Abstract
The Nicotiana sylvestris mutant, CMS, lacks the mitochondrial gene nad7 and functional complex I, and respires using low-affinity NADH (alternative) mitochondrial dehydrogenases. Here, we show that this adjustment of respiratory pathways is associated with a profound modification of foliar carbon-nitrogen balance. CMS leaves are characterized by abundant amino acids compared to either wild-type plants or CMS in which complex I function has been restored by nuclear transformation with the nad7 cDNA. The metabolite profile of CMS leaves is enriched in amino acids with low carbon/nitrogen and depleted in starch and 2-oxoglutarate. Deficiency in 2-oxoglutarate occurred despite increased citrate and malate and higher capacity of key anaplerotic enzymes, notably the mitochondrial NAD-dependent isocitrate dehydrogenase. The accumulation of nitrogen-rich amino acids was not accompanied by increased expression of enzymes involved in nitrogen assimilation. Partitioning of 15N-nitrate into soluble amines was enhanced in CMS leaf discs compared to wild-type discs, especially in the dark. Analysis of pyridine nucleotides showed that both NAD and NADH were increased by 2-fold in CMS leaves. The growth retardation of CMS relative to the wild type was highly dependent on photoperiod, but at all photoperiod regimes the link between high contents of amino acids and NADH was observed. Together, the data provide strong evidence that (1) NADH availability is a critical factor in influencing the rate of nitrate assimilation and that (2) NAD status plays a crucial role in coordinating ammonia assimilation with the anaplerotic production of carbon skeletons.
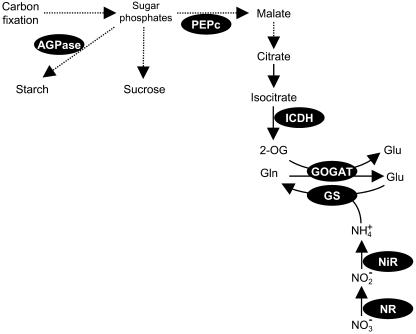
Nitrogen is often a limiting factor for plant growth and development. There is keen interest and considerable potential agronomic benefit in understanding the mechanisms that determine nitrogen use efficiency and in identifying targets for improvement. Hence, much attention has focused on the regulation of key enzymes involved in the core interaction between carbon and nitrogen metabolism (Fig. 1; Kaiser and Huber, 1994; Coschigano et al., 1998; Hirel and Lea, 2002; Hodges, 2002; Stitt et al., 2002). Numerous studies have shown that carbon and nitrogen metabolites are monitored by the cell and act together to orchestrate gene expression, thus determining transcriptome profiles that are appropriate to nutritional and metabolic status (Wang et al., 2000; Palenchar et al., 2004). Other key controls occur posttranslationally via phosphorylation of proteins such as phosphoenolpyruvate carboxylase (PEPc) and nitrate reductase (NR; Van Quy et al., 1991; Kaiser and Huber, 1994), as well as by intricate regulation of enzyme activities by metabolite effectors (Moraes and Plaxton, 2000; Smith et al., 2000).
Figure 1.
Central reactions in the integration of nitrogen assimilation and carbon metabolism. Key enzymes are shown by black ellipses. Continuous arrows represent a single enzymatic step, while broken arrows indicate pathways involving more than one reaction. GOGAT, Glu synthase.
As previously emphasized by ourselves and others, the carbon/nitrogen (C/N) interaction takes place within a context of energy use and production involving cooperation between different subcellular compartments (Foyer et al., 1994a, 2003; Hoefnagel et al., 1998; Noctor and Foyer, 1998a; Gardeström et al., 2002; Raghavendra and Padmasree, 2003; Guo et al., 2005). First, nitrate assimilation requires reductant to be supplied to both NR and nitrite reductase (NiR). Extensive work on the regulation of NR expression and activity has not yet established which factors are most important in limiting nitrate reduction in leaves (Foyer et al., 1994b; Lejay et al., 1997; Kaiser et al., 2002). Given that the KmNADH of NR may be several-fold higher than the cytosolic NADH concentration, the ability of chloroplasts or mitochondria to deliver reductant to the cytosol could be key in determining the rate of nitrogen assimilation (Krömer and Heldt, 1991; Hanning and Heldt, 1993). Work with spinach (Spinacia oleracea) leaves suggests that low photosynthetic activity may limit NR activity through decreased reductant availability, as well as through posttranslational inactivation (Kaiser et al., 2000, 2002).
Recent studies on wheat (Triticum aestivum) leaves indicate that nitrate reduction is favored by active photorespiration (Rachmilevitch et al., 2004), a process that involves complex redox cycling, in which reductant originating in the chloroplasts and mitochondria is exported to the cytosol and peroxisome (Hanning and Heldt, 1993). Studies comparing plants assimilating nitrate and ammonia suggest that the electron demand for nitrate reduction can affect intercompartmental distribution of reductant (Guo et al., 2005) and decrease mitochondrial respiration rates in the light (Cousins and Bloom, 2004). However, while nitrate reduction and mitochondrial electron transport may compete with each other for reductant under some conditions, it is unclear whether genetic modification of mitochondrial respiration holds any promise for improving nitrogen assimilation.
In addition to nitrate reduction, a second process that may be subject to redox modulation in the C/N interaction is the formation of organic acids. Compounds such as 2-oxoglutarate (2-OG) act as carbon skeletons for amino acid synthesis, and their production requires oxidation through respiratory pathways (Smith et al., 2000; Hodges, 2002; Foyer et al., 2003). However, the significance of mitochondrial events in the production of carbon skeletons for ammonia assimilation remains unresolved (Hodges, 2002; Foyer et al., 2003).
Because amino acid synthesis involves nitrate reduction occurring alongside carbon oxidation, redox status may be an important factor in the integration of the two processes. Despite important recent developments in the analysis of mitochondrial metabolism and electron transport (e.g. Vanlerberghe et al., 2002; Michalecka et al., 2003; Moore et al., 2003; Fernie et al., 2004; Rasmusson et al., 2004), there are few reports on the potential influence of the leaf mitochondrial electron transport chain over the integration of major leaf metabolic pathways.
In this study, we have addressed these questions by exploiting the Nicotiana sylvestris mutant CMS, which respires through alternative low-affinity NADH dehydrogenases (Gutierres et al., 1997; Sabar et al., 2000). Our previous work has shown that the mutant has increased respiration but decreased photosynthesis, and has indicated that this is linked to modified intracellular redox recycling (Dutilleul et al., 2003a, 2003b). It is logical, therefore, to pose the question of whether these redox adjustments impact on the integration of major NAD-dependent metabolic pathways. Here, we show that complex I deficiency is associated with enhanced rates of nitrogen assimilation, adjustment of anaplerotic pathways, and realignment of metabolite profiles to a nitrogen-rich state, and that these effects are linked to substantially increased pools of NAD and NADH.
RESULTS
Metabolite Profiling of the CMS Mutant at the Same Stage of Development as the Wild Type
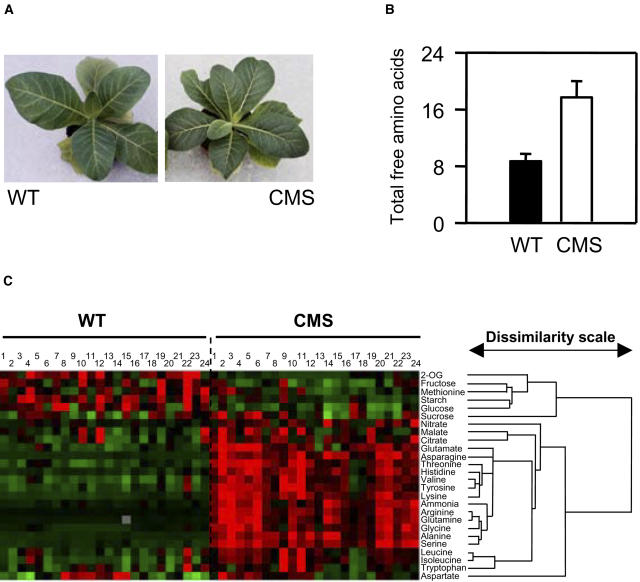
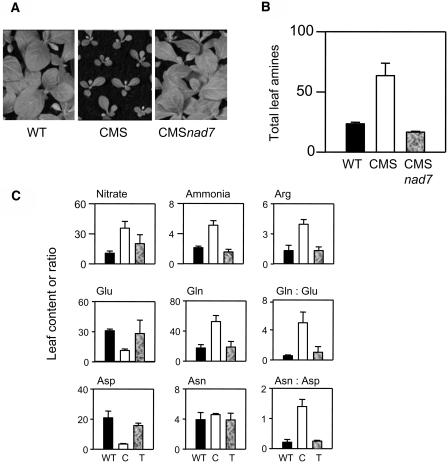
Although it grows more slowly, the CMS mutant achieves similar shoot biomass to the wild type, and it is therefore possible to compare the two lines at a similar stage of development (Fig. 2A). Metabolite analysis of adult plants revealed that complex I deficiency is associated with double the amount of total free leaf amino acids (Fig. 2B). To explore this effect further, we generated quantitative metabolite profiles of leaves from wild type and CMS (Fig. 2C). For each leaf, parallel samples were taken simultaneously and used for analysis of (1) nitrate and ammonia, (2) amino acids, (3) major organic acids, and (4) key carbohydrates. In all, metabolite signatures were generated for 24 leaves of each genotype. Analysis of these signatures by hierarchical clustering showed that metabolites clearly divided into two groups at the highest level of dissimilarity (Fig. 2C). The first group contained 2-OG and carbohydrates; the second contained nitrate, ammonia, malate, citrate, and all amino acids (except Met). Within the second group of metabolites, the classification placed Asp apart from all the others (Fig. 2C). Comparison of a color-coded heatmap with the hierarchical clustering shows that these shifts in metabolite groupings reflected different patterns between the wild type and mutant. Highest values for starch, hexoses, and 2-OG were generally found in wild-type extracts (red on left half of Fig. 2), whereas most amino acids were more abundant in CMS leaves (red on right half of Fig. 2). Hierarchical clustering of the dissimilatory index for individual leaves confirmed the clear intergenotype differences, showing that the leaves fell into two major groupings, one composed of 21 CMS leaves and the other containing all 24 wild-type leaves and three CMS leaves (data not shown). Thus, hierarchical clustering of metabolite profiles showed that clear differences in behavior of C and N metabolites could be ascribed to intergenotype differences that were extractable from the background of intragenotypic variation.
Figure 2.
Complex I deficiency in the CMS mutant is associated with readjustment of metabolite profiles toward accumulation of nitrogen-rich metabolites. A, Photograph of wild-type and CMS plants, showing shoot morphology at the developmental stage sampled to produce the data shown in Figures 2 to 8. B, Increased total free amino acids (μmol mg−1 chlorophyll) in CMS leaves. C, Hierarchical classification of metabolite profiles in individual wild-type and CMSII plants. Quantitative techniques were used to analyze amino acids, organic acids, carbohydrates, ammonia, and nitrate in samples taken in parallel from the same leaves (for details see “Materials and Methods”). The hierarchical clustering of metabolites (right) is based on 48 values, each obtained from independent extracts of 24 wild-type and 24 CMS plants. Different leaves are numbered from 1 to 24 for each gentotype. For each metabolite, degrees of red and green indicate the extent of increase or decrease relative to the median value.
The CMS Leaf Metabolite Profile Cannot Be Explained by Changes in Expression of Key Enzymes
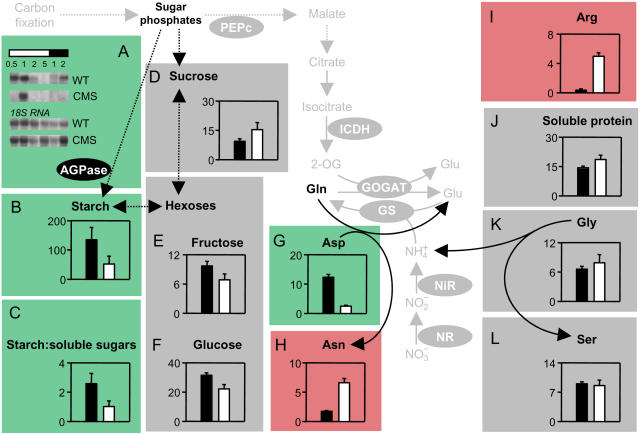
Given that total free amino acids were much higher in CMS leaves than in wild-type leaves, we investigated the factors driving accumulation of nitrogen compounds in the mutant. For this, we compared key C and N metabolites between the genotypes and aligned the data, together with transcripts, protein, and activities of important enzymes, on a metabolic map of the core C/N interaction shown in Figure 1. This analysis revealed that slight differences in ADP-Glc pyrophosphorylase (AGPase) expression in CMS (Fig. 3A) were associated with diminished starch contents (Fig. 3B) and a substantial decrease in the ratio of starch to soluble sugars (Fig. 3C). Although Suc showed a slight increase (Fig. 3D) and hexoses were somewhat lower in the mutant (Fig. 3, E and F), decreased carbohydrate in CMS mainly reflected a specific effect on starch content. Similarly, accumulation of nitrogen compounds in CMS did not occur indiscriminately. Asp decreased 5-fold as a percentage of amino acids, while Asn was increased by a similar factor (Fig. 3, G and H). Changes in the Arg pool were even more dramatic: A very minor amino acid in wild-type leaves, Arg accounted for almost 5% of total amino acids in CMS leaves (Fig. 3I). The accumulation of these nitrogen metabolites was also accompanied by enhanced soluble leaf protein on a chlorophyll basis (Fig. 3J). In stark contrast to the accumulation of Arg and the dramatic shift in Asn/Asp, the percentage of amino acids present as Gly and Ser was similar in the two genotypes (Fig. 3, K and L).
Figure 3.
Lack of mitochondrial complex I function markedly affects whole leaf C/N balance. Black bars, wild type; white bars, CMS. Color coding indicates higher in CMS (red), higher in wild type (green), no change between the genotypes (gray). A, RNA gel blots of transcripts for AGPase. The numbers above the horizontal bars indicate duration (h) into the light (white bar) or dark (black bar) period. B, Starch. C, The ratio of starch to soluble sugars (Suc + Glc + Fru). D, Suc. E, Fru. F, Glc. G, Asp. H, Asn. I, Arg. J, Soluble leaf protein. K, Gly. L, Ser. For enzyme activities and metabolites, data are means ± se of triplicate leaves. RNA blots are representative of triplicate analyses from samples taken from the same leaves as for enzyme and metabolite assays. Amino acids are expressed as % total amino acids. Carbohydrates are in μmol hexose mg−1 chlorophyll. Soluble protein is given as mg mg−1 chlorophyll.
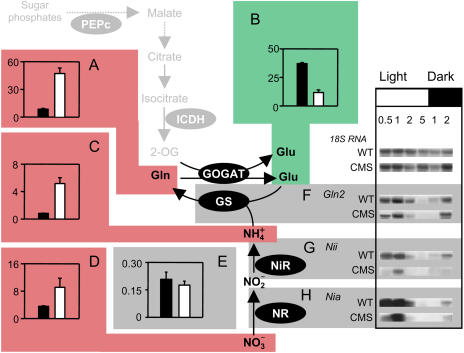
Next, we compared key features of nitrate and ammonia assimilation in CMS and wild type (Fig. 4). A striking observation was the dramatic shift from a Glu-rich amino acid profile in the wild type to one dominated by Gln in the mutant. In wild-type leaves, Gln was 8% of total amino acids (Fig. 4A) and Glu was predominant, representing 37% of the total amino acids (Fig. 4B). In CMS, however, Glu accounted for only 12% of amino acids (Fig. 4B), while Gln accumulated to account for almost half the total pool (Fig. 4A). This reorientation of the amino acid profile to one dominated by Gln was associated with marked increases in ammonia (Fig. 4C) and a less dramatic change in nitrate in CMS (Fig. 4D). No difference was observed in the maximal extractable activity of NR (Fig. 4E) or in the activation state of the enzyme (data not shown). Neither was the increase in Gln accompanied by increased abundance of transcripts for the organellar Gln synthetase (GS), GS2 (Fig. 4F). Likewise, Nia and Nii transcripts, which showed a clear day-night rhythm in both genotypes, were not increased in the mutant (Fig. 4, G and H). Thus, the accumulation of ammonia and amino acids (Fig. 2B) and the preferential enhancement of Gln, Asn, and Arg in CMS leaves (Figs. 3 and 4) were not driven by increased expression of the enzymes of nitrate and ammonia assimilation.
Figure 4.
Accumulation of nitrogen-rich metabolites in CMS is not accompanied by enhanced expression of enzymes involved in primary nitrogen assimilation. Black bars, wild type; white bars, CMS. Color coding as for Figure 3. A, Gln. B, Glu. C, Ammonia. D, Nitrate. E, Maximal extractable NR activity. F, RNA gel blots of Gln2 transcripts encoding GS2. G, RNA gel blots of Nii transcripts encoding NiR. H, RNA gel blots of Nia transcripts encoding NR. Symbols and sampling as in Figure 3. Gln and Glu are expressed as percent of total amino acids. Nitrate and ammonia are expressed in μmol mg−1 chlorophyll. In panels F to H, the numbers indicate duration in hours into the light (white bar) or dark (black bar) period.
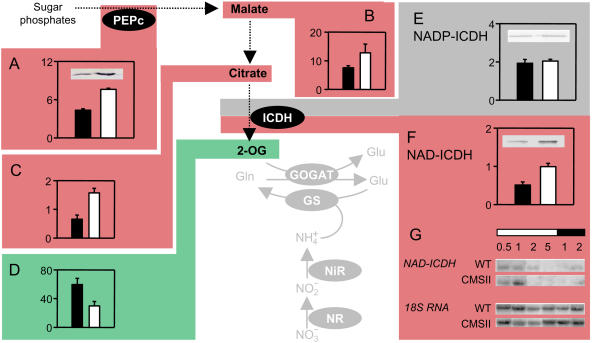
To further explore the underlying causes of the modifications in amino acid profiles, we examined the pathway that generates 2-OG for ammonia assimilation. An increase in extractable PEPc activity (Fig. 5A) was matched by increased PEPc protein (Fig. 5A, inset), and these changes were accompanied by increased malate (Fig. 5B) and citrate (Fig. 5C). Despite the increased malate and citrate, 2-OG contents were significantly lower in CMS (Fig. 5D). Low 2-OG was not accompanied by changes in total extractable NADP-isocitrate dehydrogenase (ICDH) activity (Fig. 5E). Neither were any changes observed in the abundance of the major cytosolic NADP-ICDH isoform (Fig. 5E, inset) or in transcripts encoding this isoform (data not shown). In contrast, 2-OG deficit in CMS was associated with substantially increased mitochondrial NAD-ICDH, both in terms of activity (Fig. 5F) and protein (Fig. 5F, inset). These increases were associated with an enhanced maximum in the light-dark rhythm of NAD-ICDH transcripts (Fig. 5G).
Figure 5.
CMS leaves are deficient in 2-OG despite increased maximal extractable activities of key anaplerotic enzymes. Black bars, wild type; white bars, CMS. Color coding as for Figure 3. A, PEPc activity; inset, immunoblot of PEPc protein. B, Malate. C, Citrate. D, 2-OG. E, NADP-ICDH activity; inset, immunoblot of cytosolic NADP-ICDH protein. F, NAD-ICDH activity; inset, immunoblot of mitochondrial NAD-ICDH protein. G, NAD-ICDH and 18S RNA gel blots. The horizontal bars above the blots indicate light or dark. The numbers indicate duration in hours into the light (white bar) or dark (black bar) period. RNA and immunoblots are representative of triplicate analyses of independent leaf samples. Other details as in Figure 3. Malate and citrate contents, μmol mg−1 chlorophyll; 2-OG, nmol mg−1 chlorophyll; enzyme activities, μmol mg−1 protein h−1.
Feeding 2-OG Alleviates High Amide Contents in CMS
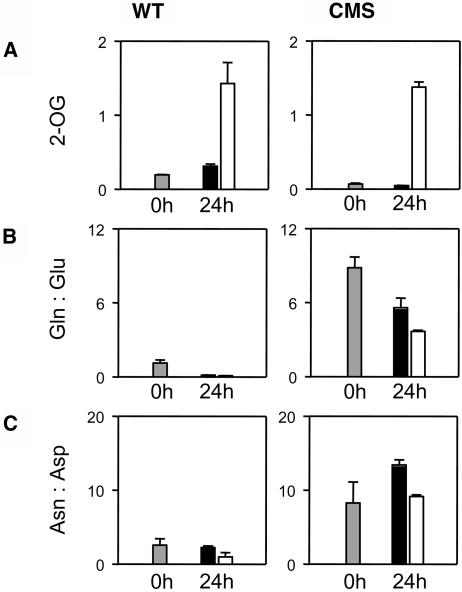
The above data show that accumulation of N-rich compounds in CMS is associated with low 2-OG. We hypothesized that this could be due to either increased demand for 2-OG in nitrogen assimilation or a restriction in 2-OG production, or to both. We therefore analyzed this question by feeding experiments. First, we fed 2-OG to leaf discs, thus complementing endogenous contents, particularly the low values in CMS (Fig. 6A). In the buffer control, a decrease in Gln:Glu in CMS discs (Fig. 6B, right) was accompanied by a rise in Asn:Asp (Fig. 6C, right), possibly reflecting net transfer of amide nitrogen from Gln to Asn during the course of the experiment. Supplementing 2-OG caused Gln:Glu to decrease further and prevented the rise in Asn:Asp (Fig. 6, B and C, right). Hence, the marked increase in amides in CMS leaves (Figs. 3, 4, and 6) could be partly reversed by supplementary 2-OG, indicating that decreased 2-OG availability is to some extent responsible for the high Gln and Asn contents in the mutant.
Figure 6.
High Gln:Glu and Asn:Asp ratios in CMS are partly alleviated by feeding 2-OG. A, Leaf 2-OG (μmol mg−1 chlorophyll). B, Gln:Glu. C, Asn:Asp. Gray bars, control values at time 0. Black bars, 24 h incubation on buffer. White bars, 24 h incubation on buffer + 2-OG.
Incorporation of 15N-Nitrate into Amines Is Stimulated in CMS
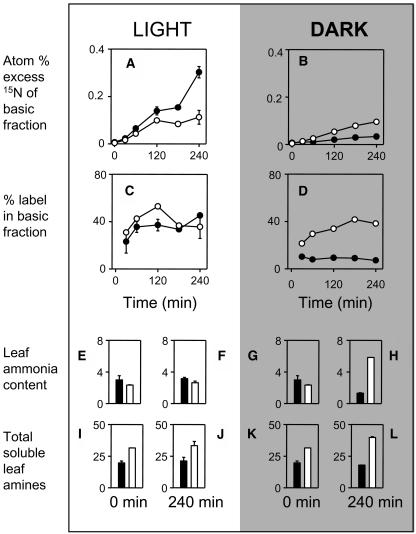
We next examined flux from nitrate into amines during experiments where leaf discs were incubated with 15N-labeled nitrate. Total uptake of 15N from nitrate by CMS leaf discs was lower than in the wild type (data not shown). Because of this, absolute labeling of the basic fraction was also significantly lower in the light (Fig. 7A). Expressed as a percentage of total 15N uptake, however, partitioning into the basic fraction was similar or higher in CMS discs, representing around 40% of the total label incorporated in the light (Fig. 7C). In the dark, despite the lower rates of total uptake, labeling of the basic fraction was markedly increased in CMS relative to wild type (Fig. 7B). Partitioning into amines remained at around 40% in CMS leaf discs (Fig. 7D) whereas darkening more or less abolished labeling of the basic fraction in wild-type discs (compare Fig. 7, C and D). This suggests that nitrate reduction is switched off in the wild type in the dark, as might be predicted, but that this does not occur in the mutant. Total amines were consistently higher in CMS than in wild-type discs (Fig. 7, I–L), but the only condition in which ammonia and amines increased during the experiment was for CMS discs in the dark (Fig. 7, H and L).
Figure 7.
Partitioning of 15N-labeled nitrate into the basic and neutral soluble fraction is enhanced in CMS leaf discs, particularly in the dark. Black symbols and bars, wild type; white symbols and bars, CMS. A and B, Atom percent excess 15N of the soluble basic fraction of leaf extracts. C and D, Percentage of total 15N absorbed partitioned into the soluble basic fraction. E to H, leaf ammonia content at time 0 (E and G) or after 4 h incubation (F and H). I to L, Total soluble leaf amines at time 0 (I and K) or after 4 h incubation (J and L). 15N data are means of two independent extracts. Error bars indicate actual values and are contained within the symbol where not apparent. Ammonia and amine contents are means ± se (μmol mg−1 chlorophyll) of four independent extracts.
Complex I Deficiency Is Associated with Increases in Pyridine Nucleotides
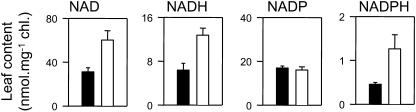
Following the observation that nitrate assimilation in CMS was particularly stimulated in the dark (Fig. 7), we assayed total leaf pyridine nucleotide pools under these conditions. While there was no change in the NADP pool between wild-type and CMS, the NADPH pool was increased in the mutant (Fig. 8). No change, however, was observed in the sum of NADP and NADPH, which in both genotypes was stable at around 18 nmol mg−1 chlorophyll. The most striking change was observed in leaf NAD and NADH, both of which were about 2-fold higher in CMS than in wild-type leaves (Fig. 8). The increases in NADH and NAD occurred without any change in the NADH/NAD ratio, which in both plants was approximately 0.2.
Figure 8.
Total pyridine nucleotide pools in CMS and wild-type leaves. Black bars, wild type; white bars, CMS. Each value is the mean ± se of three independent extracts from freeze-clamped tissue sampled after 2 h darkness.
Wild-Type Amino Acid Profiles Are Reestablished Following Complex I Restoration by Transformation of CMS
In order to determine whether amine accumulation and associated metabolite changes are a direct result of complex I deficiency caused by deletion of nad7, CMS transformants were constructed carrying an edited nad7 cDNA with a mitochondrial targeting sequence under the control of the 35S promoter. In these transformants, functional complex I is restored (Pineau et al., 2005). Reinstatement of complex I restores the wild-type growth phenotype (Fig. 9A) and reverses the amino acid accumulation observed in CMS (Fig. 9B). Furthermore, key features of the wild-type metabolite profile were also restored, including lower ammonia, Arg, Gln:Glu, and Asn:Asp than in CMS (Fig. 9C).
Figure 9.
Nuclear transformation of CMS with a nad7 cDNA reverses the CMS growth phenotype and restores wild-type amino acid profiles. WT, Wild-type; C, CMS mutant; T, CMS transformed with the nad7 cDNA. A, Shoot morphologies 6 weeks after germination. B, Restoration of wild-type amine levels by nad7 complementation in CMSnad7. Leaf material was sampled from each line in the middle of the photoperiod. C, Restoration of wild-type ammonia and amino acid profiles by nad7 complementation. Sampling as for leaf amines. Data for total amines, nitrate, and ammonia are means ± se of four to six independent extracts (μmol g−1 fresh weight). Individual amino acid contents are means of two independent extracts and are expressed as percentage total amino acids or ratios.
High Amine Contents in CMS Are Associated with Increased NAD and NADH
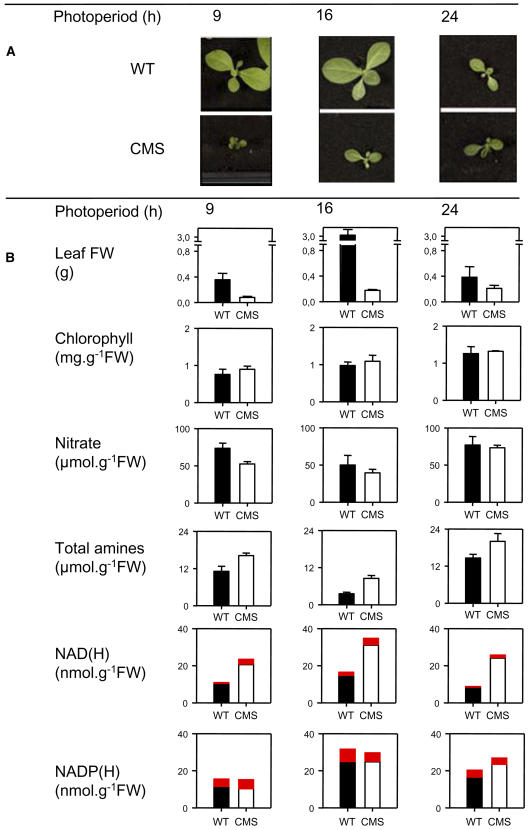
Increased amino acids are a key feature of the CMS leaf metabolite profile, which we have observed in numerous experiments. Associated with the CMS mutation is a slower growth phenotype under many conditions. To examine whether the increased amines were linked to complex I deficiency through changes in pyridine nucleotide status or through differences in growth rates, we analyzed metabolites in conditions in which growth differences were accentuated or minimized. This was achieved by growing batches of plants under conditions of different photoperiod (Fig. 10). Whereas large differences in shoot phenotype were observed at 9- and 16-h photoperiods, the difference under continuous light was much less pronounced (Fig. 10A). This observation was confirmed by shoot biomass values (Fig. 10B). In these experiments, no increases in leaf nitrate were observed, but leaf amine contents were enhanced under all conditions (Fig. 10B). Associated with the increased amines was an increase in NAD and NADH, but no change in NADP or NADPH (Fig. 10B).
Figure 10.
Enhanced amines in CMS leaves are not correlated with growth phenotype or leaf nitrate, but are associated with increases in NADH. A, Shoot morphologies at different photoperiods: 9 h (left), 16 h (middle), or continuous light (24 h, right). Photographs show plants 10 (9 h) or 7 weeks (16 and 24 h) after germination. B, Leaf fresh weight (FW), and contents of chlorophyll, nitrate, amines, and reduced and oxidized forms of pyridine nucleotides at different photoperiods. For pyridine nucleotides, the red blocks indicate reduced forms (NADH and NADPH). Assays were performed on leaf material harvested in the middle of the photoperiod from plants 11 weeks (9 h photoperiod) or 8 weeks (16 and 24 h photoperiods) after germination (three independent extracts; standard errors were in the same range as data shown in Figure 8).
DISCUSSION
The plant inner mitochondrial membrane contains, in addition to complex I, several alternative rotenone-insensitive NAD(P)H dehydrogenases (Douce et al., 1973; Møller and Palmer, 1982). These enzymes, which are internally or externally oriented, have been recently characterized at the molecular and functional levels (Michalecka et al., 2003; Moore et al., 2003; Rasmusson et al., 2004). Much remains to be discovered concerning the physiological and metabolic impact of partitioning between these dehydrogenases and complex I. Here, we have shown that this partitioning has profound consequences for the integration of C/N metabolism. Respiration through alternative dehydrogenases rather than complex I promotes nitrate assimilation, is associated with low 2-OG, and favors amino acid profiles dominated by Gln rather than Glu. These changes are accompanied by accumulation of Asn and Arg, two N-rich amino acids that are synthesized using amido nitrogen from Gln. In CMS, the three amino acids accumulate together while carbohydrate contents are much decreased.
The N. sylvestris CMS mutant carries the only well-characterized stable homeoplasmic mitochondrial DNA mutation in any plant species that results in alteration of the respiratory electron transport chain (Gutierres et al., 1997). The mutant is at least as competent as the wild type to perform respiration (Dutilleul et al., 2003a), which it does through the alternative dehydrogenases (Sabar et al., 2000). In this respect, mitochondrial electron transport in CMS is somewhat analogous to the situation in budding yeast (Saccharomyces cerevisiae), which is devoid of complex I. Moreover, it has been shown in complex I-deficient mammalian cell lines that expression of the yeast internal NADH dehydrogenase can restore respiration capacity (Seo et al., 1998; Bai et al., 2001). Thus, complex I function can be replaced by alternative NADH dehydrogenases in plant, yeast, and animal mitochondria. The CMS mutant is therefore an excellent tool in which to test the functional significance of mitochondrial complex I in plants, and it has recently been shown that expression of nad7 alone is sufficient to restore functional complex I in transformed CMS (Pineau et al., 2005).
Flux through alternative dehydrogenases is not coupled to ATP production, and the absence of complex I dictates that the rate of NADH oxidation has to increase 1.5-fold to match ATP production in the wild type (Bai et al., 2001). Indeed, it is noteworthy that both dark O2 consumption and CO2 evolution are increased approximately 1.5-fold in CMS leaves (Dutilleul et al., 2003a), suggesting that the loss of complex I leads to a compensatory increase in respiratory rates to maintain similar rates of ATP production to the wild type. Here, we show that the absence of complex I is associated with appreciable increases in leaf NAD and NADH that likely occur to satisfy the kinetic properties of the alternative dehydrogenases. The internally oriented alternative NADH dehydrogenase has an affinity for NADH that is much lower than complex I (Møller, 2001). This property means that in order to drive respiration at rates commensurate with ATP requirements, mitochondria in CMS leaves operate at a higher NADH concentration than in the wild type.
Our data suggest that switching from complex I to alternative dehydrogenase function exerts a powerful impact on regulatory processes at the crossroads of nitrogen assimilation and carbon metabolism. What mechanisms explain how respiration through alternative dehydrogenases could modulate C/N partitioning so markedly? A first possibility is that the CMS metabolite profile is driven by modified expression of key enzymes involved in the C/N interaction. Changes in expression could occur through acclimation linked to the slow-growth CMS phenotype. A second possibility is that the changes in metabolite profiles are driven by nitrate-signaling effects on enzyme expression, previously described in Nicotiana tabacum (Scheible et al., 1997). Third, CMS metabolite profiles could be independent of changes in enzyme expression. They could instead reflect secondary allosteric or substrate control effects over key dehydrogenases and reductases that arise from primary changes in NAD status. As we now discuss, only the last of these possibilities is able to provide a coherent explanation of our data.
The Main Features of the CMS Metabolite Profile Cannot Be Explained by Acclimatory Changes in Expression of Key Enzymes
With the exception of parallel changes in AGPase and starch, the CMS metabolite profile does not appear to be the result of modified enzyme capacities. The marked accumulation of nitrogen assimilation products in CMS is not accompanied by changes in NR capacity (Fig. 4). In the case of 2-OG, decreases in leaf contents in the mutant were accompanied by increases in PEPc and NAD-ICDH capacity (Fig. 5). The enhanced citrate to 2-OG ratio points to increased restriction in vivo that is not due to decreased enzyme capacity, because ICDH was either unchanged (NADP-ICDH) or enhanced (NAD-ICDH). This suggests that in CMS some factor is preventing ICDH from working optimally, and this question is further discussed below.
The CMS Metabolite Profile Is Not the Result of Enhanced Nitrate or Activated Nitrate Signaling
The metabolic signature of the CMS leaf is characterized by increased amino acids. These features have been consistently observed in our studies over the last four years and are reversible by nad7 expression. Moreover, the trend toward increased amino acids is observed irrespective of the extent of the difference in growth between CMS and the wild type, and does not show a robust correlation with leaf nitrate contents (Fig. 10). Nitrate has been implicated as an important factor coordinating the C/N interaction through signaled modulation of expression of key enzymes (Scheible et al., 1997). Nitrate is sometimes increased in CMS (Figs. 4 and 9), but this trait is associated much more weakly with the mutation than increased amines (Fig. 10). Although several CMS extracts showed up red for nitrate on a heatmap comparing multiple samples, increases in nitrate were less dramatic than effects on many amino acids (Fig. 2C). The CMS organic acid profile also provides little evidence for a strong nitrate-signaling effect. NR-deficient tobacco grown on high nitrate had similar growth rates and leaf protein to wild-type plants grown on limiting nitrate, but much more 2-OG (Scheible et al., 1997). The decreased citrate to 2-OG ratio promoted by high leaf nitrate was associated with induction of cytosolic NADP-ICDH (Scheible et al., 1997). Here we show that enrichment of nitrogen compounds in CMS is associated with a marked increase in the citrate to 2-OG ratio, no induction of NADP-ICDH, but markedly enhanced mitochondrial NAD-ICDH capacity (Fig. 5). Thus, the effects we report in CMS occur though metabolic/signaling mechanisms that are distinct from the nitrate effect previously described in N. tabacum.
Mitochondria-Driven Effects on NAD Status Exert a Major Influence on the Rate of Nitrate Assimilation
These data demonstrate that ammonia assimilation, which in source leaves is considered to be a largely chloroplastic function (Hirel and Lea, 2002), is affected by partitioning between different mitochondrial electron transport pathways. Indeed, it is striking that complex I deficiency affects the Gln:Glu and Asn:Asp ratios much more than the Gly:Ser ratio (Figs. 3 and 4). Thus, mitochondrial electron transport status, often previously considered in relation to oxidation of photorespiratory Gly (e.g. Igamberdiev et al., 1997), may play a more critical role in controlling events linked to nitrogen assimilation. Flux from nitrate into amines is enhanced in CMS, despite no change in NR capacity (Fig. 7). How might this occur? Our data strongly point to an important role for cytosolic NADH availability, in agreement with recent studies in wheat and spinach (Kaiser et al., 2000, 2002; Rachmilevitch et al., 2004). Unlike these studies, however, which focused on the chloroplast-cytosol interaction, our findings in CMS provide in vivo evidence identifying the influence of mitochondrial electron transport pathways in the regulation of nitrate assimilation. This interpretation is strongly supported by the increases in NADH in CMS leaves (Figs. 7 and 10).
Several factors must be taken into account when considering the biochemical consequences of the marked increases in NAD and NADH. One key issue is compartmentation. There are few or no direct measurements of NAD status in the different compartments of leaf cells. Studies on protoplasts and estimations from the Asp aminotransferase/malate dehydrogenase equilibrium point to a marked intracellular redox gradient, with the cytosolic NAD pool being much less reduced than the chloroplast NADP and mitochondrial NAD pools (Heineke et al., 1991; Igamberdiev and Gardeström, 2003). Indeed, the cytosolic NADH concentration has been estimated to be as low as 1 μm (Heineke et al., 1991). As this concentration is below KmNADH of NR, which is about 5 μm (Kaiser et al., 2002), even small absolute increases in cytosolic NADH should appreciably stimulate nitrate reduction. If the increase in cytosolic NADH is proportional to the global tissue increase in CMS, we should predict that nitrate assimilation would be stimulated around 2-fold. In fact, our data show that the stimulation is less than 2-fold in the light but greater than 2-fold in the dark (Fig. 8).
The data of Figure 8 demonstrate that changes in mitochondrial reductant use can markedly influence the in vivo efficiency of nitrate assimilation into amino acids. This effect, and the increase in leaf amines in CMS, stand in marked contrast to the minor increases in leaf amino acid pools produced by overexpression of NR (Foyer et al., 1994b). Much attention has previously focused on the role of phosphorylation state in switching off NR when CO2 assimilation is slow or inactive (Kaiser and Huber, 1994). However, it has been shown that expression of a constitutively active NR did not prevent inactivation of nitrate assimilation in the dark (Lejay et al., 1997). The stimulation of nitrate assimilation in CMS is not related to enhanced expression or capacity of NR or other key enzymes, but is associated with changes in NADH availability. This suggests that a key factor that switches off nitrate reduction in darkened wild-type leaves is insufficiently rapid delivery of reductant to the cytosol.
Leaf NAD Status Is a Key Factor in Coordinating the Supply of 2-OG and Ammonia to the GS-Glu Synthase Pathway
Stimulation of nitrate reduction and an overall increase in amino acids in CMS leaves were accompanied by a radical redistribution of amines within the different amino acid pools. Although the Arabidopsis (Arabidopsis thaliana) gene GLN2 has been found to encode a dual-targeted GS (Taira et al., 2004), most evidence suggests that net ammonia assimilation occurs mainly in the chloroplast, where ferredoxin-Glu synthase activity requires 2-OG (Hirel and Lea, 2002). Insufficient rates of 2-OG generation to support ammonia assimilation may lead to enhanced Gln:Glu (Novitskaya et al., 2002). Thus, one factor driving the increased Gln:Glu in CMS leaves is likely an imbalance between 2-OG delivery and ammonia production. Any limitation on 2-OG supply should become especially critical under conditions in which reduction of nitrate to ammonia is stimulated, leading to the drastic realignment in partitioning of reduced nitrogen that we have observed in CMS.
It is striking that decreased 2-OG occurred despite an up-regulation of anaplerotic pathway expression. Thus, enhanced enzyme capacity was not sufficient to restore 2-OG to wild-type values. Despite the importance often previously accorded to the cytosolic NADP-ICDH, we observed no induction of this activity. Indeed, decreasing this enzyme capacity by antisense technology in potato (Solanum tuberosum) produced only minor effects on organic acid and amino acid contents (Kruse et al., 1998). In contrast, the induction of the mitochondrial NAD-ICDH points to a crucial role for this enzyme in anaplerosis in N. sylvestris. This isoform is particularly sensitive to changes in NAD and NADH status. Igamberdiev and Gardeström (2003) have shown that the enzyme from pea (Pisum sativum) has comparable values for KmNAD and KiNADH (about 0.2 mm). However, the total mitochondrial NAD concentration is much higher than the NADH concentration, with the NADH/NAD ratio being around 0.05 to 0.25 (Igamberdiev and Gardeström, 2003), i.e. similar to the range of the whole leaf values we report here. Although extraction methods for pyridine nucleotides liberate both the protein-bound and free pools, we conclude that a 2-fold increase in both NAD and NADH is likely to increase control over 2-OG production by NAD-ICDH. This conclusion is based on the data of Igamberdiev and Gardeström (2003), which show that mitochondrial NAD in pea leaves is likely above the NAD-ICDH Km value whereas NADH concentrations are probably below or close to the enzyme's Ki value. This being so, a 2-fold increase in NAD should have relatively little effect compared to the increased restriction produced by a 2-fold increase in NADH. Such increased restriction on the in vivo function of NAD-ICDH is likely one of the factors that explain why increases in the amount of enzyme (and extractable activity, measured in the absence of NADH) are not able to restore 2-OG contents to wild-type values (Fig. 5).
CONCLUSION
Our data suggest that modulation of NADH use by the mitochondrial electron transport chain has far-reaching repercussions for the integration of carbon and nitrogen metabolism. The effects of increased NADH are observed despite no change (NR, NiR, GS2) or opposing changes (PEPc, NAD-ICDH) in the expression levels of key genes. Increases in NADH in CMS can be understood in terms of the need to drive alternative dehydrogenase function, and the accumulation of both NAD and NADH may be the result of mechanisms operating to ensure redox homeostasis and thus minimize perturbation of respiratory carbon flux. The stimulation of nitrate assimilation and the modulation of anaplerosis are likely side effects of this acclimatory response, and offer interesting perspectives for the manipulation of plant metabolism.
MATERIALS AND METHODS
Plant Growth and Sampling
Nicotiana sylvestris wild type and the CMS mutant (previously called CMSII; Gutierres et al., 1997) were grown under two culture conditions. To obtain the data shown in Figures 1 to 9, plants were grown in pots of 21 cm diameter in a constant-temperature (23°C) greenhouse room. The medium was soil with added Osmocote slow-release fertilizer (Petersfield Products) containing 15% N as ammonium nitrate, ammonium phosphates, and potassium nitrate. Plants were introduced into controlled-environment growth chambers 1 week before sampling. Day conditions were 600 μmol quanta m−2 s−1, 24°C, 14 h; night was 20°C, 10 h. Samples were taken from the youngest pair of fully expanded leaves of plants of similar shoot size and developmental stage (10 weeks after sowing for the wild type, 13 weeks after sowing for the CMS mutant). Four samples from each leaf were used for metabolite analysis; other samples of the same leaves were taken simultaneously and used for analysis of transcripts, protein abundance, and extractable enzyme activities. To ensure metabolite contents reflected in vivo values, sampling was carried out by rapid quenching using portable clamping tongs precooled in liquid N2 (Novitskaya et al., 2002). All samples were transferred from the sampling apparatus to liquid N2 then stored at −80°C until analysis.
For 15N labeling experiments (Fig. 7), discs of 5 mm diameter were cut from several leaves of each genotype grown as above, randomized, and transferred to HEPES at 21°C. For each genotype, discs were divided between four petri dishes, two containing 30 mm KNO3 (5% 15N-nitrate), and two 30 mm KCl. One dish of each set was incubated in darkness, the other at an irradiance of 600 μmol quanta m−2 s−1 (24°C in both cases). Duplicate samples, each of approximately 10 discs, were taken at the times indicated and frozen in liquid N2.
For the analyses shown in Figure 10, sets of wild type and CMS were grown at photoperiods of 9 h, 16 h, or 24 h (continuous light) at 200 μmol m−2 s−1, and supplied daily with nutrient medium containing N as nitrate.
RNA Gel Blots
Total RNA was extracted by the Trizol-chloroform procedure (Gibco-BRL). Ten micrograms of total RNA was fractionated on an agarose gel (1.2%), blotted onto nylon-based membranes (Appligène), and hybridized with 32P-labeled probes. Homologous probes were Nicotiana tabacum cytosolic NADP-ICDH (Gálvez et al., 1996), mitochondrial NAD-ICDH (Lancien et al., 1998), organellar GS (GS2; Dubois et al., 1996), NR (nia2; Vaucheret et al., 1989), NiR (NiR3; Kronenberger et al., 1993), and AGPase small subunit cloned from N. tabacum using the primers 5′ CGTGATAAGTTCCCTGGTGC 3′ and 5′ GCCAATGCCAATCGGGACAC 3′ (Miyazawa et al., 1999). Procedures for blot analysis were performed as described in Sabar et al. (2000).
Immunoblots
Soluble proteins (10 μg per lane) were separated on a 12% SDS-polyacrylamide gel and transferred to nitrocellulose membranes as described in Gutierres et al. (1997). Rabbit antisera raised against the following proteins were used: N. tabacum cytosolic ICDH (Gálvez et al., 1996), N. tabacum mitochondrial IDH (Lancien et al., 1998), and Sorghum PEPc (Osuna et al., 1996). Horseradish (Armoracia lapathifolia) peroxidase-conjugated goat anti-rabbit IgG (1:2,000 dilution) was used as a secondary antibody, and immune complexes were visualized by the color reaction of peroxidase as described in Gutierres et al. (1997).
Enzyme Assays
Frozen leaf samples were ground to a powder in liquid N2. NADP-ICDH was measured as in Gálvez et al. (1994) by extraction of powder into 100 mm potassium phosphate (pH 7.5), 14 mm β-mercaptoethanol, plus approximately 100 mg insoluble polyvinylpyrrolidone (PVP). After centrifugation, NADP reduction was monitored for a period of 10 min at 340 nm (30°C) in a reaction mixture containing 100 mm potassium phosphate (pH 7.5), 5 mm MgCl2, 0.25 mm NADP, plus 50 μL sample. The reaction was initiated by addition of isocitrate to 2.5 mm. For NAD-ICDH assay, powdered leaf tissue was extracted into 100 mm HEPES (pH 7.4), 14 mm β-mercaptoethanol, 1 mm phenyl methyl sulfonyl fluoride, 20 μm leupeptin, 20 μm chymostatin, 40% glycerol, plus approximately 100 mg insoluble PVP. Following centrifugation, the supernatant was desalted on a NAP-5 column preequilibrated with 100 mm HEPES (pH 7.4), 40% glycerol, and eluted in the same buffer. Activity was assayed in a reaction mixture (30°C) containing 100 mm HEPES buffer (pH 7.4), 5 mm MnCl2, 5 mm NAD, plus 100 μL sample. The reaction was initiated by addition of isocitrate to 10 mm. NAD reduction was monitored for 10 min at 340 nm. PEPc activity was measured according to Ferrario-Méry et al. (2002), by extracting powder into 100 mm Tris (pH 8.0), 20% glycerol, 10 mm MgCl2, 5 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 20 μm leupeptin, 16 μm chymostatin, 0.1% β-mercaptoethanol, 2 nm okadaïc acid, approximately 100 mg insoluble PVP, and clarified by centrifugation. Activity was measured in a reaction mixture (30°C) containing 50 mm HEPES (pH 8.0), 5 mm MgCl2, 5 mm NaF, 1 mm NaHCO3, 0.2 mm NADH, 10 units malate dehydrogenase, plus 50 μL sample. The reaction was initiated by addition of PEP to 3 mm, and NADH oxidation was followed for 5 min at 340 nm. To measure maximal extractable NR activity, powder was extracted into 50 mm MOPS-KOH (pH 7.6), 5 mm NaF, 1 μm sodium molybdate, 10 μm FAD, 4 μm leupeptin, 2 mm β-mercaptoethanol, 3 nm okadaic acid, 5 μm Na4EDTA, with 30 mg insoluble PVP. After centrifugation, the maximal activity was measured as nitrite formation in the presence of EDTA and absence of magnesium (Ferrario-Méry et al., 2002). Soluble proteins were quantified using the Bio-Rad assay, against a bovine serum albumin standard curve.
Metabolite Analysis
To measure pyridine nucleotides, parallel samples of equal fresh weight were ground in liquid nitrogen then extracted into 0.2 n HCl or 0.2 n NaOH. Following centrifugation, a sample of the supernatant was briefly heated, then cooled on ice and neutralized to pH 6 to 7 (acid extracts) or 7 to 8 (alkali extracts). Nucleotides were immediately assayed in the neutralized supernatant as the phenazine methosulfate-catalyzed reduction of dichlorophenolindophenol in the presence of ethanol and alcohol dehydrogenase (NAD and NADH) or Glc-6-P and Glc-6-P dehydrogenase (NADP and NADPH). NAD and NADP were calculated from rates obtained for acid extracts, NADH and NADPH from rates for alkali extracts. To determine leaf nitrate, ammonia, and total amines, samples were ground in 100 mm HCl and insoluble PVP. The extract was clarified by centrifugation. Nitrate was measured by addition of 10 μL of supernatant to 40 μL 5% salicylic acid prepared in H2SO4. After 20 min incubation at room temperature, 0.95 mL 2N NaOH was added and the absorbance measured at 410 nm against a blank containing everything but sample. Nitrate was then quantified against a KNO3 standard curve. Leaf ammonia and total amines were measured by a ninhydrin color assay as in Ferrario-Méry et al. (2002). To measure organic acids, leaf material was ground in liquid nitrogen and the powder extracted into 0.7 mL 1 m HClO4. The supernatant was clarified by centrifugation and neutralized with K2CO3 in the presence of 10 mm HEPES (pH 7.0). KClO4 was removed by centrifugation and the resulting supernatant was clarified by a charcoal treatment. The final supernatant was used for determination of organic acids by NAD(P)-linked end-point assays in the presence of appropriate dehydrogenases and coupling enzymes (Novitskaya et al., 2002). Amino acids were extracted from ground leaf powder into 80% ethanol and quantified using automated precolumn derivatization of amines with o-phthaldialdehyde, separation by reverse-phase HPLC, and fluorescence detection (Noctor and Foyer, 1998b). To determine carbohydrate contents, leaf material was ground in liquid nitrogen and the powder extracted into 1 mL 80% ethanol. After 20 min incubation at 80°C, the supernatant was clarified by centrifugation. The final supernatant was used for enzymic determination of soluble sugars and the pellet was used for determination of starch as in Foyer et al. (1994b). Chlorophyll was measured after extraction into 80% acetone, either at 652 nm or (for acid extracts) as pheophytin at 666 and 655 nm.
15N Labeling
Labeling of the neutral and basic soluble fraction was carried out on leaf discs by a similar protocol to that described by Reed et al. (1983). Samples were ground in liquid nitrogen and extracted into 0.6 mL 0.1 n HCl. Extracts were clarified by centrifugation and anions were removed from the supernatant by retention on DOWEX resin (type I strongly basic anion exchanger). The flow-through fraction containing neutral and basic compounds was dried and the 15N/14N ratio determined by isotope ratio mass spectrometry. Data were converted to atom % excess 15N as in Reed et al. (1983). Control experiments showed that nitrate was negligible in the basic and neutral fraction and that percentage recovery of amines from the DOWEX column was 97 ± 3.
Bioinformatics Analysis
Hierarchical classification of metabolites was carried out using Cluster and Tree View software (http://rana.lbl.gov/EisenSoftware.ht; Eisen et al. 1998), using a median average-centered protocol.
Acknowledgments
We are grateful to Michael Hodges for the gift of ICDH cDNA and antibodies, and for advice on measuring ICDH activities. G.N. thanks Myroslawa Miginiac-Maslow for advice on extracting and assaying pyridine nucleotides. We also thank Jean Vidal for antibodies against PEPc, Sylvie Ferrario-Méry for the gift of Nia, Nii, and Gln2 cDNAs, and Philippe Chétrit and Bernard Pineau for supplying CMSnad7 seeds.
This work was supported by joint project initiatives funded by the British Council, the U.K. Royal Society, and the French Centre National de la Recherche Scientifique and Ministry of Research. Rothamsted Research receives grant-aided support from the U.K. Biotechnology and Biological Sciences Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066399.
References
- Bai YD, Hajek P, Chomyn A, Chan E, Seo BB, Matsuno-Yagi A, Yagi T, Attardi G (2001) Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J Biol Chem 276: 38808–38813 [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Melo-Olivieira R, Lim J, Coruzzi GM (1998) Arabidopsis gls mutants and distinct ferredoxin-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins AB, Bloom AJ (2004) Oxygen consumption during leaf nitrate assimilation in a C-3 and C-4 plant: the role of mitochondrial respiration. Plant Cell Environ 27: 1537–1545 [Google Scholar]
- Douce R, Manella CA, Bonner WD (1973) The external NADH dehydrogenases of intact plant mitochondria. Biochim Biophys Acta 292: 105–116 [DOI] [PubMed] [Google Scholar]
- Dubois F, Brugiere N, Sangwan RS, Hirel B (1996) Localization of tobacco cytosolic glutamine synthetase enzymes and the corresponding transcripts shows organ- and cell-specific patterns of protein synthesis and gene expression. Plant Mol Biol 31: 803–817 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003. a) Functional mitochondrial complex I is required for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier C, Noctor G, Mathieu C, Chétrit P, Foyer CH, De Paepe R (2003. b) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Ferrario-Méry S, Valadier MH, Hodges M, Hirel B, Foyer CH (2002) Photorespiration-dependent increases in phosphoenolpyruvate carboxylase, isocitrate dehydrogenase and glutamate dehydrogenase in transformed tobacco plants deficient in Fd-GOGAT. Planta 214: 877–886 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Lescure JC, Lefebvre C, Morot-Gaudry JF, Vincentz M, Vaucheret H (1994. b) Adaptations of photosynthetic electron transport, carbon assimilation, and carbon partitioning in transgenic Nicotiana plumbaginifolia plants to changes in nitrate reductase activity. Plant Physiol 104: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G, Lelandais M, Lescure JC, Valadier MH, Boutin JP, Horton P (1994. a) Short-term effects of nitrate, nitrite and ammonium assimilation on photosynthesis, carbon partitioning and protein phosphorylation in maize. Planta 192: 211–220 [Google Scholar]
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Gálvez S, Bismuth E, Sarda C, Gadal P (1994) Purification and characterization of chloroplast NADP-isocitrate dehydrogenase from mixotrophic tobacco cells: comparison with the cytosolic isoenzyme. Plant Physiol 105: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S, Hodges M, Decottignies P, Bismuth E, Lancien M, Sangwan RS, Dubois F, LeMaréchal P, Cretin C, Gadal P (1996) Identification of a tobacco cDNA encoding a cytosolic NADP-isocitrate dehydrogenase. Plant Mol Biol 30: 307–320 [DOI] [PubMed] [Google Scholar]
- Gardeström P, Igamberdiev AU, Raghavendra AS (2002) Mitochondrial functions in the light and significance to carbon-nitrogen interactions. In CH Foyer, G Noctor, eds, Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism. Kluwer Academic Press, Dordrecht, The Netherlands, pp 151–172
- Guo S, Schinner K, Sattelmacher B, Hansen UP (2005) Different apparent CO2 compensation points in nitrate- and ammonium-grown Phaseolus vulgaris and the relationship to non-photorespiratory CO2 evolution. Physiol Plant 123: 288–301 [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chétrit P, Diolez P, Degand H, Boutry M, Vedel F, De Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning I, Heldt HW (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves. Plant Physiol 103: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Peter U, Flügge U-I, Heldt HW (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Lea PJ (2002) The biochemistry, molecular biology and genetic manipulation of primary ammonia assimilation. In CH Foyer, G Noctor, eds, Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism. Kluwer Academic Press, Dordrecht, The Netherlands, pp 151–172
- Hodges M (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J Exp Bot 53: 905–916 [DOI] [PubMed] [Google Scholar]
- Hoefnagel MHN, Atkin OK, Wiskich JT (1998) Interdependence between chloroplasts and mitochondria in the light and the dark. Biochim Biophys Acta 1366: 235–255 [Google Scholar]
- Igamberdiev AU, Bykova NV, Gardeström P (1997) Involvement of cyanide-resistant and rotenone-insensitive pathways of mitochondrial electron transport during oxidation of glycine in higher plants. FEBS Lett 412: 265–269 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Gardeström P (2003) Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim Biophys Acta 1606: 117–125 [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Huber SC (1994) Post-translational regulation of nitrate reductase in higher plants. Plant Physiol 106: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Kandlbinder A, Stoimenova M, Glaab J (2000) Discrepancy between nitrate reduction in intact leaves and nitrate reductase activity in leaf extracts: What limits nitrate reduction in situ? Planta 210: 801–807 [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Stoimenova M, Man HM (2002) What limits nitrate reduction in leaves? In CH Foyer, G Noctor, eds, Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism. Kluwer Academic Press, Dordrecht, The Netherlands, pp 63–70
- Krömer S, Heldt HW (1991) Respiration of pea leaf mitochondria and redox transfer between the mitochondrial and extramitochondrial compartment. Biochim Biophys Acta 1057: 42–50 [Google Scholar]
- Kronenberger J, Lepingle A, Caboche M, Vaucheret H (1993) Cloning and expression of distinct nitrite reductases in tobacco leaves and roots. Mol Gen Genet 236: 203–208 [DOI] [PubMed] [Google Scholar]
- Kruse A, Fieuw S, Heineke D, Müller-Röber B (1998) Antisense inhibition of cytosolic NADP-dependent isocitrate dehydrogenase in transgenic potato plants. Planta 205: 82–91 [Google Scholar]
- Lancien M, Gadal P, Hodges M (1998) Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: evidence for a heteromeric structure by the complementation of yeast mutants. Plant J 16: 325–333 [DOI] [PubMed] [Google Scholar]
- Lejay L, Quilleré I, Roux Y, Tillard P, Cliquet JB, Meyer C, Morot-Gaudry JF, Gojon A (1997) Abolition of post-transcriptional regulation of nitrate reductase partially prevents the decrease in leaf nitrate reduction when photosynthesis is inhibited by CO2 deprivation, but not in darkness. Plant Physiol 115: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalecka AM, Svensson AS, Johansson FI, Agius SC, Johanson U, Brennicke A, Binder S, Rasmusson AG (2003) Arabidopsis genes encoding mitochondrial type II NADP(H) dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol 133: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa Y, Sakai A, Miyagishima S, Takano H, Kawano S, Kuroiwa T (1999) Auxin and cytokinin have opposite effects on amyloplast development and the expression of starch synthesis genes in cultured bright yellow-2 tobacco cells. Plant Physiol 121: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Møller IM, Palmer JM (1982) Direct evidence for the presence of a rotenone-resistant NADH dehydrogenase on the inner surface of the inner membrane of plant mitochondria. Physiol Plant 54: 267–274 [Google Scholar]
- Moore CS, Cook-Johnson RJ, Rudhe C, Whelan J, Day DA, Wiskich JT, Soole KL (2003) Identification of AtNDI1, an internal non-phosphorylating NAD(P)H dehydrogenase in Arabidopsis mitochondria. Plant Physiol 133: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes TF, Plaxton WC (2000) Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures: implications for phosphoenolpyruvate carboxylase regulation during phosphate starvation, and the integration of glycolysis with nitrogen assimilation. Eur J Biochem 267: 4465–4476 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998. a) A re-evaluation of the ATP:NADPH budget during C3 photosynthesis: a potential role for nitrate assimilation and its associated respiratory activity? J Exp Bot 49: 1895–1908 [Google Scholar]
- Noctor G, Foyer CH (1998. b) Simultaneous measurement of foliar glutathione, γ-glutamylcysteine and amino acids by high-performance liquid chromatography: comparison with two other assay methods for glutathione. Anal Biochem 264: 98–110 [DOI] [PubMed] [Google Scholar]
- Novitskaya L, Trevanion S, Driscoll S, Foyer CH, Noctor G (2002) How does photorespiration modulate leaf amino acids? A dual approach through modelling and metabolite analysis. Plant Cell Environ 25: 821–836 [Google Scholar]
- Osuna L, Gonzàlez MC, Cejudo FJ, Vidal J, Echevarria C (1996) In vivo and in vitro phosphorylation of the phosphoenolpyruvate carboxylase from wheat seeds during germination. Plant Physiol 111: 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM (2004) Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol 5: R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Mathieu C, Gerard-Hirne C, De Paepe R, Chétrit P (2005) Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild-type phenotype in the Nicotiana sylvestris CMSII mutant lacking nad7. J Biol Chem 280: 25994–26001 [DOI] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ (2004) Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA 101: 11506–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Reed AJ, Canvin DT, Sherrard JH, Hageman RH (1983) Assimilation of [15N]nitrate and [15N]nitrite in leaves of five plant species under light and dark conditions. Plant Physiol 71: 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, De Kouchkovsky Y (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo BB, Kitajima-Ihara T, Chan EKL, Scheffler IE, Matsuno-Yagi A, Yagi T (1998) Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci USA 95: 9167–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Knowles VL, Plaxton WC (2000) Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures: implications for the integration of glycolysis with nitrogen assimilation. Eur J Biochem 267: 4477–4485 [DOI] [PubMed] [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Taira M, Valtersson U, Burkhardt B, Ludwig RA (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16: 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Robson CA, Yip JYH (2002) Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Quy L, Foyer CH, Champigny ML (1991) Effect of light and nitrate on wheat leaf phosphoenolpyruvate carboxylase activity. Plant Physiol 97: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vincentz M, Kronenberger J, Caboche M, Rouzé P (1989) Molecular cloning and characterization of the homologous genes for nitrate reductase in tobacco. Mol Gen Genet 216: 10–15 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]