Abstract
The present study compared the recently developed multilocus sequence typing (MLST) approach with a well-established molecular typing technique, pulsed-field gel electrophoresis (PFGE), for subspecies differentiation of Enterococcus faecalis isolates. We sequenced intragenic regions of three E. faecalis antigen-encoding genes (ace, encoding a collagen and laminin adhesin; efaA, encoding an endocarditis antigen; and salA, encoding a cell wall associated antigen) and one housekeeping gene (pyrC) of 22 E. faecalis isolates chosen largely for their temporal and geographical diversity, but also including some outbreak isolates. MLST analysis of polymorphic regions of these four genes identified 13 distinct sequence types (STs) with different allelic profiles; the composite sequences generated from the four sequenced gene fragments of individual isolates showed 98.3 to 100% identity among the 22 isolates. We also found that the allelic profiles from two sequences, ace and salA, were sufficient to distinguish all 13 STs of this study. The 13 STs corresponded to 12 different PFGE types, with one previously designated PFGE clone (a widespread U.S. clone of β-lactamase-producing isolates) being classified into two highly related STs which differed at 2 of 2,894 bases, both in the same allele. MLST also confirmed the clonal relationships among the isolates of two other PFGE clonal groups, including vancomycin resistant isolates. Thus, this pilot study with representative E. faecalis isolates suggests that, similar to PFGE, the sequence-based typing method may be useful for differentiating isolates of E. faecalis to the subspecies level in addition to identifying outbreak isolates.
Enterococci, normal gut commensals, were recognized as a causative agent of endocarditis and urinary tract infections in ca. 1900 and have also been reported as a common cause of nosocomial infections since the 1970s (25, 26). Recently, accumulation of antibiotic resistances has made enterococcal infections a life-threatening clinical challenge, and thus, the methods that distinguish an outbreak from an endogenous strain have become important for designing strategies to prevent and control outbreaks (24, 26). A number of phenotypic or genotypic typing methods (including biochemical typing, serotyping, multilocus enzyme electrophoresis [MLEE], phage typing, insertion sequence element-based typing, pulsed-field gel electrophoresis [PFGE], restriction fragment length polymorphism [RFLP] analysis, ribotyping, repetitive sequence-based PCR, arbitrary primed PCR, and random amplification of polymorphic DNA) have been applied to the epidemiological investigations of Enterococcus faecalis (2-4, 6, 14, 18, 22, 28, 35, 40, 47, 48). These molecular and epidemiological studies have provided valuable information and clarified some misconceptions regarding E. faecalis infections, such as demonstrating that some E. faecalis infections are caused by nosocomial transmission of outbreak strains rather than arising from the patient's own prehospitalization intestinal flora. Among the known enterococcal molecular typing methods, PFGE has proven to be a highly reproducible and accurate typing method, which can distinguish clonal populations, and hence is considered a “gold standard” for subspecies discrimination of E. faecalis clinical isolates. However, the results obtained by PFGE are not readily transportable, making it difficult to compare results among different laboratories and thus limiting studies involving interlaboratory comparisons.
Microbial genome sequencing programs have provided enormous data for easy identification of species-specific genes. This, together with rapid automated DNA sequencing, has generated considerable interest in DNA sequence-based typing methods. A few recent studies have used variable intragenic sequences (flanked by highly conserved regions) of either a single gene or two genes and reported their usefulness in strain differentiation. These include the hsp65 gene (which encodes a 65-kDa heat shock protein) of Mycobacterium scrofulaceum, the porB gene (which encodes an outer membrane porin protein) of Neisseria meningitidis, and repeat regions of the spaA (which encodes protein A) as well as the coa (which encodes coagulase) genes of Staphylococcus aureus and the emm gene (which encodes an M protein) of Streptococcus pyogenes (1, 33, 38, 39, 41, 42, 44, 45).
Recently, multilocus sequence typing (MLST) was developed for identification of clonal complexes within bacterial populations and has been used successfully for molecular epidemiological analysis of N. meningitidis, Streptococcus pneumoniae, S. aureus, S. pyogenes, and Campylobacter jejuni (7, 9, 11, 12, 21). MLST typically characterizes isolates of bacteria using ∼400 to 500 bp of intragenic sequences of six to seven housekeeping genes or loci, and thus, MLST is similar in principle to MLEE, but with greater sensitivity due to its ability to detect neutral genetic variations. This method distinguishes strains based on the observed allelic variations in the nucleotide sequences of several loci, rather than the degree of sequence variation in any single gene or locus. MLST and other the DNA sequence-based typing methods have been suggested as offering advantage over other techniques because (i) the data are objective and readily comparable between laboratories, (ii) the data can be stored in a shared central database to provide a broader resource for epidemiological studies, and (iii) evolutionary genetic analyses can be performed. At the same time, DNA sequencing is expensive when compared to PFGE and considerable technical skill and knowledge in sequence analysis is critical for typing the isolates (32).
In the present study, we have evaluated the discriminatory ability of a sequence-based typing method and compared the results to those obtained with PFGE. Internal fragments of three antigen-encoding genes that were detected with E. faecalis-infected patient sera and an internal fragment of one pyrimidine biosynthesis gene were used for the MLST analysis. The MLST generated sequence types were found to be comparable to types or clones identified with PFGE.
MATERIALS AND METHODS
Bacterial isolates.
A total of 22 E. faecalis isolates from different geographic locations were chosen for this study, primarily because they had been previously studied by us using one or more other typing techniques (Table 1). A collection of eight β-lactamase-producing (Bla+) isolates that were recovered from five cities in the United States (four isolates from the same hospital) during an 11-year period (1981 through 1991), three additional Bla+ isolates from a single hospital in Argentina (isolated between June and September of 1989), and one Bla+ isolate from Lebanon (isolated in 1989) that were previously analyzed by PFGE and MLEE were included (27-29, 34, 48). Six other isolates were vancomycin resistant, including five clinical isolates from a single hospital (of which three isolates were part of a single outbreak) in the United States (5) and one nonclinical isolate collected from Spain isolated from chicken products (37). Two isolates with high-level aminoglycoside resistance were from Thailand (14, 28), and two commonly used laboratory isolates of E. faecalis OG1RF (ATCC 47077) and JH2-2 were also included in this study (14, 17, 22, 23, 30).
TABLE 1.
Sequence-based typing results for various E. faecalis isolates in this study
| Laboratory name (alternative designation[s])a | Clinical source; origin; yr of isolation or collection | Reference(s) | MLEE typeb | PFGE typeb | Sequence type (allelic profile for ace, efaA, pyrC, salA) |
|---|---|---|---|---|---|
| TX4002 (OG1RF, ATCC 47077) | Laboratory strain; <1978c | 14, 22, 48 | 25 | VIII | ST-1 (A, A, A, A) |
| TX2621def | Blood; Houston, Tex.; 1996 | 5 | HH6 | ST-2 (C, C, B, B) | |
| TX2619def | Urine; Houston, Tex., 1996 | 5 | HH9 | ST-3 (B, C, D, E) | |
| TX0855 (BE83) | Urine; Bangkok, Thailand; 1980 | 14, 28 | B-1 | ST-4 (E, F, A, A) | |
| TX2783 (B343)d | Chicken product; Logrono, Spain; 1998 | 37 | BR-1 | ST-5 (D, I, B, A) | |
| TX0614 (E228)gh | Urine; Richmond, Va.; 1990 | 22, 40, 48 | 20 | 5 | ST-6 (C, E, B, H) |
| TX0615 (E278)gh | Foot ulcer; Richmond, Va.; 1990 | 40, 48 | 20 | 5 | ST-6 (C, E, B, H) |
| TX0616 (E340)gh | Urine; Richmond, Va.; 1990 | 40, 48 | 20 | 5 | ST-6 (C, E, B, H) |
| TX0617 (E366)gh | Sputum; Richmond, Va.; 1991 | 40, 48 | 20 | 5 | ST-6 (C, E, B, H) |
| TX0638 (DEL)g | Wilmington, Del.; 1986 | 22, 29, 48 | 20 | 5 | ST-7 (C, E, B, I) |
| TX0669 (PENN, PA)g | Blood; Philadelphia, Pa.; 1983 | 29, 48 | 17 | 5 | ST-7 (C, E, B, I) |
| TX0921 (HH22)f | Houston, Tex.; 1981 | 22, 29, 48 | 20 | 5 | ST-7 (C, E, B, I) |
| TX4000 (JH2-2) | Laboratory strain; <1974c | 14, 22, 48 | 17 | VII | ST-8 (D, B, B, G) |
| TX0630 (HG6280)gi | Blood; Buenos Aires, Argentina; 1989 | 22, 27, 29, 48 | 16 | 19 | ST-9 (G, G, A, C) |
| TX0631 (HG10528)gi | Gastric fluid; Buenos Aires, Argentina; 1989 | 27, 48 | 16 | 19 | ST-9 (G, G, A, C) |
| TX0633 (HG9829)gi | Subdiaphragmatic abscess; Buenos Aires, Argentina; 1989 | 27, 48 | 16 | 19 | ST-9 (G, G, A, C) |
| TX0645 (Beirut)g | Blood; Beirut, Lebanon; 1989 | 29, 48 | 7 | 7 | ST-10 (F, D, E, D) |
| TX0860 (BE88) | Catheter tip; Bangkok, Thailand; 1980 | 14, 28, 48 | 18 | B-3 | ST-11 (G, H, C, F) |
| TX2486de | Urine; Houston, Tex.; 1994 | 5, 22 | 1 | ST-12 (H, C, B, B) | |
| TX2487ade | Urine; Houston, Tex.; 1994 | 5 | 1 | ST-12 (H, C, B, B) | |
| TX2490de | Urine; Houston, Tex.; 1994 | 5 | 1 | ST-12 (H, C, B, B) | |
| TX0635 (WH245)g | Urine; West Haven, Conn.; 1986 | 29, 34, 48 | 5 | 8 | ST-13 (I, G, A, D) |
As designated in previous studies.
The original MLEE and PFGE pattern names in the earlier publication(s) are used.
Isolated at unknown time prior to the year shown.
Vancomycin resistant isolate.
Isolates TX2619, TX2621, TX2486, TX2487a, and TX2490 were from the same hospital.
Isolates TX2619 and TX2621 were not tested for vancomycin resistance in the previous study (5) and hence were not reported as vancomycin-resistant strains.
β-Lactamase-producing isolate.
Isolates TX0614, TX0615, TX0616, and TX0617 were from the same hospital.
Isolates TX0630, TX0631, and TX0633 were from the same hospital.
PFGE.
PFGE was performed with some modifications of a previously described method (14, 28). Agarose plugs containing genomic DNA were digested with SmaI (Gibco, BRL, Gaithersburg, Md.), and electrophoresis was carried out using a clamped homogeneous electric field (CHEF-DRII device; Bio-Rad Laboratories, Richmond, Calif.), with ramped pulse times beginning with 5 s and ending with 45 s, at 200 V for 26 h. The gels were stained with ethidium bromide (0.4 μg/ml) for 30 min and photographed. The PFGE patterns were interpreted using the criteria suggested by Tenover et al. (46), with closely and possibly related patterns being designated as belonging to a single clone. PFGE pattern names that were presented in earlier publications are used here.
Genomic DNA isolation; PCR; and DNA sequencing of ace, efaA, pyrC, and salA.
Genomic DNA was extracted from E. faecalis isolates freshly streaked from freezer vials and cultured in brain heart infusion broth. Genomic DNA was isolated by the hexadecyltrimethyl ammonium bromide method as described previously (22). We chose internal regions of three E. faecalis antigen-encoding genes (ace, efaA, and salA) and one housekeeping gene (pyrC) for sequencing. These four loci were chosen primarily because all these genes were well studied in our laboratory (8, 19, 31, 43), and the sequence diversity of the region coding for the A domain of ace has been recognized in our earlier study (31). The open reading frame (ORF) sizes of the four chosen loci are listed in Table 2. The internal gene fragments of ace, which encode a collagen and laminin adhesin (959 bp [890 bp for one isolate due to a 69-bp deletion] [31, 36]); efaA, which encodes an endocarditis antigen (693 bp [20]); pyrC, which encodes a dihydroorotase (320 bp [19]); and salA, which encodes a cell wall-associated antigen (F. Teng, B. E. Murray, and G. M. Weinstock, unpublished data) (919 or 922 bp, due to a 3-bp in-frame deletion in some isolates) were amplified using the optimized buffer B (1× buffer: 60 mM Tris-HCl [pH 8.5], 15 mM ammonium sulfate, 2 mM MgCl2) obtained from Invitrogen (San Diego, Calif.). PCR was performed in volumes of 50 μl, with an initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min 30 s (72°C for 45 s for pyrC) and a final extension of 72°C for 7 min. The PCR primers used for amplification and sequencing of all four genes are listed in Table 2. Our initial attempts with ace amplification primers were unsuccessful for three strains, namely, TX2486, TX2487a, and TX2490, subsequently found to be due to variations in the ace gene at the reverse primer region. The ace amplicon for these strains was amplified using ace forward PCR primer (Table 2) and reverse primer with the sequence 5′-ATTTAATTTTTGAATTGGTTCACTAAGCAG-3′ (located at positions 1896 to 1867, relative to the start codon). The PCR amplicons were purified using the Wizard PCR DNA Cleanup System (Promega Corporation, Madison, Wis.). Sequencing of both strands of the amplified fragments was achieved using an Applied Biosystems Prism 377 automated DNA sequencer using the Taq Dye-Deoxy terminator method (PE Applied Biosystems, Foster City, Calif.). Sequences were assembled using SeqMan program of DNASTAR software (Lasergene, Madison, Wis.).
TABLE 2.
PCR and sequencing primers used in this study
| Gene | ORF size (bp) | Deoxynucleotide primera
|
Amplicon size (bp) | |||
|---|---|---|---|---|---|---|
| Function(s) | Type | Sequence (5′→3′) | Locationb | |||
| ace | 2,025c | PCR, sequencing | Forward | GAGCAAAAGTTCAATCGTTGAC | 99-120 | 1,003 |
| PCR, sequencing | Reversed | GTCTGTCTTTTCACTTGTTTCT | 1101-1080 | |||
| Sequencing | Forward | CCAAATTGAGCGAGACTATC | 510-529 | |||
| Sequencing | Forward | CACTTGCCGAGTTTGAGC | 719-736 | |||
| Sequencing | Reverse | GAGAACTATTGGTGATAAGCG | 424-404 | |||
| efaA | 924 | PCR, sequencing | Forward | CGTTAGCTGCTTGCGGGAATC | 47-67 | |
| PCR, sequencing | Reverse | CCATACTACGTTTATCGACAC | 781-761 | 735 | ||
| Sequencing | Forward | CCACAATATTTAACAAGTGCC | 364-384 | |||
| Sequencing | Forward | CAAACTACATGAGGAAGCC | 537-555 | |||
| Sequencing | Reverse | GTTTGTTCTTGACCGGCAC | 398-380 | |||
| pyrC | 1,284 | PCR, sequencing | Forward | CGGGTAGTAAAGCTGCTGC | 227-245 | 361 |
| PCR, sequencing | Reverse | CTTCTCCTTCATGCATCACAC | 586-566 | |||
| salA | 1,449 | PCR, sequencing | Forward | CATTAACAAGCGTAGCGTTG | 44-63 | 922 |
| PCR, sequencing | Reverse | GCCTTTTTCAGGAGTCGTTG | 1005-986 | |||
| Sequencing | Forward | GATGCTGTCTTAGATGCAG | 364-382 | |||
| Sequencing | Forward | GCACAACGTTCTGAAGAACAAG | 613-634 | |||
| Sequencing | Reverse | CACTTGGACATCGCGTGC | 339-322 | |||
ace, efaA, pyrC, and salA primers were designed from E. faecalis strain V583 database sequences (The Institute for Genomic Research).
Numbering for all the primers is given relative to the start codon (ATG for ace, efaA, and pyrC; TTG for salA) for the respective genes.
ace ORF size represented here is for E. faecalis strain V583 (36). ace gene size varies in different E. faecalis strains due to variation in the number of repeats of the B domain (31).
See text for the reverse primer used for the selected strains.
Sequence analysis.
Multiple sequence alignments of the 22 isolates for the four gene fragments (ace, efaA, pyrC, and salA) were done by the Jotun Hein method (16) using the MegAlign program of DNASTAR software. A 2,894-bp (for 10 strains), 2,891-bp (for 11 strains), 2,822-bp (for one strain) nucleotide composite sequence (derived from four concatenated gene fragments) was also aligned by the MegAlign program. Phylogenetic trees (cladograms) based on the matrix of pairwise sequence divergence were constructed using the MegAlign program.
Allele and sequence type (ST) assignment.
In order to identify the nucleotide variation in the above gene fragments, sequences from the different isolates were compared to the corresponding sequences in the well-studied E. faecalis strain OG1RF (30). Each gene sequence differing by one or more nucleotides was considered to be a different allele (no weight was given to the degree of sequence divergence between alleles, although the alleles that differed by a single nucleotide were denoted as single nucleotide variants [SNVs]), and the distinct allelic sequences were assigned an arbitrary letter designation in the order of the number of base pairs that varied with respect to the OG1RF sequence. The alleles at the four loci provided the allelic profile, which defined the ST for each isolate. Isolates showing identical allelic profiles in the four gene fragments were assigned to the same ST, and those with a difference in their allelic profile were assigned to a different ST. The relatedness among the strains was analyzed by constructing a dendrogram based on the matrix of pairwise differences in the allelic sequences by the unweighted pair group method with arithmetic averages (UPGMA) method.
RESULTS AND DISCUSSION
Among known phylogenetic typing methods, PFGE and MLST, which are based on multiple sites and loci scattered around the chromosome, have consistently been shown to be capable of discriminating isolates at the subspecies level, although in different ways. PFGE is particularly useful for distinguishing strains circulating within a geographical location (microvariation) and is based on the selected variable regions of the genome (9, 15). MLST is based on variations that accumulate slowly and appears more suitable for long-term and global epidemiology (macrovariation) (9, 10, 15). However, in both methods, the relative rates of nucleotide substitutions and/or recombination occurring in nature set the limitations of phylogenetic analysis, and there are insufficient experiential data to establish these rates for enterococci.
Choice of loci.
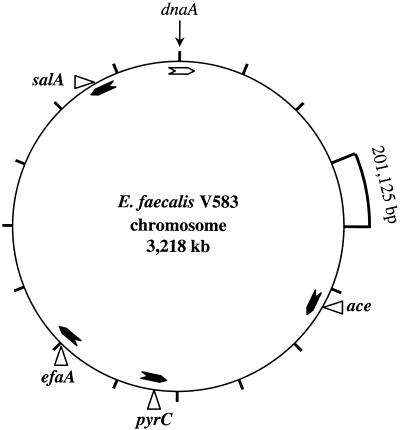
Since it is impractical to sequence large or multiple regions of the chromosome, we chose loci we had previously studied (ace, efaA, pyrC, and salA) and that had been shown to be present in all E. faecalis strains tested; indeed, hybridization with ace and efaA probes had proven useful in differentiating E. faecalis from other species (8, 43). On the basis of the E. faecalis V583 complete genome sequence (available at http://www.tigr.org [The Institute for Genomic Research, Rockville, Md.]), we derived the physical map shown in Fig. 1, which illustrates that the loci chosen were spread around the chromosome and the distance between any two loci is ranged from 328 to 1,355 kb.
FIG. 1.
Chromosomal locations of the sequenced loci. The locations are marked on a constructed physical map of E. faecalis V583 using the complete genome sequence (available at http://www.tigr.org/ [The Institute for Genomic Research]). The 3,218-kb genome is divided into 16 segments, with each segment representing 201,125 bases. The ORF coding for chromosomal replication factor (dnaA) was identified based on identity with dnaA sequences of S. pyogenes and S. aureus and positioned at nucleotide one. The arrowheads inside the circle represent the ORF orientations of the four loci and are marked at the respective positions.
PFGE fingerprinting.
PFGE with SmaI yielded <20 visible bands for each isolate (data not shown). As per the recommendations described by Tenover et al. (46), including closely and possibly related patterns, a total of 12 PFGE types, 3 of which contained three or more isolates, were recognized. Seven of the twelve Bla+ isolates were classified as belonging to the same clonal group (PFGE pattern 5); two of these strains showed an identical pattern (TX0614 and TX0615), while others (TX0616, TX0617, TX0638, TX0669, and TX0921) showed patterns with three to six band differences, depending on which two isolates were being compared. Among three Argentinean Bla+ isolates, TX0630 and TX0633 showed an identical PFGE pattern, while TX0631 showed a pattern almost identical to these two (classified as PFGE pattern 19) (27). Similarly, three of the five vancomycin-resistant Houston isolates were classified as clonally related (PFGE pattern 1). These results are in agreement with our earlier studies (5, 14, 18, 22, 28, 29, 37, 48).
MLST typing.
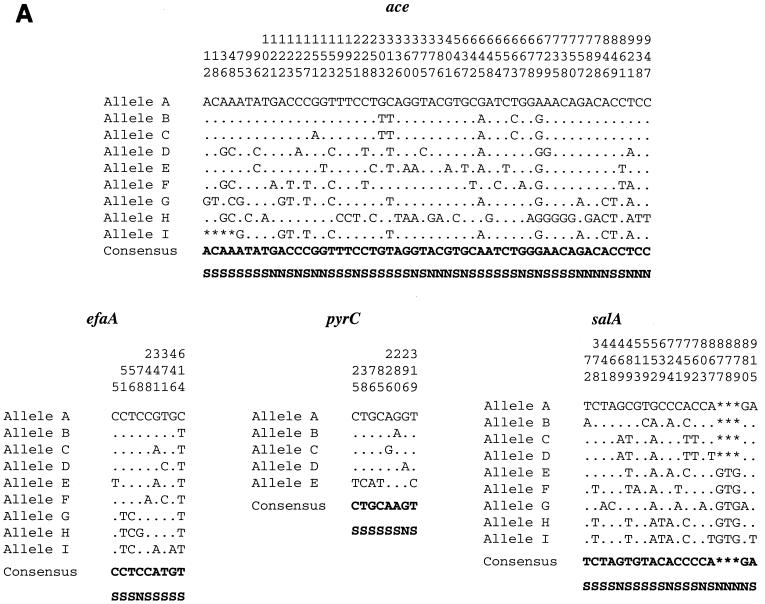
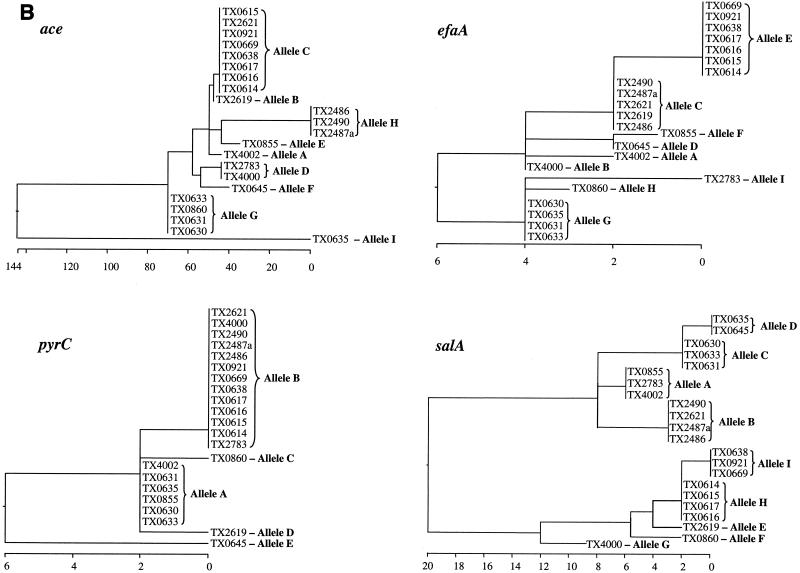
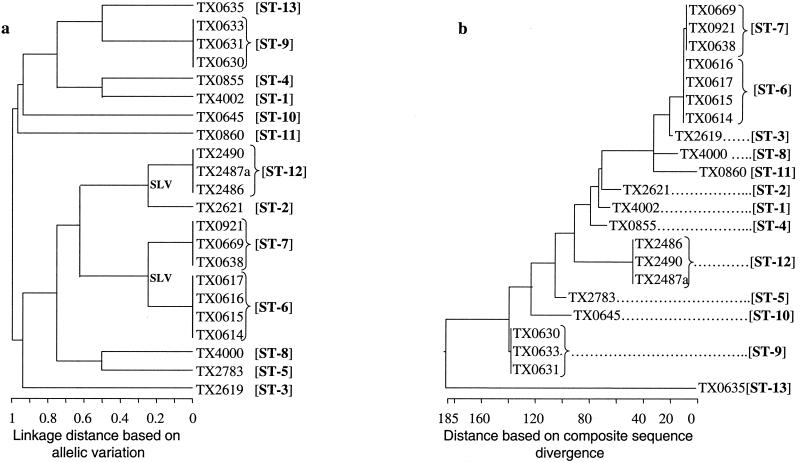
The length of the loci used for allele assignment in our MLST scheme was between 959 bp (ace) and 320 bp (pyrC). The total length of the four sequenced loci of this study approximates the total length of six loci common to several other MLST studies. Variable sites of different alleles of the four sequenced loci are presented in Fig. 2A, and the phylogenetic trees based on the matrix of pairwise divergence in these sequences are shown in Fig. 2B. Nine unique alleles were identified for three of the loci, while only five alleles were identified for the pyrC locus, likely because of the smaller region evaluated (Table 3; Fig. 2). Allelic profiles of the four loci identified 13 STs among the 22 isolates (Table 1). A dendrogram drawn from allelic profiles by the UPGMA method demonstrated four (ST-6, ST-7, ST-9, and ST-12) of the 13 STs contain more than one isolate (Fig. 3a). ST-6 and ST-7 as well as ST-2 and ST-12 differed by variation at a single locus.
FIG. 2.
Allelic variation of the four sequenced loci. (A) Variable sites identified in all the four gene fragments. The nucleotide sites that are identical in all the alleles are not shown. The nucleotides present in each of the variable sites of allele A (E. faecalis OG1RF) are shown. Only those sites that differ are shown for all the other alleles. The position of each variable site within the sequenced fragment is shown in the numbers above the nucleotide, read vertically. The consensus sequence is shown on the bottom. The variations that are synonymous (S) and nonsynonymous (N) are also shown. (B) Cladograms of the four loci sequenced. Phylogenetic trees were based on the matrix of pairwise sequence divergence in the sequences (generated by the Jotun Hein method of the DNASTAR software package). The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events.
TABLE 3.
Genetic variation in four sequenced loci
| Gene | Fragment size (bp) | No. of alleles | No. of variable sites | No. of synonymousa base substitutions | SNVb alleles |
|---|---|---|---|---|---|
| ace | 959c | 9 | 54 | 34 | B, C |
| efaA | 693 | 9 | 9 | 8 | A, B; B, C, D; C, E; D, F; G, H |
| pyrC | 320 | 5 | 8d | 7 | A, B, C, D |
| salA | 919, 922e | 9 | 21f | 14 | C, D |
Synonymous base substitutions are nucleotide changes which did not result in amino acid change.
SNVs are alleles that differ by a single nucleotide.
In strain TX0635, a 69-nt in-frame deletion was observed.
Five of eight variable sites were observed in a single allele.
A 3-bp in-frame deletion is observed in 12 of 22 strains.
Of 21 variable sites, 18 nt are base substitutions and the remaining 3 nt are deletions.
FIG. 3.
Phylogenetic trees of genetic relationships among 22 E. faecalis strains based on sequences of four gene fragments. (a) The dendrogram is based on the matrix of pairwise differences in the allelic sequences as determined by the UPGMA method. Linkage distances are indicated by scale at the bottom. (b) The cladogram is based on the matrix of pairwise sequence divergence in the concatenated composite sequences (generated by the Jotun Hein method of the DNASTAR software package). The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events.
The percentage of variable sites in a given locus ranged from 1.3 (efaA) to 5.6 (ace). A 69-bp in-frame deletion was detected in the ace locus of a Bla+ isolate (allele I) from Connecticut. Eight of nine alleles of the efaA locus differed by a single nucleotide, and they were designated SNV alleles (Table 3). Similarly, four of the five alleles of the pyrC locus are SNV alleles, and the fifth allele has five nucleotide changes (Bla+ isolate from Lebanon). In the salA locus, a 3-bp in-frame deletion was observed in 55% of the isolates. The nucleotide changes which alter the amino acid sequence are indicated as nonsynonymous base substitutions and the nucleotide changes which do not alter the amino acid sequence are indicated as synonymous base substitutions (Fig. 2A).
Composite sequence-based typing.
In order to determine the overall divergence of the sequenced gene fragments of the four loci studied, these sequences were spliced together to obtain a concatenated composite sequence for each of the isolates. For calculating the percentage of identity or divergence, in-frame insertions or deletions were not taken into account. A cladogram created from the matrix of pairwise sequence divergence of composite sequences identified 13 phylogenetic lineages (Fig. 3b), and these were identical to the 13 STs obtained from the allelic profiles. The identity between the 22 composite sequences was found to be between 98.3 and 100%. The composite sequence of ST-6 isolates from Richmond, Va. (TX0614, TX0615, TX0616, and TX0617, which showed 100% identity among the four isolates) have 99.93% identity (2,892 of 2,894 bases) with the composite sequence of ST-7 isolates from three different states (TX0638, TX0669, and TX0921, which were in turn 100% identical to each other).
Congruence between PFGE, MLST, and composite sequence-based typing methods.
Analysis of the clusters in the phylogenetic trees generated by the methods described above identified 13 different genotypes versus 12 different PFGE types among the 22 E. faecalis isolates tested. Both sequence-based typing methods, i.e., MLST with four loci as well as the composite sequence alignment, confirmed the clonal relationships among isolates of PFGE patterns 1, 5, and 19. TX0921 (Bla+) obtained from Texas in 1981 showed the identical MLST type as the Pennsylvania Bla+ strain (TX0669) isolated in 1983 and also strain TX0638 (Bla+) isolated in Delaware in 1986, confirming our previous finding of widespread dispersion of this clone in the United States (29). Sequence-based typing further confirmed that the three Bla+ clones from South America and a single isolate from Lebanon as well as the single isolate from Connecticut were different from each other.
Although there was an agreement among all three analyses, one apparent exception was the recognition of two different STs (containing multiple isolates) by sequence-based typing among isolates characterized as belonging to PFGE pattern 5. As pointed out earlier, four of these seven Bla+ PFGE type 5 isolates (ST-6) obtained from Richmond, Va., in 1990 to 1991 are identical at all four loci, while the other three PFGE type 5 Bla+ isolates (isolated 5 to 10 years earlier in different states) were classified as ST-7, which differs only by variation of two synonymous bases at the salA locus (i.e., these are single-locus variants [SLVs]); all other isolates differed from these Bla+ isolates by two or more bases at this locus, as well as differing at other loci. While most MLST studies that deal with six or more loci would have clustered these single locus variants as a single clonal group, we did not do so because of the smaller number of loci studied. The ST-6 and ST-7 group of Bla+ isolates has previously been considered clonally related based on PFGE and repetitive sequence-based PCR (22). This difference may be related to the use of a somewhat broader assignment of PFGE types (i.e., up to even six fragment differences were seen, depending on which two isolates were compared), while counting even a single base change in assigning the alleles of MLST.
Among other possibly related STs, three vancomycin-resistant outbreak isolates (ST-12) from Houston, Tex. (1994) are SLVs of another Houston vancomycin resistant 1996 isolate (ST-2), the only other SLV of this study. However, the ST-2 isolate has only 98.9% overall composite sequence identity with ST-12 isolates due to 33 scattered base changes in the ace locus. These vancomycin-resistant SLVs also show a relatively similar PFGE pattern when compared to the other isolates of this study. This, together with the identity at three of the four loci, raises the possibility that the extensive differences in ace may be due to horizontal exchange, as has been suggested to occur during conjugative transposition (49).
The other possibly related composite sequences were those of the Connecticut Bla+ ST-13 isolate and the South American Bla+ ST-9 isolates; these differed by a 69-bp deletion at the ace locus and a single base change at the salA locus. However, the PFGE patterns of these two STs differed substantially.
Results from a prior study of MLEE found general agreement for most isolates tested by both PFGE and MLEE (Table 1). The MLEE study assigned the same ET to six of seven Bla+ isolates (all ST-6 isolates and two of three ST-7 isolates of this study), consistent with the PFGE pattern assignments. However, one ST-7 Bla+ isolate (TX0669, also PFGE type 5) was assigned a different ET, which was also the ET of Chile isolates in that study as well as another isolate JH2-2 (TX4000), which differed by PFGE (type VII) and MLST (ST-8). These various results, taken together, suggest the possibility of cross-contamination in the MLEE study and misassignment of the ET type of TX0669.
The phylogenetic trees of both the MLST (linkage distance derived from allelic variation [Fig. 3a]) and the composite sequence alignment (linkage distance derived from sequence divergence [Fig. 3b]) confirmed the similar clustering of clonal and related isolates, in addition to differentiating the nonrelated (as defined by other techniques) isolates. However, an apparent difference was observed in interrelationships of possibly related isolates such as ST-2 and ST-12 that differed extensively at a single locus. The difference in the two schemes is related to the different criteria that were used to generate linkage distances, i.e., in MLST, for a given locus, equal weight was given for an allele with a single base change or multiple base changes, while in the composite sequence-based typing weight was given to every base change. At this stage, we are not able to draw any conclusion on the epidemiological significance of standard MLST approach versus the composite sequence approach due to our inadequate knowledge on relative rates of mutations/recombinations in enterococci.
Evaluation of minimum number of loci needed for subtyping.
We also examined the possibility that the sequence variation of fewer loci might be adequate for subspecies typing. The allelic profiles from two antigenic genes, namely, ace and salA, were found to be sufficient to distinguish all 13 STs. Similarly, the allelic profiles from a second pair of genes, ace and efaA, distinguished the 12 STs that match with the 12 PFGE types. These results indicate that the combined mutation rates of the ace, efaA, and salA polymorphic regions are comparable to the overall chromosome mutation rates that were detected by PFGE. Strains with different ace genes may have the same salA or efaA genes, or vice versa, as a result of differences in the rates of selection pressure or horizontal recombination as suggested for por and opa genes of Neisseria gonorrhoeae (50). Similar to this idea, others have also applied sequence typing to one or two genes and successfully differentiated most isolates of N. meningitidis, N. gonorrhoeae, S. aureus, and S. pyogenes (1, 33, 38, 39, 41, 42, 45, 50). A recent MLST study also used an antigen-encoding gene, in addition to housekeeping genes, to distinguish a meningococcal outbreak (13).
We also identified the most polymorphic regions by examining the distribution of variable sites in the sequenced fragments of all four loci. Although the variations in ace are distributed evenly in the complete sequenced region, omission of 200 bases at the 3′ end would not have affected the number of ace alleles identified. Designing the ace reverse PCR primer in the region between bp 910 and 880 (with reference to the start codon) would allow amplification of a PCR product in all 22 strains, thus resolving the problem of using different reverse amplification primers for some isolates (see Materials and Methods). Similarly, the first 377 bp of the sequenced region of salA has only one nucleotide variation, and omission of this 377-bp salA sequence did not affect the allele profile of this study.
In summary, we studied both micro- and macrovariation of E. faecalis isolates by sequencing three antigen-encoding genes and one housekeeping gene. Our results demonstrated that this sequence-based typing method was comparable to PFGE typing in differentiating E. faecalis at the subspecies level, including identification of outbreak isolates. DNA sequencing of ace and salA gene fragments appears to be as efficient as sequencing of all four genes for distinguishing isolates included in this study.
Acknowledgments
This work was supported by NIH grant AI47923 from the Division of Microbiology and Infectious Diseases to B. E. Murray.
REFERENCES
- 1.Beall, B., R. Facklam, T. Hoenes, and B. Schwartz. 1997. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J. Clin. Microbiol. 35:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caprioli, T., F. Zaccour, and S. S. Kasatiya. 1975. Phage typing scheme for group D streptococci isolated from human urogenital tract. J. Clin. Microbiol. 2:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiew, Y. F., and L. M. Hall. 1998. Comparison of three methods for the molecular typing of Singapore isolates of enterococci with high-level aminoglycoside resistances. J. Hosp. Infect. 38:223-230. [DOI] [PubMed] [Google Scholar]
- 4.Cocconcelli, P. S., D. Porro, S. Galandini, and L. Senini. 1995. Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett. Appl. Microbiol. 21:376-379. [DOI] [PubMed] [Google Scholar]
- 5.Coque, T. M., J. F. Tomayko, S. C. Ricke, P. C. Okhuysen, and B. E. Murray. 1996. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob. Agents Chemother. 40:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Descheemaeker, P., C. Lammens, B. Pot, P. Vandamme, and H. Goossens. 1997. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int. J. Syst. Bacteriol. 47:555-561. [DOI] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duh, R. W., K. V. Singh, K. Malathum, and B. E. Murray. 2001. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb. Drug Resist. 7:39-46. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordillo, M. E., K. V. Singh, and B. E. Murray. 1993. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J. Clin. Microbiol. 31:1570-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulding, J. N., J. V. Hookey, J. Stanley, W. Olver, K. R. Neal, D. A. A. Ala'Aldeen, and C. Arnold. 2000. Fluorescent amplified-fragment length polymorphism genotyping of Neisseria meningitidis identifies clones associated with invasive disease. J. Clin. Microbiol. 38:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hein, J. 1990. Unified approach to alignment and phylogenies. Methods Enzymol. 183:626-645. [DOI] [PubMed] [Google Scholar]
- 17.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn, I., L. G. Burman, S. Haeggman, K. Tullus, and B. E. Murray. 1995. Biochemical fingerprinting compared with ribotyping and pulsed-field gel electrophoresis of DNA for epidemiological typing of enterococci. J. Clin. Microbiol. 33:2812-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X., G. M. Weinstock, and B. E. Murray. 1995. Generation of auxotrophic mutants of Enterococcus faecalis. J. Bacteriol. 177:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe, A. M., P. A. Lambert, and A. W. Smith. 1995. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect. Immun. 63:703-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malathum, K., K. V. Singh, G. M. Weinstock, and B. E. Murray. 1998. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J. Clin. Microbiol. 36:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda, A. G., K. V. Singh, and B. E. Murray. 1992. Determination of the chromosomal size of three different strains of Enterococcus faecalis and one strain of Enterococcus faecium. DNA Cell Biol. 11:331-335. [DOI] [PubMed] [Google Scholar]
- 24.Mundy, L. M., D. F. Sahm, and M. Gilmore. 2000. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 27.Murray, B. E., H. A. Lopardo, E. A. Rubeglio, M. Frosolono, and K. V. Singh. 1992. Intrahospital spread of a single gentamicin-resistant, beta-lactamase-producing strain of Enterococcus faecalis in Argentina. Antimicrob. Agents Chemother. 36:230-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, B. E., K. V. Singh, S. M. Markowitz, H. A. Lopardo, J. E. Patterson, M. J. Zervos, E. Rubeglio, G. M. Eliopoulos, L. B. Rice, F. W. Goldstein, S. G. Jenkins, G. M. Caputo, N. Nasnas, L. S. Moore, E. S. Wong, and G. Weinstock. 1991. Evidence for clonal spread of a single strain of beta-lactamase-producing Enterococcus (Streptococcus) faecalis to six hospitals in five states. J. Infect. Dis. 163:780-785. [DOI] [PubMed] [Google Scholar]
- 30.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nallapareddy, S. R., K. V. Singh, R.-W. Duh, G. M. Weinstock, and B. E. Murray. 2000. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect. Immun. 68:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., I. Crisostomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-de-Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson, J. E., K. V. Singh, and B. E. Murray. 1991. Epidemiology of an endemic strain of beta-lactamase-producing Enterococcus faecalis. J. Clin. Microbiol. 29:2513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pryce, T. M., R. D. Wilson, and J. K. Kulski. 1999. Identification of enterococci by ribotyping with horseradish-peroxidase-labelled 16S rDNA probes. J. Microbiol. Methods 36:147-155. [DOI] [PubMed] [Google Scholar]
- 36.Rich, R. L., B. Kreikemeyer, R. T. Owens, S. LaBrenz, S. V. Narayana, G. M. Weinstock, B. E. Murray, and M. Hook. 1999. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 274:26939-26945. [DOI] [PubMed] [Google Scholar]
- 37.Robredo, B., C. Torres, K. V. Singh, and B. E. Murray. 2000. Molecular analysis of Tn1546 in vanA-containing Enterococcus spp. isolated from humans and poultry. Antimicrob. Agents Chemother. 44:2588-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacchi, C. T., A. P. Lemos, A. M. Whitney, C. A. Solari, M. E. Brandt, C. E. Melles, C. E. Frasch, and L. W. Mayer. 1998. Correlation between serological and sequencing analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin. Diagn. Lab. Immunol. 5:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders, N. A., G. Hallas, E. T. Gaworzewska, L. Metherell, A. Efstratiou, J. V. Hookey, and R. C. George. 1997. PCR-enzyme-linked immunosorbent assay and sequencing as an alternative to serology for M-antigen typing of Streptococcus pyogenes. J. Clin. Microbiol. 35:2689-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seetulsingh, P. S., J. F. Tomayko, P. E. Coudron, S. M. Markowitz, C. Skinner, K. V. Singh, and B. E. Murray. 1996. Chromosomal DNA restriction endonuclease digestion patterns of beta-lactamase-producing Enterococcus faecalis isolates collected from a single hospital over a 7-year period. J. Clin. Microbiol. 34:1892-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shopsin, B., M. Gomez, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 38:3453-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, K. V., T. M. Coque, G. M. Weinstock, and B. E. Murray. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323-331. [DOI] [PubMed] [Google Scholar]
- 44.Swanson, D. S., X. Pan, and J. M. Musser. 1996. Identification and subspecific differentiation of Mycobacterium scrofulaceum by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. J. Clin. Microbiol. 34:3151-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, Y. W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorisdottir, A. S., L. L. Carias, S. H. Marshall, M. Green, M. J. Zervos, C. Giorgio, L. A. Mermel, J. M. Boyce, A. A. Medeiros, H. Fraimow, and L. B. Rice. 1994. IS6770, an enterococcal insertion-like sequence useful for determining the clonal relationship of clinical enterococcal isolates. J. Infect. Dis. 170:1539-1548. [DOI] [PubMed] [Google Scholar]
- 48.Tomayko, J. F., and B. E. Murray. 1995. Analysis of Enterococcus faecalis isolates from intercontinental sources by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres, O. R., R. Z. Korman, S. A. Zahler, and G. M. Dunny. 1991. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol. Gen. Genet. 225:395-400. [DOI] [PubMed] [Google Scholar]
- 50.Viscidi, R. P., J. C. Demma, J. Gu, and J. Zenilman. 2000. Comparison of sequencing of the por gene and typing of the opa gene for discrimination of Neisseria gonorrhoeae strains from sexual contacts. J. Clin. Microbiol. 38:4430-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]