50 μg/ml for the times indicated.
Measurement of Pre-B Cell Receptor Responsiveness. Primary human bone marrow pre-B cells were enriched by using immunomagnetic MACS beads, as described (7). Pre-B lymphoblastic leukemia cells were cultured with 10% FCS in RPMI medium 1640 for the times and under the conditions indicated. After preincubation, cells were washed and stained with Fluo-3 dye (Calbiochem) for 30 min. Changes of cytosolic Ca2+ were measured by laser scans by using confocal microscopy (8, 9). After 10–30 sec of measurement, antibodies against human μ-chains (Jackson ImmunoResearch) were added to bone marrow pre-B cells or preincubated pre-B lymphoblastic leukemia cells (in the presence or absence of LFM-A13). Cytosolic Ca2+ concentrations were determined as described (8).
Flow Cytometry. Surface expression of IgM and CD19 on pre-B lymphoblastic leukemia cell lines and primary leukemia cells was monitored by using antibodies from BD Biosciences after the incubation times indicated. Preapoptotic or dead cells were identified by staining with annexin V and propidium iodide (BD Biosciences).
Sequence Analysis of BTK and Semiquantitative RT-PCR. In a search for BTK isoforms and somatic mutations of the BTK gene, BTK cDNA fragments covering the entire coding region or genomic DNA fragments of the BTK gene were amplified by using 5′-ATCCCAACAGAAAAAGAAAACAT-3′ (BTK exon 2 forward), 5′-GTTGCTTTCCTCCAAGATAAAAT-3′ (BTK exon 8 reverse), 5′-ATCTTGAAAAAGCCACTACCG-3′ (BTK exon 8 forward), 5′-TGATACGTCATTATGTTGTGTGTT-3′ (BTK exon 12 forward), 5′-TGATACGTCATTATGTTGTGTGTT-3′ (BTK exon 13 forward), 5′-ATCATGACTTTGGCTTCTTCAAT-3′ (BTK exon 14 reverse), 5′-CTCA A ATATCCAGTGTCTCAACA-3′ (BTK exon 14 forward), 5′-CTTTAACAACTCCTTGATCGTTT-3′ (BTK exon 17 reverse), and 5′-GGATTCTTCATCCATGACATCTA-3′ (BTK exon 19 reverse) were used. For normalization of cDNA and genomic DNA amounts, 5′-TTAGCACCCCTGGCCAAG-3′ (GA PDH forward), 5′-CTTACTCCTTGGAGGCCATG-3′ (GAPDH reverse), 5′-AACTACAAGACCGCCCCTTT-3′ (COX6B forward), and 5′-GCAGCCAGTTCAGATCTTCC-3′ (COX6B reverse) were used.
BTK amplification products were sequenced as described (6). Sequences of BTK isoforms are available from GenBank/EMBL (accession nos. AM051275–AM051286).
Retroviral Expression of a Kinase-Deficient BTK Splice Variant. All aberrant BTK splice variants identified lack a functional kinase domain (see Fig. 6, which is published as supporting information on the PNAS web site). Therefore, we generated a cDNA fragment of human BTK comprising the entire coding region but lacking the C-terminal portion of the kinase domain (exons 15–19; BTK-ΔK). After digestion with NotI (New England Biolabs), the PCR product was ligated into the retroviral S11IN expression vector (10). This vector is based on the retroviral plasmid SFβ11 (kindly provided by Christopher Baum, Hannover Medical School, Hannover, Germany) in which a multicloning site with NotI, EcoRI, and BamHI restriction sites was introduced, followed by an internal ribosome entry site (IRES) NEO cassette.
293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FCS (Invitrogen), 2 mM l-glutamine (Invitrogen), penicillin G (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen). 293T cells were cotransfected with 10 μg of the helper plasmid pHIT60, 10 μg of pczVSV-G envelope (11), and 10 μg of S11IN (as a control vector) or S11-BTK-ΔK-IN by using Fugene 6 (Roche, Basel), following the manufacturer's instructions. Both vectors are based on SF11 (10) with the 3′ LTR of the spleen focus-forming virus and internal ribosome entry site (IRES). Alternatively, the MIG-GFP (12) and MIG-BTK-IRES-GFP vectors were used for retroviral delivery of full-length BTK. For construction of the MIG-BTK-IRES-GFP vector, the cDNA encoding human full-length BTK was cloned into the MIG vector (12). Twenty-four hours after transfection, the medium was changed for Iscove's modified medium. Forty-eight hours after transfection, supernatants were filtered through a 0.45-μm filter and used to infect pre-B acute lymphoblastic leukemia cell lines on plates coated with 2 μg/cm2 retronectin (Takara Shuzo, Otsu, Japan; ref. 13). Plates were preloaded three times with fresh supernatant (14), and subsequently 2.5 × 105 cells were added to each well. Forty-eight hours after infection, cells expressing the S11IN vector were selected by using G418 to a final concentration of 0.5 mg/ml.
Cell Sorting. GFP-expressing cells or annexin V+ and annexin V- cells were sorted by using a FACStar 440 cell sorter. Cells were sorted under sterile conditions and either kept under cell culture conditions or subjected to RNA isolation and RT-PCR analysis.
Amplification of Double-Strand Recombination Signal Sequence Breaks by Ligation-Mediated PCR. Genomic DNA was isolated from ≈2.5 × 106 leukemia cells carrying retroviral expression vectors for either GFP alone or GFP and BTK and ligated to a blunt-end linker using T4 DNA ligase (Invitrogen) at 14°C overnight. The linker was constructed by annealing the oligonucleotides 5′-TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACATG-3′ and 3′ amino (C7)-GACGAGCTTAAGTTCGAAGATTGCTACATGCCCCT-5′, and protruding 3′ overhangs were removed by 3′→5′ exonuclease activity of the Klenow fragment of Escherichia coli DNA polymerase I (Invitrogen). Ligation-mediated PCR (15) was carried out with modifications as described (16). In two seminested rounds of amplification at an annealing temperature of 59°C, recombination signal sequence (RSS) intermediates with a DNA double-strand break at the 5′ heptamer of Jκ1 gene segments were amplified (see Fig. 7, which is published as supporting information on the PNAS web site) by using 5′-GTAATTAACATTCAGTCTACTTTC-3′ as external forward primer and 5′-TAACATTCAGTCTACTTTCTAAAA-3′ as internal forward primer together with 5′-TCCCCGTACATCGTTAGAAG–3′ as reverse primer specific for DNA-ligated linker molecules. To amplify RSS intermediates with a DNA double-strand break at the 5′ heptamer of Jλ7 gene segments, 5′-TTCTCACTTCTTCCATGGTGAC-3′ and 5′-ACTTCTTCCATGGTGACAGTCT-3′ were used in two rounds of PCR amplification as described above.
γ Radiation. Pre-B lymphoblastic leukemia cells were irradiated with 10-Gy γ radiation at the Clinic for Radiotherapy and Radiological Oncology, Heinrich-Heine-Universität. Twenty-four hours after radiation, leukemia cells were stained with annexin V and propidium iodide to separately sort preapoptotic or surviving cells.
Results and Discussion
Rare Occurrence of Somatic Mutations of the BTK Gene in B Cell Precursor Leukemia Cells. Pre-B lymphoblastic leukemia cells frequently exhibit defective expression of the pre-B cell receptor-related linker molecules SLP65 (1). Comparing single slp65-/- and slp65-/-/btk-/- double mutant mice, Btk was identified as a critical cofactor in preventing leukemic transformation of murine B cell precursors (3). We therefore searched for somatic mutations of the BTK gene in 12 cases of B cell precursor leukemia including four cases with MLL-AF4, three cases with E2A-PBX1, four cases with BCR-ABL1, and one case with a TEL-AML1 gene rearrangement. Because the majority of the BTK gene mutations in the germline leading to X-linked agammaglobulinemia have been found in the BTK kinase domain (see http://bioinf.uta.fi/BTKbase/BTKbasebrowser.html), we focused our analysis on this region using PCR primers for BTK exons 12–19. Deleterious somatic mutations of the BTK gene were found in 1 of 12 cases. In this case, the leukemia cells in a female patient carry an MLL-AF4 gene rearrangement and deleterious mutations of the BTK gene on both alleles (both X chromosomes). One BTK allele is inactivated by a mutation Lys→Stop at codon 420 (AAA→TAA), whereas the second allele harbors a frameshift mutation due to a 1-bp deletion in codon 386. In the 11 other cases studied, no clonal replacement mutations of the BTK gene were detected. This analysis does not exclude deleterious mutations in other regions of the BTK gene, yet it indicates that inactivation of the BTK gene by somatic mutation is rare in B cell precursor leukemia cells.
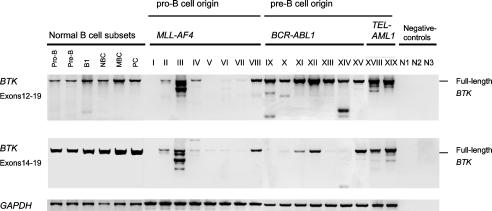
Expression of Kinase-Deficient Splice Variants of BTK in Pre-B Lymphoblastic Leukemia Cells. We next examined BTK mRNA expression in 29 human leukemias, including 12 cell lines and 17 primary cases. In a PCR strategy covering the entire coding region of BTK, we identified 14 aberrant mRNA splice variants (Table 1). As a control, BTK cDNA fragments were amplified from normal human pro- and pre-B cells (isolated from bone marrow) and mature B cell subsets isolated from peripheral blood (Fig. 1). At least one aberrant splice variant was coamplified with full-length BTK in 21 of 29 cases (Table 1). In only 4 of 29 cases, full-length BTK was exclusively expressed. Of note, absence of BTK expression was found in 4 of 29 cases, which all harbor an MLL-AF4 rearrangement. In the remaining cases of primary B lymphoid leukemia carrying an MLL-AF4 gene rearrangement, three exhibit very low expression of full-length BTK or high expression of aberrant splice variants, whereas one shows exclusive expression of full-length BTK (Fig. 1). In contrast, full-length BTK could be detected (not shown) in all three leukemia cell lines with an MLL-AF4 fusion gene (BEL1, RS4;11, SEM).
Fig. 1.
BTK mRNA expression in primary B cell precursor leukemia cells. Using PCR primers for exons 12, 14, and 19, two regions of the BTK mRNA were analyzed by RT-PCR in normal human pro-B cells, pre-B cells, B1 cells, naïve B cells (NBC), memory B cells (MBC), and plasma cells (PC), as well as in eight primary cases of B cell precursor leukemia with MLL-AF4 gene rearrangement, seven primary cases with BCR-ABL1 and two primary cases with TEL-AML1 fusion genes. As a loading control for all samples, a GAPDH cDNA fragment was amplified.
The aberrant BTK splice variants all have in common that they encode a truncated BTK kinase domain (Table 1 and Fig. 6). From in vitro kinase assays of BTK mutants derived from BTK-deficient X-linked agammaglobulinemia patients, it is known that truncation of the BTK kinase domain at amino acid 520 and even replacement mutations in the distal portion of the kinase domain result in a complete loss of BTK kinase activity (4). Hence, only full-length BTK but none of the truncated BTK splice variants identified here should exhibit BTK kinase activity (Table 1, Fig. 6). We conclude that BTK function is compromised at least in the B cell precursor leukemias that either express only kinase-deficient BTK or no BTK at all. In all these cases, the leukemia cells harbor an MLL-AF4 gene rearrangement.
In many leukemia cases with other gene rearrangements, however, concomitant expression of full-length BTK was also detected. Among the 14 splice variants, 3 were recurrently amplified, and two previously described variants of BTK (17) were also detected in this analysis. Although a splice variant lacking exon 15 was consistently and specifically amplified from leukemia cells carrying a BCR-ABL1 fusion gene (Table 1), the expression of other recurrent splice variants was not linked to a specific chromosomal rearrangement (Table 1). Of note, aberrant splicing of BTK in the leukemia cells not only leads to the expression of defective BTK transcripts. The splice mechanism itself also seems to be deranged as cryptic splice sites were frequently used (9 of 29 cases), and splice site slippage leading to small nucleotide insertions or deletions was observed in 4 of 29 cases (Table 1).
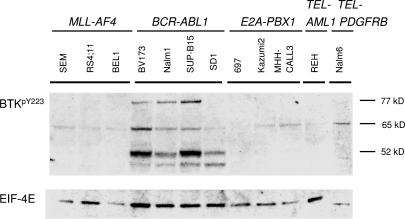
Because BTK activity is mainly regulated by tyrosine phosphorylation, we investigated the expression of tyrosine-phosphorylated BTK protein in B cell precursor leukemia cell lines carrying MLL-AF4, E2A-PBX1, BCR-ABL1, TEL-AML1, and TEL-PDGFRB gene rearrangements. Constitutive tyrosine phosphorylation of full-length BTK was observed only in BCR-ABL1+ pre-B lymphoblastic leukemia cells (three of the four cell lines tested; Fig. 2). Whereas BTK lacking exon 15 alone (52 kDa) was heavily phosphorylated specifically in BCR-ABL1+ pre-B lymphoblastic leukemia cells, another recurrent BTK variant lacking exons 15 and 16 (65 kDa) was also tyrosine-phosphorylated in other leukemias (Fig. 2).
Fig. 2.
Tyrosine phosphorylation of BTK and kinase-deficient splice variants in B cell precursor leukemia cells. Using an antibody against phosphorylated BTKY233, tyrosine-phosphorylation of BTK (77 kDa), and BTK isoforms was analyzed by Western blot in 12 B cell precursor leukemia cell lines. The defining gene rearrangements of the B cell precursor leukemia cases analyzed are indicated. Western analysis of EIF4E expression was used as a loading control.
Expression of Kinase-Deficient BTK Prevents Pre-B Cell Receptor-Driven Differentiation of B Cell Precursor Leukemia Cells. BTK kinase deficiency in humans leading to X-linked agammaglobulinemia results in a breakdown of pre-B cell receptor signals and a differentiation block at the pre-B cell stage (4). In normal pre-B cells, engagement of the pre-B cell receptor using a μ-chain-specific antibody (arrows, Fig. 3A Left) induces a strong Ca2+ signal, which is sensitive to BTK kinase inhibition by LFM-A13 (Fig. 3A Left).
Fig. 3.
Expression of kinase-deficient BTK suppresses pre-B cell receptor-dependent Ca2+ signals and differentiation. Normal pre-B cells were sorted from bone marrow of four healthy donors using antibodies against CD19 and Vpre-B. Sorted pre-B cells were incubated in cell culture medium in the presence or absence of the BTK kinase inhibitor LFM-A13 for 12 h (A Left). The pre-B lymphoblastic leukemia cell line 697 was transduced with a retroviral expression vector for kinase-deficient BTK (BTK-ΔK) or a control vector (A Right). Using an anti-μ-chain antibody (arrows), Ca2+ release in response to pre-B cell receptor engagement was measured by laser-scanning microscopy (A). To elucidate the function of kinase-deficient BTK splice variants (BTK-ΔK) on differentiation, BCR-ABL1+ pre-B lymphoblastic leukemia SUP-B15 cells were transduced with either a control vector or a BTK-ΔK expression vector. In both cases, SUP-B15 cells were induced to differentiate by inhibition of BCR-ABL1 kinase activity through 10 μmol/liter STI571 for 4 days. Thereafter, leukemia cells were stained by using antibodies against CD19 and IgM (B). To clarify the effect of BTK-ΔK expression on STI571-induced differentiation, outgrowth of differentiating subclones in the presence or abscence of BTK-ΔK expression was analyzed by flow cytometry (B). Dead cells were identified and excluded from analysis by propidium iodide uptake.
Because BTK kinase activity is required for the transduction of pre-B cell receptor-dependent Ca2+ signals, the kinase-deficient BTK splice variants (BTK-ΔK) expressed in pre-B lymphoblastic leukemia cells likely cannot contribute to pre-B cell receptor-dependent Ca2+ signals. However, kinase-deficient forms of BTK may also act as a linker in the signal transduction of B lymphocytes (6, 18). To investigate the function of BTK-ΔK, we expressed kinase-deficient BTK using a retroviral expression system in 697 cells that exhibit active pre-B cell receptor signaling (Fig. 3A Right). 697 cells transduced with a control vector responded by vigorous Ca2+ release upon pre-B cell receptor engagement. However, 697 cells carrying the BTK-ΔK expression vector entirely lost pre-B cell receptor responsiveness (Fig. 3A Right). We conclude that expression of BTK-ΔK suppresses Ca2+ signals in response to pre-B cell receptor stimulation.
We next tested whether BTK-ΔK also interferes with pre-B cell receptor-driven differentiation. As shown by us and others (16, 19), inhibition of oncogenic kinase activity in BCR-ABL1+ leukemia cells by STI571 induces differentiation in a minor population of cells that survive in the absence of BCR-ABL1 kinase activity. Therefore, STI571-induced differentiation in BCR-ABL1+ pre-B lymphoblastic leukemia cells was used as a model to study the effect of kinase-deficient BTK on differentiation. To this end, BCR-ABL1+ pre-B lymphoblastic leukemia cells were transduced either with a retroviral control vector or a retroviral expression vector for BTK-ΔK. Inhibition of BCR-ABL1 kinase activity in BCR-ABL1+ pre-B lymphoblastic leukemia cells carrying a control vector induced the outgrowth of a small fraction of differentiating IgM+ subclones (5% after 2 days; Fig. 3B). However, upon forced expression of BTK-ΔK, <0.5% of BCR-ABL1+ pre-B lymphoblastic leukemia cells differentiated into IgM+ subclones (Fig. 3B).
We conclude that BTK-ΔK can act as a dominant-negative form of BTK with respect to (i)Ca2+ release in response to pre-B cell receptor engagement and (ii) pre-B cell receptor-driven differentiation.
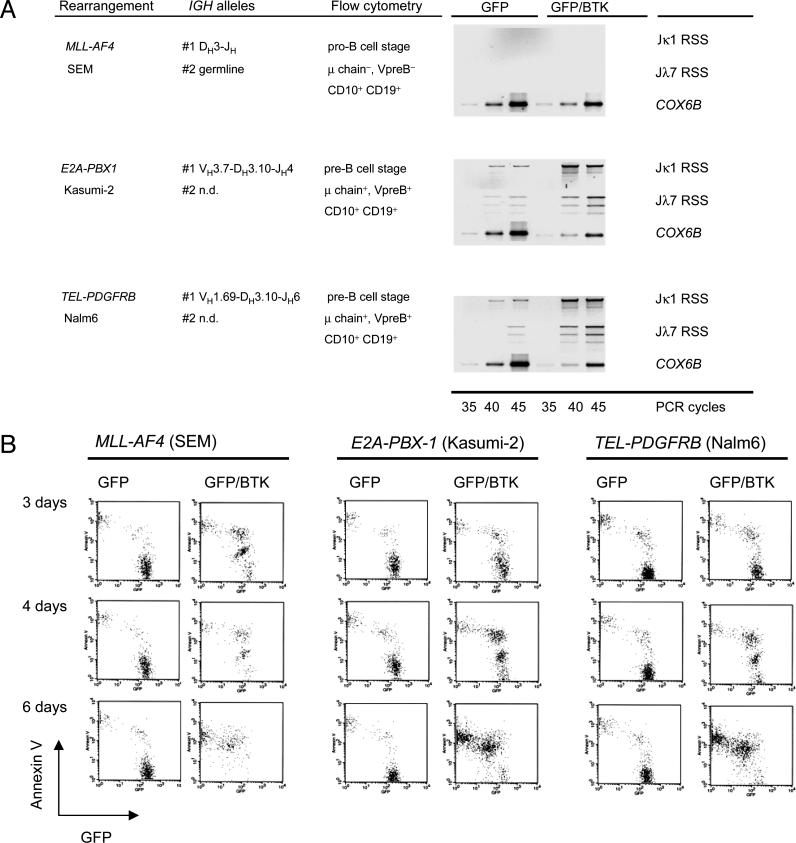
Reconstitution of Functional BTK Expression Induces Differentiation and Confers Propensity to Apoptosis in B Cell Precursor Leukemia Cells. Given that kinase-deficient BTK splice variants can act in a dominant-negative way, we next investigated whether induced expression of functional BTK can rescue normal B cell differentiation in B cell precursor leukemia cells. To this end, B cell precursor leukemia cells carrying an MLL-AF4, E2A-PBX1, or TEL-PGFRB gene rearrangement were transduced with a retroviral vector expressing functional BTK together with GFP or GFP alone as a control. GFP-expressing cells carrying either the control or the BTK expression vector were sorted and analyzed separately. We next tested whether BTK-transduced pre-B cell leukemia cells initiated Ig light-chain gene rearrangement. Rearrangement of Ig κ and λ light-chain genes and subsequent replacement of surrogate light chains with conventional κ or λ light chains represent a hallmark of pre-B cell differentiation into mature B cells. Genomic DNA was isolated from GFP+ or GFP+/BTK+ leukemia cells and subjected to ligation-mediated PCR for DNA double-strand break intermediates at RSS within the IGK or IGL loci (Fig. 7). Locus-specific DNA double-strand break intermediates were amplified at RSS flanking Jκ1 and Jλ7 gene segments, which would indicate ongoing Vκ-Jκ1 or Vλ-Jλ7 gene rearrangement, respectively. Although Jκ1-RSS and Jλ7-RSS breaks can be detected at low levels in two pre-B lymphoblastic leukemia cell lines transduced with the control vector, the frequency of Jκ1-RSS and Jλ7-RSS DNA double-strand breaks was increased in leukemia cells transduced with the BTK-IRES-GFP expression vector (Fig. 4A). This indicates that overexpression of functional BTK can partially relieve the differentiation block of acute lymphoblastic leukemia cells that were arrested at the pre-B cell stage. However, MLL-AF4+ acute lymphoblastic leukemia cells, which are arrested at the pro-B cell stage do not initiate Ig light-chain gene rearrangement in response to BTK overexpression. This might indicate that the initiation of Ig light-chain gene rearrangement requires expression not only of functional BTK but also of a functional pre-B cell receptor, which is present in pre-B lymphoblastic leukemia but missing in pro-B lymphoblastic leukemia cells carrying an MLL-AF4 gene rearrangement (Fig. 4A). Although reconstitution of BTK expression initiated Ig light-chain gene rearrangement in three of four cell lines tested, we could not detect κ or λ light chains on the surface of BTK-GFP-transduced leukemia cells (not shown). Attempting to establish permanent lines from the control GFP- or BTK-GFP-transduced leukemia cells, we noted that reconstitution of expression of functional BTK substantially increases sensitivity of the leukemia cells to apoptosis. Comparing acute lymphoblastic leukemia cells carrying control-GFP or BTK-GFP vectors, the BTK-transduced cells underwent apoptosis spontaneously after 6 days in cell culture (Fig. 4B).
Fig. 4.
Reconstitution of BTK expression in acute lymphoblastic leukemia cells initiates differentiation and induces propensity to apoptosis. Acute lymphoblastic leukemia cell lines carrying either an MLL-AF4 (SEM), E2A-PBX1 (Kasumi-2), or TEL-PDGFRB (Nalm6) gene rearrangement were characterized by sequence analysis of their IGH loci and flow cytometry. The leukemia cells were transduced either with a retroviral MIG_GFP or a MIG_BTK-IRES-GFP expression vector. GFP-expressing cells were sorted and genomic DNA was extracted from the sorted cells. Amounts of genomic DNA were normalized by amplification of a genomic fragment of the COX6B gene. Genomic DNA from cells transduced with the control MIG_GFP-vector or MIG_GFP-IRES-BTK was subjected to ligation with a double-stranded DNA-linker of known sequence. To detect DNA double-strand breaks within the IGK and IGL loci, linker-ligated DNA was subjected to ligation-mediated PCR by using linker-specific primers together with primers binding to recombination signal sequences flanking the Jκ1- (Jκ1 RSS) or the Jλ7-gene segment (Jλ7 RSS). Sorted GFP-expressing leukemia cells were also kept under cell culture conditions for 3, 4, and 6 days, and preapoptotic cells were labeled with annexin V. After 6 days, almost all BTK-transduced leukemia cells underwent apoptosis, whereas viability of leukemia cells transduced with the GFP-control vector remained unchanged (B).
Dominant-Negative BTK Splice Variants Can Protect Pre-B Lymphoblastic Leukemia Cells Against Radiation-Induced Apoptosis. BTK-mediated propensity to apoptosis was observed in MLL-AF4+, E2A-PBX1+, and TEL-PDFRB+ acute lymphoblastic leukemia cells irrespective of pre-B cell receptor expression (Fig. 4).
An earlier study invoked BTK kinase activity as a sensitizer to γ radiation-induced apoptosis in chicken DT-40 lymphoma cells (20). Therefore, kinase-deficient dominant-negative BTK splice variants may have a protective effect against γ radiation-induced apoptosis in human acute lymphoblastic leukemia cells. To test this hypothesis, we γ irradiated E2A-PBX1+ acute lymphoblastic leukemia cells with 10 Gy. Twenty-four hours after irradiation, >90% of the cells had already undergone apoptosis, as assessed by propidium iodide uptake. Among the remaining cells, we sorted annexin V+ preapoptotic cells and annexin V- viable cells and compared BTK splice variant expression between preapoptotic and surviving leukemia cells. In two different PCR approaches, the entire kinase domain (using primers for exons 12–19) and at a higher level of resolution, a hotspot region of aberrant splicing comprising exons 14–17 was amplified. Consistent with a protective effect against γ radiation-induced apoptosis, surviving leukemia cells exhibit preferential expression of kinase-deficient dominant-negative BTK splice variants (Fig. 5A). Sequence analysis of these splice variants revealed that some of the BTK splice variants identified here in the context of resistance to γ radiation-induced apoptosis were recurrently amplified from various leukemia subentities (see Table 1). However, additional BTK splice variants were also amplified (Fig. 5B), which again all have in common their lack of a functional kinase domain.
Fig. 5.
Acute lymphoblastic leukemia cells surviving γ radiation are selected for the expression of dominant-negative BTK isoforms. The pre-B lymphoblastic leukemia cell line Kasumi-2 carrying a E2A-PBX1 gene rearrangement were γ irradiated with 10 Gy. After 24 h, >90% of the leukemia cells were apoptotic. Among the remaining cells, annexin V+ preapoptotic cells and annexin V- surviving cells were sorted separately. Sorted cells were subjected to RT-PCR analysis for BTK splice variant expression, and cDNA amounts were normalized by amplification of a GAPDH cDNA fragment. Splice variants were detected by amplification of the entire BTK kinase domain by using primers for exons 12 and 19 (low resolution; A Top) or for exons 14 and 17 (high resolution, A Bottom). PCR products of this amplification were subjected to sequence analysis (B).
Conclusion
Taking these findings together, we conclude that somatic mutation of the BTK gene is rare in B cell precursor leukemia cells. However, in the majority of cases, the leukemia cells exhibit deranged expression of BTK. Although in four of eight primary cases with an MLL-AF4 gene rearrangement BTK expression is missing, B cell precursor leukemia cells typically exhibit coexpression of full-length BTK with kinase-deficient dominant-negative BTK splice variants. These dominant-negative BTK splice variants can interfere with pre-B cell receptor-dependent signal transduction, induce a differentiation block, and prevent Ig κ or λ light-chain gene rearrangement. Although overexpression of full-length BTK in B lymphoid leukemia cells induces propensity to apoptosis, expression of dominant-negative BTK splice variants can protect against radiation-induced apoptosis.
Supplementary Material
Acknowledgments
We thank Klaus Rajewsky and Martin Krönke for continuous support and discussions and Stefanie Jauch and Peter Wurst for excellent technical assistance. N.F. is supported by a fellowship from the German José Carreras Leukemia Foundation, and F.K. is supported by the Studienstiftung des deutschen Volkes and the Köln Fortune program of the Faculty of Medicine of the University of Cologne. M.M. is supported by the Deutsche Forschungsgemeinschaft through the Emmy Noether Program and through Grants MU1616/2-1, MU1616/3-1 (to M.M.), the German José Carreras Leukemia Foundation (grant to M.M.), the Ministry of Science and Research for North Rhine-Westphalia through the Stem Cell Network NRW (to M.M.), and the Deutsche Krebshilfe (through the grant Molecular Mechanisms of Malignant Lymphoma, to M.M.).
Author contributions: N.F., M.M., and F.K. designed research; N.F., P.R., B.N.B.S., S.L., M.S., and M.M. performed research; P.W., H.J., W.-K.H., and H.H. contributed new reagents/analytical tools; N.F., M.M., F.K., and J.D.R. analyzed data; and M.M. wrote the paper.
Abbreviations: BTK, Bruton's tyrosine kinase; RSS, recombination signal sequence; IRES, internal ribosome entry site, BTK-ΔK, kinase-deficient BTK isoform.
Data deposition: The sequences reported in this paper have been deposited in the GenBank/EMBL database (accession nos. AM051275–AM051286).
References
- 1.Jumaa, H., Bossaller, L., Portugal, K., Storch, B., Lotz, M., Flemming, A., Schrappe, M., Postila, V., Riikonen, P. & Pelkonen, J. (2003) Nature 423, 452-456. [DOI] [PubMed] [Google Scholar]
- 2.Klein, F., Feldhahn, N., Harder, L., Wang, H., Wartenberg, M., Hofmann, W. K., Wernet, P., Siebert, R. & Müschen, M. (2004) J. Exp. Med. 199, 673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersseboom, R., Middendorp, S., Dingjan, G. M., Dahlenborg, K., Reth, M., Jumaa, H. & Hendriks, R. W. (2003) J. Exp. Med. 198, 91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holinski-Feder, E., Weiss, M., Brandau, O., Jedele, K. B., Nore, B., Backesjo, C. M., Vihinen, M., Hubbard, S. R., Belohradsky, B. H. & Smith, C. I. (1998) Pediatrics 101, 276-284. [DOI] [PubMed] [Google Scholar]
- 5.van Dongen, J. J., Langerak, A. W., Bruggemann, M., Evans, P. A., Hummel, M., Lavender, F. L., Delabesse, E., Davi, F., Schuuring, E. & Garcia-Sanz, R. (2003) Leukemia 17, 2257-2317. [DOI] [PubMed] [Google Scholar]
- 6.Feldhahn, N., Klein, F., Mooster, J. L., Hadweh, P., Sprangers, M., Wartenberg, M., Bekhite, M. M., Hofmann, W. K., Herzog, S., Jumaa, H., et al. (2005) J. Exp. Med. 201, 1837-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müschen, M., Lee, S., Zhou, G., Feldhahn, N., Barath, V. S., Chen, J., Moers, C., Krönke, M., Rowley, J. D. & Wang, S. M. (2002) Proc. Natl. Acad. Sci. USA 99, 10014-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldhahn, N., Schwering, I., Lee, S., Wartenberg, M., Klein, F., Wang, H., Zhou, G., Wang, S. M., Rowley, J. D. & Hescheler, J. (2002) J. Exp. Med. 196, 1291-1305. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Klein, F., Feldhahn, N., Lee, S., Wang, H., Ciuffi, F., von Elstermann, M., Toribio, M. L., Sauer, H., Wartenberg, M., Barath, V. S., et al. (2003) Proc. Natl. Acad. Sci. USA. 100, 6747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildinger, M., Abel, K.L., Ostertag, W. & Baum, C. (1999) J. Virol. 73, 4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietschmann, T., Heinkelein, M., Heldmann, M., Zentgraf, H., Rethwilm, A. & Lindemann, D. (1999) J. Virol. 73, 2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pear, W. S., Miller, J. P., Xu, L., Pui, J. C., Soffer, B., Quackenbush, R. C., Pendergast, A. M., Bronson, R., Aster, J. C., Scott, M. L., et al. (1998) Blood 92, 3780-3792. [PubMed] [Google Scholar]
- 13.Hanenberg, H., Xiao, X. L., Dilloo, D., Hashino, K., Kato, I. & Williams, D. A. (1996) Nat. Med. 2, 876-882. [DOI] [PubMed] [Google Scholar]
- 14.Hanenberg, H., Hashino, K., Konishi, H., Hock, R. A., Kato, I. & Williams, D. A. (1997) Hum Gene Ther. 8, 2193-2206. [DOI] [PubMed] [Google Scholar]
- 15.Schlissel, M., Constantinescu, A., Morrow, T., Baxter, M. & Peng, A. (1993) Genes Dev. 7, 2520-2532. [DOI] [PubMed] [Google Scholar]
- 16.Klein, F., Feldhahn, N., Mooster, J. L., Sprangers, M., Hofmann, W. K., Wernet, P., Wartenberg, M. & Müschen, M. (2005) J. Immunol. 174, 367-375. [DOI] [PubMed] [Google Scholar]
- 17.Middendorp, S., Dingjan, G. M., Maas, A., Dahlenborg, K. & Hendriks, R. W. (2003) J. Immunol. 171, 5988-5996. [DOI] [PubMed] [Google Scholar]
- 18.Goodman, P. A., Wood, C. M., Vassilev, A. O., Mao, C. & Uckun, F. M. (2003) Leukemia Lymphoma 44, 1011-1008. [DOI] [PubMed] [Google Scholar]
- 19.Muljo, S. A. & Schlissel, M. S. (2003) Nat. Immunol. 4, 31-37. [DOI] [PubMed] [Google Scholar]
- 20.Uckun, F. M., Waddick, K. G., Mahajan, S., Jun, X., Takata, M., Bolen, J. & Kurosaki, T. (1996) Science 273, 1096-1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.