Abstract
The role of the eukaryotic release factor 1 (eRF1) in translation termination has previously been established in yeast; however, only limited characterization has been performed on any plant homologs. Here, we demonstrate that cosuppression of eRF1-1 in Arabidopsis (Arabidopsis thaliana) has a profound effect on plant morphology, resulting in what we term the broomhead phenotype. These plants primarily exhibit a reduction in internode elongation causing the formation of a broomhead-like cluster of malformed siliques at the top of the inflorescence stem. Histological analysis of broomhead stems revealed that cells are reduced in height and display ectopic lignification of the phloem cap cells, some phloem sieve cells, and regions of the fascicular cambium, as well as enhanced lignification of the interfascicular fibers. We also show that cell division in the fascicular cambial regions is altered, with the majority of vascular bundles containing cambial cells that are disorganized and possess enlarged nuclei. This is the first attempt at functional characterization of a release factor in vivo in plants and demonstrates the importance of eRF1-1 function in Arabidopsis.
Protein synthesis is an essential process for all living organisms. Translation of mRNA into protein basically consists of three stages: (1) initiation, involving the assembly of the ribosomal subunits at the 5′ end of an mRNA, (2) elongation, the process of tRNA-mediated decoding of the mRNA to form a polypeptide chain, and (3) termination, during which a stop codon signals the end of translation, and the ribosomal subunits dissociate from the mRNA. Each stage requires specific accessory proteins or factors.
The signal that indicates the end of a polypeptide is the presence of an in-frame stop codon (UAA, UGA, or UAG) at the ribosomal A-site. In eukaryotes, termination of protein synthesis is carried out by two classes of release factors: eukaryotic release factor 1 (eRF1) and eRF3 (Frolova et al., 1994; Kisselev and Frolova, 1995; Stansfield et al., 1995; Zhouravleva et al., 1995; Frolova et al., 1996). The stop codon is recognized by eRF1, which binds the ribosome and promotes hydrolysis of the ester bond linking the polypeptide chain with the tRNA in the ribosomal P-site (Tate and Caskey, 1974; Caskey, 1980; Frolova et al., 1994). Because eRF1 has a number of structural and functional similarities to a tRNA molecule, it has been hypothesized to enter the ribosomal A-site and catalyze the termination reaction through tRNA mimicry (Ito et al., 1996; Song et al., 2000). Interaction between eRF1 and eRF3 C-terminal regions results in a conformational rearrangement of either eRF1 or both factors in the complex (Frolova et al., 1998). The function of eRF3 is to stimulate the activity of eRF1 through the hydrolysis of GTP (Zhouravleva et al., 1995), although the presence of eRF3 in human cells has been shown to be nonessential for termination (Le Goff et al., 1997).
Homologs of eRF1 have been identified in a wide range of eukaryotes, including human, frog (Xenopus laevis), nematode (Caenorhabditis elegans), ciliate (Tetrahymena theromophila), and fungus (Podospora anserina). Despite considerable differences between these organisms, a high degree of sequence similarity exists among homologs. Complementation studies have shown that this sequence similarity also translates into functional similarity. Xenopus eRF1 cDNA expressed in a yeast (Saccharomyces cerevisiae) mutant carrying a temperature-sensitive eRF1 allele, eRF1 (ts), rescued the conditional-lethal phenotype of the mutant at 36°C (Tassan et al., 1993). Other studies using human, rabbit, and Syrian hamster eRF1 homologs have also been shown to complement eRF1 (ts) in yeast (Urbero et al., 1997; Karamyshev et al., 1999).
To date, three eRF1 genes (eRF1-1, eRF1-2, and eRF1-3) have been identified in Arabidopsis (Arabidopsis thaliana) by sequence homology (Brown et al., 1995), and all three genes have been shown to possess release factor activity (Chapman and Brown, 2004). In this article, we describe an unusual phenotype resulting from cosuppression of eRF1-1 in Arabidopsis. Cosuppressed plants exhibit a reduction in internode elongation and are affected in radial cell division. Based on these findings, we discuss the implications of down-regulating a release factor and explore the possibility of an alternative function for eRF1-1.
RESULTS
The Broomhead Phenotype
Homozygous Arabidopsis transgenic lines were produced that overexpress an endogenous eRF1-1 cDNA (GenBank accession no. H37361). The eRF1-1 overexpression lines predominantly produced phenotypically normal plants, and we refer to these transgenics as nonphenotype plants. However, a limited percentage of plants (3%–60%) across multiple transgenic lines displayed an abnormal broomhead phenotype (Fig. 1A). The most distinctive feature of the phenotype is the reduction in internode spacing of the inflorescence stems, creating a broomhead-like structure. Compared to wild-type plants, broomhead plants are reduced in height (Table I), produce malformed siliques, and synthesize anthocyanin in nearly all mature plant tissues (Fig. 1, B and C). No significant difference in time to flowering was observed between broomhead and wild-type plants (Table I). However, broomhead plants flowered for longer, and there was an increase in the total number of inflorescence meristems formed and a reduction in number of seeds produced per pod (Table I). During juvenile rosette development, wild-type and broomhead plants are identical in appearance (Fig. 1, D and E). The onset of the broomhead phenotype primarily occurs during reproductive development, although the more severe transgenics exhibit signs of the phenotype in the adult vegetative stage through the production of anthocyanin in the veins of the rosette leaves (Fig. 1, F and G). In less severe transgenics, the onset of the phenotype is preceded by the production of anthocyanin in the inflorescence stem below the bud. As the phenotype progresses, the synthesis of anthocyanin subsequently occurs in the rosette and cauline leaves.
Figure 1.
Wild-type and broomhead phenotypes. A, A mature wild-type plant (left) and a broomhead plant exhibiting a severe phenotype (right). B, Wild-type inflorescence showing regular spacing of flowers along the internode. C, Reduced internode elongation in the broomhead phenotype. Wild-type (D) and broomhead (E) plants are identical during juvenile rosette development. However, at the late adult vegetative stage, broomhead transgenics (G) are distinguishable from wild-type plants (F) by the production of anthocyanin in the veins of the rosette leaves.
Table I.
Developmental characteristics of wild-type and broomhead plants
| Parameter | Wild Typea | Broomheada |
|---|---|---|
| Plant height (cm) | 37.1 ± 1.8 | 18.5 ± 6.5 |
| Days to flowering | 27.9 ± 1.3 | 28.3 ± 1.9 |
| Days to termination of flowering | 47.1 ± 1.2 | 64.5 ± 2.0 |
| Total number of inflorescence meristems | 5.3 ± 1.1 | 15.8 ± 5.2 |
| Number of seeds per pod | 60.8 ± 6.3 | 34.0 ± 23.3 |
Results shown are the average ± sd taken from 17 wild-type and 19 broomhead plants.
Typically, only a small percentage of plants in a specific line show the broomhead phenotype, and not all of these plants display the phenotype to the same degree. Differences in plant height, number of inflorescences produced, total seed production, and the height of the primary bolt can be observed within a single homozygous line (Fig. 2, A–D). The broomhead structures also vary in the degree of clustering, and sometimes broomhead plants revert to producing wild-type inflorescences (Fig. 2, E–H). Plants displaying the most severe phenotype terminate growth of the primary inflorescence at the rosette level. A decrease in phenotype severity is correlated with an increase in the height of the primary inflorescence and the corresponding production of broomhead structures further away from the rosette.
Figure 2.
Variation in the broomhead phenotype at the whole plant level and in the broomhead-like structures. A to D, Broomhead plants showing growth retardation of the primary bolt at the rosette level and the production of numerous auxiliary bolts (A), prominent broomhead-like structures on the inflorescence apices (B), an intermediate severity phenotype and the presence of multiple auxiliary bolts (C), and a weak phenotype with broomhead-like structures appearing later in development (D). E to H, Inflorescence stems showing a severe reduction in internode elongation (E) and a reversion of the phenotype with newly formed inflorescences exhibiting an increase in internode elongation (F–H).
The Broomhead Phenotype Is Caused by eRF1-1 Cosuppression
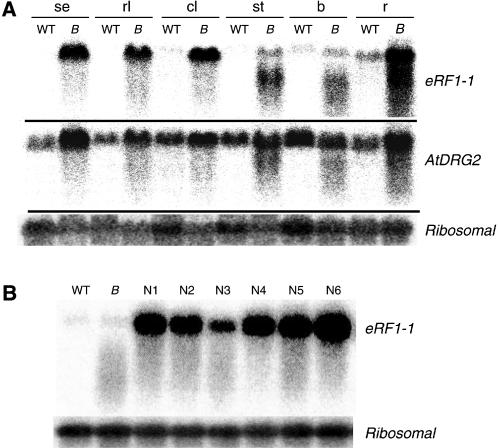
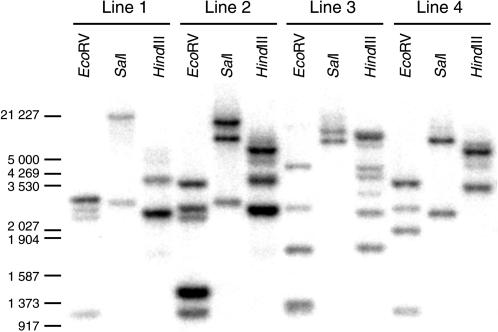
Expression levels of the endogenous eRF1-1 gene as well as the transgene were determined by northern analysis performed on a variety of tissues from both broomhead and wild-type plants. In most tissues examined, eRF1-1 transcript levels were significantly higher in the transgenics than in the wild type (Fig. 3A). However, in the stem and bud tissue where the broomhead phenotype is most visible, significant degradation of the eRF1-1 transcript was observed. To exclude the possibility of poor RNA quality, the northern was reprobed with a second unrelated gene, AtDRG2, encoding a cytoplasmic G-protein (At4g39520). Although some degradation was observed in all broomhead samples, most of the AtDRG2 transcripts remained intact (Fig. 3A). Probing with a ribosomal gene also failed to reveal any significant degradation products. This suggests that the degradation of eRF1-1 in the stem and bud tissues of broomhead plants is transcript specific. The possibility that the phenotype was due to an insertional effect was ruled out by southern analysis on four independent broomhead lines that clearly shows different chromosomal integration sites for each line (Fig. 4). The degradation observed in broomhead plants was specific to plants showing the phenotype as eRF1-1 transcript levels were up-regulated in nonphenotype transgenic plants from six different lines with no significant degradation products observed (Fig. 3B).
Figure 3.
Northern analysis of wild-type, broomhead, and nonphenotype tissues. A, Northern blot of total RNA prepared from different tissues (se, seedlings; rl, rosette leaves; cl, cauline leaves; st, stem; b, buds; r, roots) of wild-type (WT) and broomhead (B) plants. The membrane was hybridized with a radiolabeled eRF1-1 probe and subsequently stripped and hybridized with an AtDRG2 probe. B, Total RNA was isolated from bud tissue of nonphenotype transgenics (N1–N6) in six independent eRF1-1 overexpressor lines. Wild-type and broomhead RNA bud samples were included as controls. A ribosomal probe for both blots was used to verify equal sample loading.
Figure 4.
Southern-blot analysis of four independent broomhead lines. Genomic DNA was extracted from broomhead lines and digested with EcoRI, SalI, or HindIII, transferred to a membrane, and probed with a radiolabeled eRF1-1 probe.
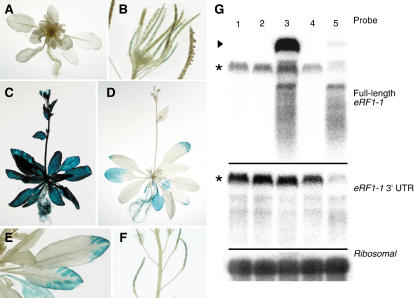
To investigate the link between transgene degradation and the broomhead phenotype, an eRF1-1:β-glucuronidase (GUS) in-frame gene fusion was introduced into Arabidopsis plants under the control of the 35S promoter of cauliflower mosaic virus (CaMV 35S). A number of lines carrying this hybrid construct showed the characteristic broomhead phenotype, and homozygous lines were selected and analyzed in further detail. All severe broomhead phenotype transgenics failed to show GUS activity anywhere in the plant (Fig. 5A). These plants did not bolt above the rosette level, and no viable seed was produced. However, broomhead plants from less severe lines showed GUS activity in the developing seeds (Fig. 5B). This is in agreement with other cosuppression reports where gene silencing is reset during seed development (Hart et al., 1992; Deborne et al., 1994; Kunz et al., 1996; 2001). Staining of the nonphenotype plants was less consistent; approximately half of the plants exhibited extensive GUS staining (Fig. 5C), while the other half remained largely unstained (Fig. 5D). GUS activity in the mostly unstained plants was only apparent in the older rosette leaves either at the tips or in the area surrounding the veins (Fig. 5E) and in developing seeds (Fig. 5F). This GUS staining pattern is suggestive of a downward systemic silencing signal, as has been reported previously during cosuppression of GFP in tobacco (Nicotiana tabacum; Voinnet et al., 1998).
Figure 5.
GUS staining pattern and northern analysis of eRF1-1:GUS transgenic Arabidopsis plants. A, An eRF1-1:GUS transgenic plant exhibiting a severe broomhead phenotype and failing to show any GUS activity. B, A broomhead cluster from a less severe transgenic line revealing GUS activity in developing seeds. C and D, Nonphenotype transgenics showing GUS activity in all plant tissues (C) and minimal GUS activity in rosette leaves (D). E, Close-up of GUS staining pattern in rosette leaves. F, A mostly GUS negative nonphenotype plant exhibiting GUS activity in developing seeds. G, Northern analysis of total RNA isolated from bud tissue of wild-type plants (lane 1), CaMV 35S-GUS (transgenic control, lane 2), and eRF1-1:GUS transgenics (lanes 3, 4, and 5). eRF1-1:GUS transgenics were grouped according to their GUS expression pattern and phenotype: GUS positive nonphenotype plants (lane 3), GUS negative nonphenotype plants (lane 4), and GUS negative broomhead plants (lane 5). The cauline leaves of nonphenotype transgenics were stained at the time of bud collection to ensure the separation into GUS positive and negative samples. The membrane was hybridized with a probe corresponding to the full-length eRF1-1 transcript to allow detection of both the eRF1-1:GUS transgene and the endogenous eRF1-1 gene. The membrane was then stripped and rehybridized to the 3′ untranslated region (UTR) of the eRF1-1 transcript to only enable detection of the endogenous gene. Hybridization with a ribosomal probe was used to show equal loading in each lane. An arrowhead indicates the eRF1-1:GUS transcript and the endogenous eRF1-1 is indicated by an asterisk.
Northern analysis confirmed the presence of the eRF1-1:GUS transcript in the blue nonphenotype plants (Fig. 5G, top panel, lane 3). In contrast, only a faint signal was observed for the transgene in broomhead transgenic bud tissue (Fig. 5G, top panel, lane 5), and no transcript was detected in GUS negative nonphenotype bud tissue (Fig. 5G, top panel, lane 4). When the endogenous levels of eRF1-1 were detected using the 3′ untranslated region of the gene (absent in the eRF1-1:GUS transgene) as a probe, only the broomhead tissue displayed down-regulation of the transcript (Fig. 5G, middle panel). This suggests that posttranscriptional silencing of both the transgene and endogenous gene occurs in broomhead plants, even though down-regulation of eRF1-1 is not complete. The presence of a single band followed by a smear in the blue nonphenotype and broomhead RNA samples (Fig. 5G, top panel, lanes 3 and 5) may be indicative of a cosuppression event, although the blue nonphenotype samples still exhibit high levels of the transgene and wild-type levels of the endogenous eRF1-1 gene. It is possible that cosuppression has been triggered in these plants but has not yet had the chance to progress very far. The absence of the eRF1-1:GUS transgene in GUS negative nonphenotype plants is also consistent with a silencing effect, although it appears that only the transgene is silenced in these plants, as the endogenous eRF1-1 transcript levels were unaffected.
eRF1-1 Family Member Transcripts Are Down-Regulated in Broomhead Tissue
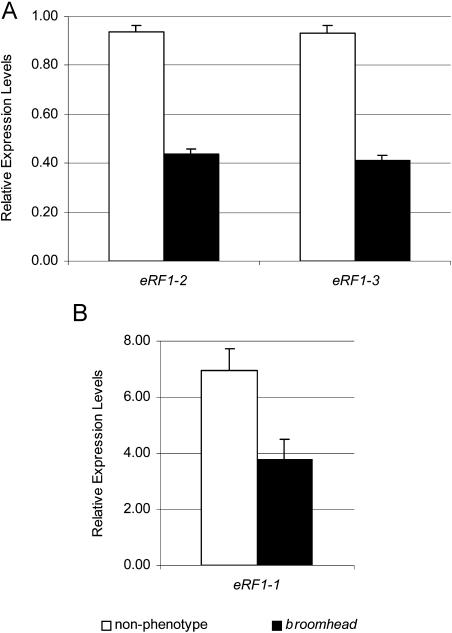
eRF1-1 is part of a gene family that includes two other members, eRF1-2 (At1g12920) and eRF1-3 (At3g26618). At the nucleotide level, these family members show 76% (eRF1-2) and 77% (eRF1-3) identity to eRF1-1 and 87% identity to each other. To determine if either the eRF1-2 or eRF1-3 endogenous gene is affected in broomhead phenotype plants, quantitative real-time PCR was performed on bud tissue with specific primers for each family member and compared to wild-type controls. In wild-type and nonphenotype transgenics, eRF1-2 and eRF1-3 transcript levels were almost identical (Fig. 6A, white bars). However, in broomhead transgenics, eRF1-2 and eRF1-3 transcript levels were down-regulated to approximately half of the wild-type levels (Fig. 6A, black bars). When total eRF1-1 (endogenous gene and transgene) transcript levels were quantified, an average of 7-fold and 3.8-fold overexpression was detected in the nonphenotype and broomhead transgenics, respectively (Fig. 6B). This shows down-regulation of eRF1-1 in broomhead plants relative to nonphenotype plants, although not below wild-type levels. Down-regulation of eRF1-2 and eRF1-3 indicates that the silencing effect is not restricted to eRF1-1 but also affects the family members, most likely due to their high sequence homology.
Figure 6.
Relative expression levels of eRF1-1, eRF1-2, and eRF1-3 family members in nonphenotype and broomhead transgenic plants. eRF1-1, eRF1-2, and eRF1-3 transcript levels were quantified in wild-type, broomhead, and nonphenotype plants using quantitative real-time PCR. The graphs show the relative values, with respect to wild-type plants, of eRF1-2 and eRF1-3 (A) and eRF1-1 (B) in broomhead plants (black bars) and nonphenotype plants (white bars). Values shown are means ± se.
Cellular Architecture of the Broomhead Phenotype
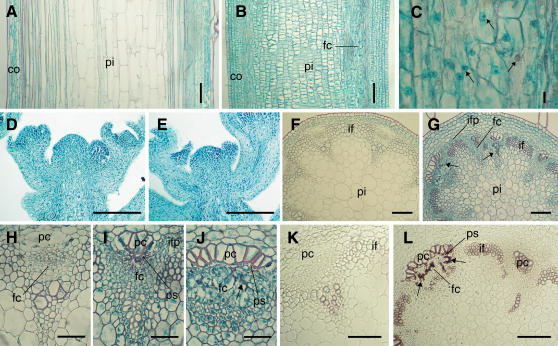
To study the broomhead phenotype at the anatomical level, histological analyses were performed on longitudinal and transverse sections of the broomhead inflorescence stem and the corresponding region in wild-type plants. Microscopic examination revealed that a reduction in cell elongation rather than cell division was responsible for the decrease in plant height. In general, the average pith cell length was reduced approximately 10-fold in broomhead inflorescence stems (17.33 μm) relative to the wild type (174.5 μm; Fig. 7, A and B), and the cells were more cytoplasmically dense. No morphological aberrations were observed in the apical meristem, suggesting that cell division in this region is unaffected (Fig. 7, D and E). However, the majority of cambial cells were disorganized and hypertrophic and contained enlarged nuclei (Fig. 7C). The nuclei were visualized with safranin-O, which stains chromosomes and nucleoli in addition to lignified tissue. Further analysis of transverse stem sections (Fig. 7, F–J) revealed that most fascicular cambial regions exhibit this disorganized cell growth, although a few fascicular cambial regions appeared more active and contained an increased number of cell layers (Fig. 7I). Several layers of interfascicular fiber precursor cells were also identified in broomhead stems that were absent in the wild-type stems (Fig. 7, F and G). In addition, ectopic lignification of the phloem cap cells and some phloem sieve cells as well as enhanced lignification of the interfascicular fibers were observed in broomhead stems (Fig. 7, G, I, and J). Some areas of the fascicular cambium that had become disorganized also showed evidence of lignification (Fig. 7, G and J). To confirm that the safranin-O stain was in fact detecting the presence of ectopic lignin, wild-type and broomhead stem sections were treated with the lignin-specific stain phloroglucinol-HCl (Fig. 7, K and L). As expected, broomhead stem sections stained pink/red around the walls of the phloem cap cells and phloem sieve cells and also in some regions of the disorganized fascicular cambium (Fig. 7L), indicating the presence of lignin. Compared to wild-type sections, enhanced lignification was also observed in the interfascicular fibers. These results confirm the presence of enhanced and ectopically lignified cells in broomhead plants, as observed with the safranin-O stain.
Figure 7.
Anatomy of inflorescence stems and apical meristems of wild-type and broomhead plants. A and B, Longitudinal sections of the mid-region of wild-type (A) and broomhead (B) inflorescence stems. C, Enlargement of the fascicular cambial region in B showing disorganization of the cambium. Enlarged nuclei are marked by arrows. D and E, Longitudinal sections of wild-type (D) and broomhead (E) apical meristems. F and G, Transverse sections of the mid-region of wild-type (F) and broomhead (G) stems. The arrows indicate lignification of the disorganized fascicular cambial region. H and I, Enlargement of the vascular bundles from F and G. H, Wild type; I and J, broomhead vascular bundles. The fascicular cambial region displays active, controlled cell division in I and disorganized cell growth with enlarged nuclei in J. The arrow in J indicates lignification of the disorganized fascicular cambial region. K and L, Phloroglucinol-HCl staining of stem sections through the mid-region of wild-type (K) and broomhead (L) plants. Phloroglucinol-HCl stains lignified tissue red/pink. Ectopic lignification in the cambial region of broomhead plants is indicated by arrows. pi, pith; co, cortex; fc, fascicular cambium; if, interfascicular fibers; pc, phloem cap cells; ps, phloem sieve cells; ifp, interfascicular fiber precursors. Bars = 100 μm in A, B, D to G, K, and L, 50 μm in H to J, and 10 μm in C.
eRF1-1 Expression Is Localized to the Vascular Tissues and Actively Growing Regions
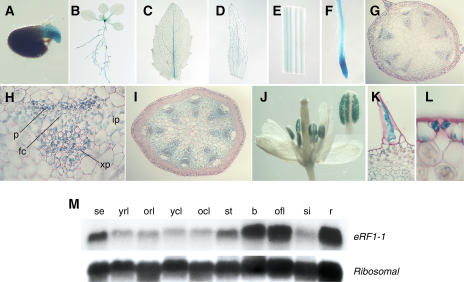
To examine the endogenous expression pattern of eRF1-1, approximately 2 kb of the eRF1-1 promoter was fused to the GUS reporter gene, and a number of Arabidopsis transgenic lines carrying the construct were analyzed. Under normal growth conditions, GUS expression was detected at all developmental stages, including germination and seedling development, and in mature plants (Fig. 8, A–F). The most obvious expression was seen in the vascular tissues at all growth stages, particularly in the phloem and also in the cambium, xylem parenchyma, and interfascicular parenchyma cells (Fig. 8, G and H). However, in the elongating stem region just below the buds, a more uniform pattern of expression was observed across all cells types (Fig. 8I). Other areas of GUS positive staining included pollen grains, trichomes, and guard cells (Fig. 8, J–L). Younger and more actively growing tissues, i.e. germinating seeds, the shoot apical meristem, and root meristem, exhibited the strongest GUS staining (Fig. 8, A, B, and F). These results are consistent with the expression pattern observed in the northern analysis of wild-type plants (Fig. 8M), where high levels of the eRF1-1 transcript were seen in closed and open buds and in the roots. The basal level of eRF1-1 expression in the rosette and cauline leaves also correlates with the low level of GUS activity detected in the leaf veins. Promoter-GUS fusions of the two family members (eRF1-2 and eRF1-3) were also constructed, and Arabidopsis transgenic lines were produced and characterized. The patterns of GUS expression found in these lines were identical to the ones observed for eRF1-1 (data not shown), indicating that there is no complementarity in expression patterns between family members and suggesting a possible redundancy in function.
Figure 8.
eRF1-1 expression pattern as revealed by staining of eRF1-1 promoter-GUS transgenic lines and northern analysis. eRF1-1 expression was detected in germinating seeds (A), the vascular tissues of young seedlings (B), rosette leaf veins (C), cauline leaf veins (D), the inflorescence stem (E), roots (F), pollen grains (J), trichomes (K), and guard cells (L). The shoot apical meristem (B) and the root meristem (F) also show GUS activity. G, Transverse section through the mid-region of an inflorescence stem revealing GUS expression is mainly restricted to the vascular bundles and the interfascicular region. H, Enlargement of a vascular bundle from G, displaying GUS expression in phloem (p), fascicular cambium (fc), xylem parenchyma (xp), and interfascicular parenchyma (ip) cells. I, Transverse section through the elongating region of an inflorescence stem showing a more uniform pattern of expression across all cell types. M, Northern blot of total RNA isolated from different tissues (se, seedlings; yrl, young rosette leaves; orl, old rosette leaves; ycl, young cauline leaves; ocl, old cauline leaves; st, stem; b, buds; ofl, open flowers; si, siliques; r, roots) of wild-type plants. The membrane was hybridized with a radiolabeled eRF1-1 probe and then subsequently stripped and rehybridized with a ribosomal probe to show equal sample loading.
DISCUSSION
The Broomhead Phenotype Is a Result of Gene Silencing
Our evidence indicates that the broomhead phenotype in Arabidopsis lines engineered to overexpress eRF1-1 is a result of gene silencing. First, northern analysis on broomhead plants revealed that eRF1-1 transcripts were partially degraded in tissues showing the phenotype. In contrast, those plants not exhibiting a phenotype displayed overexpression of the gene with no significant degradation products. A similar degradation smear below the full-length transcript on a northern has been reported for transgenic N. benthamiana plants in which the LUCIFERASE gene had been silenced (Kjemtrup et al., 1998). In addition, the occurrence of truncated, polyadenylated, and nonpolyadenylated RNAs corresponding to 5′ and 3′ regions of posttranscriptionally silenced genes has been previously described (Smith et al., 1990; Goodwin et al., 1996; Lee et al., 1997; Metzlaff et al., 1997; Tanzer et al., 1997; Han and Grierson, 2002), suggesting that transgene degradation is gene specific.
Second, all broomhead plants containing the 35S-eRF1-1:GUS gene fusion failed to show GUS activity in any plant tissues, except for the developing seeds (Fig. 5A). This indicates that at least the transgene is being silenced in broomhead phenotype plants, and the GUS activity in the developing seeds represents the post-meiotic resetting of the silencing event and has been described in a number of reports (Hart et al., 1992; Deborne et al., 1994; Kunz et al., 1996, 2001). Northern analysis revealed that in addition to the transgene being down-regulated in broomhead plants, the endogenous eRF1-1 transcript was also down-regulated relative to wild-type plants (Fig. 5G). Taken together, the reduction in transgene and endogenous eRF1-1 transcript levels provides strong evidence for cosuppression.
Third, quantitative real-time results revealed that the transcript levels for family members eRF1-2 and eRF1-3 were also reduced relative to nonphenotype and wild-type plants. It has been suggested that at least 60% to 70% identity in the coding sequence is the threshold necessary to produce posttranscriptional gene silencing of homologous genes (Kunz et al., 2001). As both family members satisfy this requirement with 76% and 77% identity to eRF1-1, it is likely that eRF1-2 and eRF1-3 are also targets of posttranscriptional gene silencing in broomhead plants. Whether down-regulation of all three family members is needed to produce the broomhead phenotype is not yet clear, although considering the functional redundancy among family members, it seems likely that all three eRF1 genes would need to be silenced in order to produce the broomhead phenotype.
The failure to detect overall lower levels of the eRF1-1 transcript in 35S-eRF1-1 broomhead transgenics does not necessarily imply that all the eRF1-1 transcripts are functional and able to be translated. In fact, one of the models proposed to explain how posttranscriptional gene silencing is induced is via the production of aberrant RNA from the homologous transgene. The aberrant RNA is suggested to act as a template for the degradation of itself and other homologous RNAs. Because we were unable to distinguish between the endogenous eRF1-1 transcript and the transgene, due to their similarity in length and sequence, we could not confirm whether or not the endogenous gene was down-regulated. However, based on the findings that (1) a significant portion of the eRF1-1 transcript is already partially degraded in broomhead plants, (2) both family members are down-regulated, and (3) 35S-eRF1-1:GUS broomhead transgenics show down-regulation of the endogenous eRF1-1 transcript, it seems likely that the broomhead phenotype is a result of cosuppression. In addition to the molecular evidence presented so far, the phenotypic characteristics of the broomhead phenotype, such as the variability within and between lines, the low percentage of progeny showing a phenotype, and the differences in the onset of silencing, are also consistent with other reports of transgenic plants showing cosuppresion effects (Elkind et al., 1990; Napoli et al., 1990; de Carvalho et al., 1992; Hart et al., 1992; Zhang et al., 1992; Kunz et al., 1996; Palauqui et al., 1996).
Down-Regulation of eRF1-1 Affects Cell Elongation and Radial Cell Division
By down-regulating an Arabidopsis eukaryotic release factor, we have produced an unexpected yet interesting phenotype that is predominantly defective in inflorescence stem cell elongation. The majority of broomhead transgenics resemble wild-type plants in the vegetative stage of development, but during reproductive growth, the inflorescence stems cease to elongate properly, producing a cluster of mostly malformed siliques. Histological analysis on the broomhead stem revealed that the clustering of siliques is a result of a reduction in cell elongation. In addition, ectopic lignification of the phloem cap cells, some phloem sieve cells, and regions of the disorganized fascicular cambium was observed as well as enhanced lignification of the interfascicular fibers. This is in contrast to what occurs in wild-type plants, where lignin deposition is limited to xylem and fully elongated interfascicular parenchyma cells (Dharmawardhana et al., 1992; Altamura et al., 2001).
Several Arabidopsis mutants that are defective in cell expansion and display ectopic lignification have been reported (Caño-Delgado et al., 2000; Zhong et al., 2000). The majority contains disruptions in genes involved in cell wall synthesis, such as cellulose synthase (CESA1 and CESA3), endo-1,4-β-glucanase, and a chitinase-like gene (Arioli et al., 1998; Nicol et al., 1998; Zhong et al., 2000; Ellis et al., 2002; Caño-Delgado et al., 2003), although disruption of the C-subunit of a putative vacuolar ATPase (Schumacher et al., 1999) and overexpression of AtMYB61, encoding for a transcription factor (Newman et al., 2004), also produced similar phenotypes. As a number of mutants (eli1-1, rsw1-1, and det3) exhibited a reduction in cellulose levels as well as increased levels of defense-related genes, such as VEGETATIVE STORAGE PROTEIN 1 and PLANT DEFENSIN 1.2, it was suggested that changes in cell wall integrity can stimulate lignification and defense responses through jasmonate and ethylene-mediated pathways (Ellis et al., 2002; Caño-Delgado et al., 2003). Despite not having a described role in cell wall synthesis, eRF1-1 may indirectly affect this process when down-regulated. However, it seems unusual that only specific cell types are ectopically lignified in broomhead plants, whereas the majority of ectopic lignification mutants, e.g. elp1, eli1, and det3, exhibit a more random pattern of lignification in the pith and surrounding areas. A possible explanation for this may be that certain cells in broomhead plants are more severely affected by a decrease in eRF1-1 levels than other cell types and, therefore, only stimulate a defense response, resulting in lignin deposition in these cells. The fact that the pith cells are not lignified, yet are inhibited in elongation, may be a consequence of lignification of the surrounding tissues.
In addition to the cell elongation defect, cell division is also affected in broomhead plants. In some fascicular cambial regions, cell division is more prolific, although in the majority of vascular bundles, the cambium exhibits disorganized cell growth. This disorganization of the cambium is also associated with a corresponding enlargement of the nuclei, suggesting that the cell cycle has been disrupted such that DNA replication has become uncoupled from cell division. However, it is also possible that the nuclei appear enlarged because they are decondensed and actively transcribing. The fact that some fascicular cambial regions appear to be more active, while others exhibit disorganized cell growth and contain enlarged nuclei, even within the same stem section, may be directly related to the level of eRF1-1 down-regulation.
Endogenous Expression of eRF1-1
The eRF1-1 promoter-GUS experiments revealed that the expression pattern of eRF1-1 is primarily observed in the vascular tissues and the actively growing and elongating zones of Arabidopsis plants. The presence of GUS activity in the meristems and elongating zones appears consistent for a protein involved in translation, as it would be expected that protein synthesis is up-regulated to accommodate for new cell growth in these tissues. However, the observation that GUS activity was mostly restricted to the vascular tissues in the inflorescence stems and leaves was unexpected, although this may be due to a lack of sensitivity in the GUS staining method. Previously it has been shown that a number of promoter-GUS transgenics exhibit preferential staining in the vascular tissues (Gallagher, 1992). This has been attributed to the presence of smaller, more cytoplasmically dense cells, such as the cambial cells and companion cells in the phloem, which would concentrate the GUS stain, thus leading to the appearance of a stronger expression pattern. However, it is interesting that the areas in the stem that are most affected in broomhead plants, i.e. the vascular bundles and interfascicular region, are also the areas where high levels of eRF1-1 are observed, suggesting that the vascular staining observed in our GUS studies does reflect the true expression pattern of eRF1-1. Furthermore, it would make sense that a reduction in eRF1-1 levels would have the most dramatic effect on cells where it is normally expressed, therefore leading the morphological aberrations described earlier in the vascular tissues and interfascicular region. The finding that the cells in the apical meristem stained positive for GUS activity but did not appear to be affected in broomhead plants may reflect the fact that the apical meristem often escapes the effects of posttranscriptional gene silencing (Voinnet et al., 1998; Foster et al., 2002).
The Broomhead Phenotype: Disruption of Release Factor Activity?
It has been previously established that a reduction in eRF1 protein levels decreases the efficiency of translation termination and therefore causes an increase in the translational readthrough of proteins (Stansfield et al., 1996). In yeast cells, complete deprivation of eRF1 results in a loss of cell viability (Inge-Vechtomov et al., 2003). Based on these data and the fact that all three Arabidopsis eRF1s have been shown to possess release factor activity (Chapman and Brown, 2004), it seems reasonable to suggest that the broomhead phenotype is due to a decrease in the efficiency of translation termination. However, it is interesting to note that eRF1 has also been suggested to perform additional functions, at least in yeast, to that of translation termination. For instance, analysis of multiprotein complexes has revealed that eRF1 is not only associated with other translational components but is also found in complexes containing nontranslational proteins (Gavin et al., 2002; Ho et al., 2002). In addition, eRF1 has also been shown to directly interact with other proteins, some of which have functions outside of translation and could implicate eRF1 in additional processes. These include protein phosphatase 2A, Upf1, Itt1, Mtt1, and a myosin light chain protein (Andjelković et al., 1996; Czaplinski et al., 1998, 2000; Urakov et al., 2001; Valouev et al., 2004).
Further support for an alternative function comes from the various yeast eRF1 mutants that are recessive and have allele-specific pleiotropic effects such as temperature sensitivity (Inge-Vechtomov and Andrianova, 1975; Ter-Avanesyan et al., 1982a), osmotic sensitivity (Ter-Avanesyan et al., 1982a), and either partial or complete respiration deficiency (Ter-Avanesyan et al., 1982b). A role for eRF1 in yeast cell cycle regulation and cytoskeleton organization has also been suggested by several authors (Tikhomirova and Inge-Vechtomov, 1996; Borkhsenius and Inge-Vechtomov, 1997; Borchsenius et al., 2000; Stevenson et al., 2001), and inactivation of eRF1 has been associated with myelodysplastic syndrome and acute myeloid leukaemia in humans (Guenet et al., 2000; Dubourg et al., 2002). However, none of these studies were able to conclude whether the phenotypes were due to increased translational readthrough or disruption of eRF1 in an unrelated process.
So far, the most compelling evidence for an additional role for eRF1 has come from a study by Valouev et al. (2002), which demonstrated that the phenotype of an eRF1 yeast mutant was independent of its function in translation. In this study, it was shown that repression of eRF1 resulted in an increase in the level of non-sense codon readthrough and the accumulation of unbudded cells with 2C and higher DNA content, indicating an uncoupling of DNA replication from budding. However, repression of eRF3 (also necessary for the termination reaction) caused a different effect on cell morphology at similar levels of non-sense codon readthrough. This implies that the cellular phenotype of the yeast eRF1 mutant was not due to impairment of the termination process and therefore suggests a role for eRF1 in cytoskeleton organization and cell cycle regulation. The similarity between this study and our observations, which showed enlarged nuclei in the disorganized fascicular cambial cells, further adds to the possibility that eRF1 may function in other processes.
In conclusion, we have established that cosuppression of eRF1-1 in Arabidopsis has a dramatic effect on plant morphology, ultimately causing a reduction in internode elongation. The broomhead phenotype is the first report of any functional characterization of a release factor in vivo in Arabidopsis. However, the specific effects down-regulating eRF1 has on certain cell types, e.g. the cambial cells and the ectopically lignified cells, allow for speculation as to whether Arabidopsis eRF1-1 also functions in additional processes to that of translation termination. Examining the subcellular localization of eRF1-1, measuring the ploidy levels in broomhead plants, and screening for interacting proteins in Arabidopsis will provide further insight into the functions of eRF1-1 and may establish whether or not translation termination is its only role.
MATERIALS AND METHODS
Plant Material and Growth Conditions
For all phenotypic analyses, Arabidopsis (Arabidopsis thaliana) seeds were sown on soil and stratified for 2 to 5 d at 4°C. Trays of seed were then transferred to a temperature-controlled growth room and grown at 21°C under 50 μE/m2/s of long-day light (16/8 light/dark).
Plasmid Constructs
The eRF1-1 overexpression construct was produced by inserting the full-length eRF1-1 cDNA (GenBank accession no. H37361) into the EcoRI and BamHI sites of the binary vector pSOV2 (Mylne and Botella, 1998). The eRF1-1:GUS fusion was constructed by amplifying the coding sequence of eRF1-1 using the primers 5′-CGTCTAGAATGGGAGACAAAAACGATGAC-3′ and 5′-CGGGATCCCCTTCCGAATCATCATCAAGAGC-3′ and cloning the fragment in frame with the GUS coding region into the XbaI and BamHI sites of the binary vector pAOV-intronGUS. In the resulting construct, eRF1-1 is fused to the N-terminal region of GUS. pAOV-intronGUS is derived from the vector pAOV (Mylne and Botella, 1998) and has the GUS gene containing an intron as described by Ohta et al. (1990) cloned into the BamHI and SacI sites of the vector.
eRF1-1, eRF1-2, and eRF1-3 promoter-GUS fusions were generated by cloning approximately 2 kb of sequence upstream of each of the translational start sites into the promoterless vector pAOV-intronGUSΔ35S. This vector is based on pAOV-intronGUS but lacks the CaMV 35S promoter. The primers used to clone all three promoter regions are as follows: eRF1-1, 5′-CGCTCGAGGGCTGATATGCGACAACCATAAAG-3′ and 5′-CGAAGCTTCTTAACAATAGAACAAAAACC-3′; eRF1-2, 5′-CGAAGCTTCTTAGTTTATTGACCAGCCAGTTG-3′ and 5′-CGAAGCTTTTTTAGCTTCTCTCGAGGTCG-3′; eRF1-3, 5′-CGCTCGAGCTGGCTCACCGAAGTTCGCGTAAG-3′ and 5′-CGATCGATGTTTTTCCTCCTTCAAGC-3′.
Wild-type Arabidopsis plants (ecotype Columbia) were transformed using the floral dip method (Clough and Bent, 1998) and selection of T1 seeds was achieved by spraying seedlings with 0.4% BASTA (Hoechst).
RNA Analysis
Tissue samples were collected and immediately frozen in liquid nitrogen. RNA was extracted as described by Etheridge et al. (1999). Northern analysis was performed with 10 μg (per lane) of total RNA using standard procedures (Sambrook et al., 1989). After the initial hybridization, all blots were stripped and rehybridized with a wheat ribosomal probe (Gerlach and Bedbrook, 1979) to test for equal loading. Hybridization was performed in Church buffer (Church and Gilbert, 1984) at 65°C overnight, and the blots were washed at high stringency in 2× SSC, 0.1% SDS for 15 min at room temperature, then in 2× SSC, 0.1% SDS for 15 min at 55°C, and finally in 0.2× SSC, 0.1% SDS for 15 min at 55°C.
Southern Analysis
Genomic DNA was isolated from 2-week-old broomhead seedlings as previously described (Etheridge et al., 1999). Three micrograms of DNA was digested with EcoRV, SalI, or HindIII, and samples were electrophoresed on a 0.8% agarose gel, then transferred to a Hybond-N+ nylon membrane (Amersham Biosciences) by capillary transfer (Sambrook et al., 1989). The blot was then hybridized with an eRF1-1 radiolabeled DNA probe and washed at high stringency as described for the northern analysis.
Quantitative Real-Time PCR
Three biological replicates were performed on three independent broomhead lines. Total RNA was extracted from bud tissue (Promega SV Total RNA Extraction kit), and 1 μg was used to synthesize cDNA using reverse transcriptase and random hexamers in a reaction volume of 20 μL. Prior to quantitative PCR, cDNA samples were diluted 5-fold in water and then amplified using the ABI PRISM 7700 sequence detector and SYBR Green Master mix (Applied Biosystems). Gene-specific primers were designed to regions of low homology between the gene family members using Primer Express 1.5 software (Applied Biosystems). Primers used and locus identifiers are as follows: eRF1-1 forward, 5′-ACTGCCTTTGATTCCGAGGA-3′; eRF1-1 reverse, 5′-GCGATGGTGAGGATTTGATTG-3′; eRF1-2 forward, 5′-TTGATTTCTCCTTCTCCATCTTCG-3′; eRF1-2 reverse, 5′-CAGATCTCGATGTTCGTATCCG-3′; eRF1-3 forward, 5′-TTAAAGAACAACACGACCGGAGA-3′; eRF1-3 reverse, 5′-GCATCATGGAAATTGCTTTGGT-3′. Locus IDs are as follows: eRF1-1 (At5g47880); eRF1-2 (At1g12920); eRF1-3 (At3g26618). Expression detected from three β-actin genes of Arabidopsis, β-actin-2 (At3g18780), β-actin-7 (At5g09810), and β-actin-8 (At1g49240) with universal actin forward primer 5′-AGTGGTCGTACAACCGGTATTGT-3′ and specific reverse primers 5′-GATGGCATGGAGGAAGAGAGAAAC-3′, 5′-GAGGAAGAGCATTCCCCTCGTA-3′, and 5′-GAGGATAGCATGTGGAACTGAGAA-3′, respec-tively, were used as combined internal standards to normalize small differences in template amounts.
Histological Analysis
Stem sections from areas of reduced internode elongation in broomhead plants and the corresponding regions in wild-type plants were selected and fixed in ethanol:acetic acid (3:1) overnight. Samples were then dehydrated through a tertiary-butyl alcohol series, and flakes of Paraplast plus (Sigma-Aldrich) were added to the final step. Once the flakes dissolved at room temperature, liquefied Paraplast plus was added, and samples were transferred to an oven at 58°C. The medium was replaced after approximately 8 h, and samples were put back in the oven overnight. Stem samples were positioned in Paraplast plus, and microtome sections of 10 μm were cut, mounted on slides, and stained with safranin-O and fast green. Stained sections were photographed under bright-field illumination. To specifically detect the presence of lignin, deparaffinized and rehydrated tissue sections were stained for 5 min with phloroglucinol (2.5% [w/v] phloroglucinol in 6 n HCl) and rinsed in water. The slides were then mounted in 50% glycerol, 6 n HCl and visualized under a light microscope.
Detection of GUS Activity
Whole plant specimens and small tissue sections were incubated at room temperature in chloroform for 15 min, then rinsed approximately two to three times in rinse solution (staining solution lacking 5-bromo-4-chloro-3-indolyl-β-d glucuronic acid). GUS stain (1 mm 5-bromo-4-chloro-3-indolyl-β-d glucuronic acid, 4 mm potassium ferro-/ferricyanide, 0.1% Silwet L-77, and 100 mm sodium phosphate buffer, pH 7.0) was added and allowed to vacuum infiltrate the tissue for 15 min, twice. Samples were then incubated at 37°C overnight and destained in 70% ethanol the following day. Stem tissue for sectioning was transferred to ethanol:acetic acid (3:1) after staining in the GUS solution and then prepared in the same way as samples described for histological analysis. Microtome sections of 15 μm were mounted on slides and stained with safranin-O.
Acknowledgments
We thank Dr. Richard Moyle and Professor Robert Birch for critically commenting on the manuscript and past and present members of the Plant Genetic Engineering Laboratory for helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062695.
References
- Altamura MM, Possenti M, Matteucci A, Baima S, Ruberti I, Morelli G (2001) Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytol 151: 381–389 [Google Scholar]
- Andjelković N, Zolnierowicz S, Van Hoof C, Goris J, Hemmings BA (1996) The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J 15: 7156–7167 [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Tchourikova AA, Inge-Vechtomov SG (2000) Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr Genet 37: 285–291 [DOI] [PubMed] [Google Scholar]
- Borkhsenius AS, Inge-Vechtomov SG (1997) The role of SUP35 and SUP45 genes in controlling Saccharomyces cell cycle. Dokl Akad Nauk 353: 553–556 [PubMed] [Google Scholar]
- Brown CM, Quigley FR, Miller WA (1995) Three eukaryotic release factor one (eRF1) homologs from Arabidopsis thaliana Columbia (accession nos. U40217, U40218, X69374, X69375) (PGR95–123). Plant Physiol 110: 336 [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado AI, Metzlaff K, Bevan MW (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405 [DOI] [PubMed] [Google Scholar]
- Caskey CT (1980) Peptide-chain termination. Trends Biochem Sci 5: 234–237 [Google Scholar]
- Chapman B, Brown C (2004) Translation termination in Arabidopsis thaliana: characterisation of three versions of release factor 1. Gene 341: 219–225 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Majlesi N, Banerjee T, Peltz SW (2000) Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA 6: 730–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborne FD, Vincentz M, Chupeau Y, Vaucheret H (1994) Co-suppression of nitrate reductase host genes and transgenes in transgenic tobacco plants. Mol Gen Genet 243: 613–621 [DOI] [PubMed] [Google Scholar]
- de Carvalho F, Gheysen G, Kushnir S, Van Montagu M, Inzé D, Castresana C (1992) Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J 11: 2595–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE (1992) Characterization of vascular lignification in Arabidopsis thaliana. Can J Bot 70: 2238–2244 [Google Scholar]
- Dubourg C, Toutain B, Hèlias C, Henry C, Lessard M, Le Gall JT, Le Treut A, Guenet L (2002) Evaluation of ETF1/eRF1, mapping to 5q31, as a candidate myeloid tumor suppressor gene. Cancer Genet Cytogenet 134: 33–37 [DOI] [PubMed] [Google Scholar]
- Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87: 9057–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge N, Trusov Y, Verbelen JP, Botella JR (1999) Characterization of ATDRG1, a member of a new class of GTP-binding proteins in plants. Plant Mol Biol 39: 1113–1126 [DOI] [PubMed] [Google Scholar]
- Foster TM, Lough TJ, Emerson SJ, Lee RH, Bowman JL, Forster RL, Lucas WJ (2002) A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14: 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis JE, Philippe M, Justesen J, Kisselev L (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703 [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341 [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Simonsen JL, Merkulova TI, Litvinov DY, Martensen PM, Rechinsky VO, Camonis JH, Kisselev LL, Justesen J (1998) Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus insect cells and complex formation between the factors. Eur J Biochem 256: 36–44 [DOI] [PubMed] [Google Scholar]
- Gallagher SR (1992) GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego
- Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7: 1869–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J, Chapman K, Swaney S, Parks TD, Wernsman EA, Dougherty WG (1996) Genetic and biochemical dissection of transgenic RNA-mediated virus resistance. Plant Cell 8: 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenet L, Henry C, Toutain B, Dubourg C, Le Gall JY, David V, Le Treut A (2000) Eukaryotic translation termination factor gene (ETF1/eRF1) maps at D5S500 in a commonly deleted region of chromosome 5q31 in malignant myeloid diseases. Cytogenet Cell Genet 88: 82–86 [DOI] [PubMed] [Google Scholar]
- Han YH, Grierson D (2002) Relationship between small antisense RNAs and aberrant RNAs associated with sense transgene mediated gene silencing in tomato. Plant J 29: 509–519 [DOI] [PubMed] [Google Scholar]
- Hart CM, Fischer B, Neuhaus JM, Meins F (1992) Regulated inactivation of homologous gene expression in transgenic Nicotiana sylvestris plants containing a defense-related tobacco chitinase gene. Mol Gen Genet 235: 179–188 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov S, Zhouravleva G, Philippe M (2003) Eukaryotic release factors (eRFs) history. Biol Cell 95: 195–209 [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov SG, Andrianova VM (1975) A new type of supersuppressor in yeast. In N Dubinin, D Goldfarb, eds, Molecular Mechanisms of Genetic Processes. Wiley, New York, pp 181–186
- Ito K, Ebihara K, Uno M, Nakamura Y (1996) Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc Natl Acad Sci USA 93: 5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev AL, Karamysheva ZN, Ito K, Matsufuji S, Nakamura Y (1999) Overexpression and purification of recombinant eRF1 proteins of rabbit and Tetrahymena thermophila. Biochemistry (Mosc) 64: 1391–1400 [PubMed] [Google Scholar]
- Kisselev LL, Frolova LY (1995) Termination of translation in eukaryotes. Biochem Cell Biol 73: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, Thompson WF, Robertson D (1998) Gene silencing from plant DNA carried by a Geminivirus. Plant J 14: 91–100 [DOI] [PubMed] [Google Scholar]
- Kunz C, Schöb H, Leubner-Metzger G, Glazov E, Meins F (2001) β-1,3-Glucanase and chitinase transgenes in hybrids show distinctive and independent patterns of posttranscriptional gene silencing. Planta 212: 243–249 [DOI] [PubMed] [Google Scholar]
- Kunz C, Schöb H, Stam M, Kooter JM, Meins F (1996) Developmentally regulated silencing and reactivation of tobacco chitinase transgene expression. Plant J 10: 437–450 [Google Scholar]
- Lee KY, Baden C, Howie WJ, Bedbrook J, Dunsmuir P (1997) Post-transcriptional gene silencing of ACC synthase in tomato results from cytoplasmic RNA degradation. Plant J 12: 1127–1137 [Google Scholar]
- Le Goff X, Philippe M, Jean-Jean O (1997) Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol Cell Biol 17: 3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzlaff M, O'Dell M, Cluster PD, Flavell RB (1997) RNA-mediated RNA degradation and chalcone synthase A silencing in Petunia. Cell 88: 845–854 [DOI] [PubMed] [Google Scholar]
- Mylne J, Botella JR (1998) Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol Biol Rep 16: 257–262 [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM (2004) Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 37: 239–250 [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a beta-glucoronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31: 805–813 [Google Scholar]
- Palauqui J-C, Elmayan T, de Borne FD, Crété P, Charles C, Vaucheret H (1996) Frequencies, timing, and spatial patterns of co-suppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol 112: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Bird CR, Ray J, Schuch W, Grierson D (1990) Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet 224: 477–481 [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D (2000) The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Stansfield I, Eurwilaichitr L, Akhmaloka, Tuite MF (1996) Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol Microbiol 20: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14: 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson LF, Kennedy BK, Harlow E (2001) A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA 98: 3946–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer MM, Thompson WF, Law MD, Wernsman EA, Uknes S (1997) Characterization of post-transcriptionally suppressed transgene expression that confers resistance to tobacco etch virus infection in tobacco. Plant Cell 9: 1411–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan JP, Le Guellec K, Kress M, Faure M, Camonis J, Jacquet M, Philippe M (1993) In Xenopus laevis, the product of a developmentally regulated mRNA is structurally and functionally homologous to a Saccharomyces cerevisiae protein involved in translation fidelity. Mol Cell Biol 13: 2815–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate WP, Caskey CT (1974) Mechanism of peptide chain termination. Mol Cell Biochem 5: 115–126 [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Inge-Vechtomov SG, Shubochkina EA (1982. a) Expression of sup1 and sup2 suppressor mutations in the yeast S. cerevisiae. Influence of hypertonic conditions. Genetika 18: 215–222 [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Zimmermann J, Inge-Vechtomov SG, Sudarikov AB, Smirnov VN, Surguchov AP (1982. b) Ribosomal recessive suppressors cause a respiratory deficiency in yeast Saccharomyces cerevisiae. Mol Gen Genet 185: 319–323 [DOI] [PubMed] [Google Scholar]
- Tikhomirova VL, Inge-Vechtomov SG (1996) Sensitivity of sup35 and sup45 suppressor mutants in Saccharomyces cerevisiae to the anti-microtubule drug benomyl. Curr Genet 30: 44–49 [DOI] [PubMed] [Google Scholar]
- Urakov VN, Valouev IA, Lewitin EI, Paushkin SV, Kosorukov VS, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD (2001) Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol Biol 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbero B, Eurwilaichitr L, Stansfield I, Tassan JP, Le Goff X, Kress M, Tuite MF (1997) Expression of the release factor eRF1 (Sup45p) gene of higher eukaryotes in yeast and mammalian tissues. Biochimie 79: 27–36 [DOI] [PubMed] [Google Scholar]
- Valouev IA, Kushnirov VV, Ter-Avanesyan MD (2002) Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil Cytoskeleton 52: 161–173 [DOI] [PubMed] [Google Scholar]
- Valouev IA, Urakov VN, Kochneva-Pervukhova NV, Smirnov VN, Ter-Avanesyan MD (2004) Translation termination factors function outside of translation: Yeast eRF1 interacts with myosin light chain, Mlc1p, to effect cytokinesis. Mol Microbiol 53: 687–696 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95: 177–187 [DOI] [PubMed] [Google Scholar]
- Zhang H, Scheirer DC, Fowle WH, Goodman HM (1992) Expression of antisense or sense RNA of an ankyrin repeat-containing gene blocks chloroplast differentiation in Arabidopsis. Plant Cell 4: 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ripperger A, Ye Z-H (2000) Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol 123: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov SG, Kisselev L, Philippe M (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14: 4065–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]