Abstract
The ospC genes of 20 southern Borrelia strains were sequenced. The strains consisted of B. burgdorferi sensu stricto, B. andersonii, B. bissettii, one undescribed genospecies, MI-8, and one probably new Borrelia species, TXW-1. A high degree of similarity exists between B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii. Lateral transfers of the ospC gene probably occurred between B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii. Internal gene recombination appears to occur among them. The highest degree of genetic diversity among them was observed in the two variable domains (V1 and V2), semivariable domain (SV), and the species-specific epitopes (between amino acids 28 and 31). Differences in ospC sequences among southern strains reflect diversity at the strain and genospecies levels. MI-8, which was recognized as an undescribed genospecies in our previous reports, remains distinguishable in our current analysis of ospC genes and is distinct from B. burgdorferi sensu stricto. Interestingly, another undescribed southern isolate, TXW-1, was not amplified under various PCR conditions. Compared to European B. burgdorferi sensu stricto strains, American B. burgdorferi sensu stricto strains show greater genetic heterogeneity. Southern B. burgdorferi sensu stricto, B. andersonii, and B. bissettii isolates were intermixed with each other in the phylogenetic trees. In the derived trees in our work, at least one southeastern strain of B. burgdorferi, MI-2, most closely aligns with a so-called invasive cluster that possesses many proven human-invasive strains. Transmission experiments show that MI-2 and the strains in this group of southern spirochetes are able to infect mice and hamsters and that the typical vector of Lyme disease, Ixodes scapularis, can acquire the spirochetes from infected mammals. Currently, strain MI-2 appears to be the only southern isolate among the 20 we analyzed that clusters with an OspC invasive group and thus might be invasive for humans.
Lyme disease is the most common vector-borne disease in humans in North America and Eurasia. There were 122,651 Lyme disease cases reported from 50 states and the District of Columbia from 1990 through 1999 in the United States (17), and about 50,000 cases are estimated to occur in Europe annually (40). Human Lyme disease may involve multiple organs or tissues, resulting in skin, cardiac, neurological, and musculoskeletal disorders. Multiple erythema migrans and Lyme arthritis are more common in the United States than in Europe, whereas neuroborreliosis occurs frequently in European patients. Borrelial lymphocytoma and acrodermatitis chronica atrophicans are well documented in European Lyme disease patients but rarely recognized among Lyme disease patients in the United States.
The causative agent of Lyme disease, Borrelia burgdorferi sensu lato, is genetically highly divergent (8, 30, 35, 36, 40, 49; T. Lin, J. H. Oliver, Jr., T. M. Kollars, Jr., and K. L. Clark, Proc. VIII Int. Conf. Lyme Borreliosis Other Emerging Tick-Borne Dis., p. 8, 1999). Different Borrelia genospecies sometimes have been associated with distinct clinical manifestations of Lyme disease, although there is often an overlap of symptoms. Lyme arthritis is often related to B. burgdorferi sensu stricto, neuroborreliosis is often associated with B. garinii, and acrodermatitis chronica atrophicans may occur in Lyme disease patients infected with B. afzelii (5, 8, 13, 39, 47, 66).
Eleven genospecies or genomic groups within the B. burgdorferi sensu lato complex have been described based on their phenotypic and genetic characteristics since the discovery of B. burgdorferi in 1982 (11) and its subsequent description (32). Three of them, B. burgdorferi sensu stricto (8), B. andersonii (6, 37, 47), and B. bissettii (6, 47, 48), have been identified in the United States. B. burgdorferi sensu stricto is widely distributed in the northeastern, southeastern, mid-Atlantic, and north central states and northern California (17, 35; Lin et al., Proc. VIII Int. Conf. Lyme Borreliosis Other Emerg. Tick-Borne Dis.). It is the only genospecies of B. burgdorferi sensu lato that has been cultured from humans with Lyme borreliosis in North America. B. andersonii and B. bissettii were isolated mainly from ticks and small mammals from New York (2, 4), California (48), Colorado (53), and the southeastern United States (35, 39; Lin et al., Proc. VIII Int. Conf. Lyme Borreliosis Other Emerg. Tick-Borne Dis.). B. andersonii occurs in the eastern half of the United States and appears to exist primarily in an enzootic cycle involving cottontail rabbits and Ixodes dentatus. B. bissettii occurs in the western and southern United States but infrequently in the northern region. It appears to be maintained in several enzootic transmission cycles in California and the southeastern United States and involves several tick species, including Ixodes spinipalpis and the human-biting I. pacificus in the west, and Ixodes minor and the human-biting I. scapularis in the eastern half of the United States (10, 42; J. H. Oliver, Jr., Proc. II Int. Symp. Lyme Dis. Emerg. Re-emerg. Dis., p. 83-94, 1997).
Various phenotypic and genotypic techniques have been used to study variability and to identify genospecies in the B. burgdorferi sensu lato complex. Analysis of the ospC gene has yielded valuable information. The ospC gene, located in the 26-kb circular plasmid, has a high degree of heterogeneity (30, 51, 68). Its diversity has been described among and within genospecies (29, 30, 68, 70), and lateral transfer of the ospC gene and internal gene recombination are reported to increase the variability among ospC sequences (20, 30, 38). Interestingly, the divergent sequences of ospC and the deduced amino acid sequences reported from B. burgdorferi sensu lato have been correlated with the distribution of the genospecies (29, 68). A species-specific motif was found in the conserved amino terminus of the OspC protein (30, 70), and intergenic transfer and internal recombination have not been reported in the motif region except for a few B. valaisiana isolates, which might have exchanged a complete ospC gene from B. garinii or B. afzelii (63).
Sequence analysis of ospC and its deduced amino acids appears to be a useful tool for identification of Borrelia species and for evaluating the divergence between and within Borrelia species but not for assignment of Borrelia genospecies. We used this approach to analyze some representative southern Borrelia strains and to evaluate relationships among them and other genospecies.
MATERIALS AND METHODS
Spirochete isolates and culture conditions.
The 20 Borrelia strains described in this study were isolated from ticks (I. scapularis, I. dentatus, I. minor, or Dermacentor variabilis) and animal reservoirs (cotton mouse [Peromyscus gossypinus], cotton rat [Sigmodon hispidus], and eastern woodrat [Neotoma floridana]) from Missouri, Georgia, Florida, South Carolina, and Texas. All analyzed isolates were third passage. Table 1 lists all Borrelia strains that were investigated and their origins. The cultures were incubated at 34°C for 1 to 2 weeks (to the stationary phase of growth) and checked under dark-field microscopy to ensure purification, and cells were counted before they were harvested.
TABLE 1.
Origins of B. burgdorferi sensu lato strains in this study
| Isolate | Host | Source | Location | Passage no. | Genospecies |
|---|---|---|---|---|---|
| MI-2 | Peromyscus gossypinus | Bladder, ear clip | Merritt Island, Brevard County, Fla. | 3 | Borrelia burgdorferi sensu stricto |
| MI-5 | Sigmodon hispidus | Bladder, ear clip | Merritt Island, Brevard County, Fla. | 3 | Borrelia burgdorferi sensu stricto |
| SI-1 | Peromyscus gossypinus | Bladder | Sapelo Island, McIntosh County, Ga. | 3 | Borrelia burgdorferi sensu stricto |
| SM-1 | Peromyscus gossypinus | Ear clip | St. Marys, Camden County, Ga. | 3 | Borrelia burgdorferi sensu stricto |
| SCI-2 | Peromyscus gossypinus | Ear clip | St. Catherines Island, Liberty County, Ga. | 3 | Borrelia burgdorferi sensu stricto |
| MI-6 | Sigmodon hispidus | Ear clip | Merritt Island, Brevard County, Fla. | 3 | Borrelia bissettii |
| MI-9 | Peromyscus gossypinus | Ear clip | Merritt Island, Brevard County, Fla. | 3 | Borrelia bissettii |
| SCW-30h | Ixodes minor nymph | Carolina wren | Wedge Plantation, Charleston County, S.C. | 3 | Borrelia bissettii |
| SCGT-8a | Ixodes minor male | Woodrat | Georgetown County, S.C. | 3 | Borrelia bissettii |
| SCGT-10 | Neotoma floridana | Ear clip | Georgetown County, S.C. | 3 | Borrelia bissettii |
| AI-1 | Sigmodon hispidus | Bladder, ear clip | Amelia Island, Nassau County, Fla. | 3 | Borrelia bissettii |
| FD-1 | Sigmodon hispidus | Bladder, ear clip | Favor Dykes, Flagler County, Fla. | 3 | Borrelia bissettii |
| MOK-3a | Ixodes dentatus nymph | Rabbit | Koch Farm, Bollinger County, Mo. | 3 | Borrelia andersonii |
| MOD-1 | 2 Ixodes dentatus nymphs | Rabbit | Dowd Farm, Bollinger County, Mo. | 3 | Borrelia andersonii |
| MOD-5 | Ixodes dentatus nymph | Rabbit | Dowd Farm, Bollinger County, Mo. | 3 | Borrelia andersonii |
| MOS-1b | Ixodes dentatus larva | Rabbit | Swinton, Stoddard County, Mo. | 3 | Borrelia andersonii |
| SI-10 | Ixodes scapularis female | Drag | Sapelo Island, McIntosh County, Ga. | 3 | Borrelia andersonii |
| BC-1 | Ixodes dentatus nymph | Drag | Macon, Bibb County, Ga. | 3 | Borrelia andersonii |
| MI-8 | Sigmodon hispidus | Ear clip | Merritt Island, Brevard County, Fla. | 3 | Borrelia sp. |
| TXW-1 | Dermacentor variabilis male | Coyote | Webb County, Tex. | 3 | Borrelia sp. |
Extraction of spirochete DNA.
Whole DNA was isolated without separating plasmid and chromosomal DNAs. Briefly, 30 ml of stationary-phase cultures were washed with 0.01 M phosphate-buffered saline (PBS) (pH 7.2) with 5 mM MgCl2 three times at 10,000 rpm (12,096 × g) for 10 min. Washed pellets of the spirochetes were resuspended in 250 μl of TES (50 mM Tris [pH 8.0], 50 mM EDTA, 15% [wt/vol] sucrose). Subsequently, an equal volume of 5 M NaCl as well as 0.01% (vol/vol) sodium deoxycholate were added to the cell suspension. Samples were placed on ice for 30 min. The partially lysed cell suspension was then centrifuged at 10,000 rpm (12,096 × g) for 10 min. After discarding the supernatant fraction, the pellet was resuspended in 250 μl of TES, followed by addition of 250 μl of 10% sodium dodecyl sulfate (SDS) and 3 μl of RNase.
After a 30-min incubation at 37°C, 50 μl of proteinase K (20 mg/ml) was added, and the tubes were incubated at 50°C for 30 min. This solution was subsequently phenol extracted twice, phenol-chloroform-isoamyl alcohol (25:24:1) extracted once, and chloroform-isoamyl alcohol (24:1) extracted once. After the addition of 1/10 (vol/vol) volume of 3.0 M sodium acetate (pH 5.2), 2 volumes of cold ethanol were added to precipitate the nucleic acid. The DNA pellets were resuspended in 50 μl of TE buffer (10 mM Tris [pH 7.6], 1 mM EDTA). DNA samples were stored at −20°C until required for PCR experiments.
Amplification of ospC genes.
A 627- to 645-bp fragment of ospC was amplified by using a pair of primers, ospC3 (5′-AAGTGCAGATATTAATGACTTTA-3′) and ospC4 (5′-TTTTTTGGACTTTCTGCCACA-3′), based on sequence data in GenBank and previously used primers (38). PCRs were performed in volumes of 50 μl containing 10 mM Tris-HCl (pH 9.0 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM dATP, 200 μM dGTP, 200 μM dCTP, 200 μM dTTP, 1.5 U of Taq DNA polymerase in storage buffer A (Promega, Madison, Wis.), 1 μM each primer, and 10 ng of extracted DNA. Reactions were performed in a GeneAmp PCR System 9700 (PE Biosystems) for 35 cycles of denaturation at 93°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 1 min, and final extension was carried out at 72°C for 9 min. The PCR products were detected in a 1% agarose gel in 0.5× Tris-borate-EDTA containing 0.5 μg of ethidium bromide/ml. The gel was photographed with an Eagle Eye II system (Stratagene).
Purification of PCR products, sequence analysis, and phylogenetic analysis.
The PCR products were purified by the Wizard PCR prep DNA purification system (Promega). The DNA sequence of ospC was determined by using an ABI Prism (model 377). Sequences were aligned manually and by using Clustal W software (28), and phylogenetic trees were constructed by neighbor joining (52), maximum likelihood (22), parsimony methods (62), and unweighted pair group method with arithmetic means analysis (UPGMA) (58) by using phylogenetic analysis using parsimony (PAUP) (62), phylogeny inference package PHYLIP (22), and molecular evolutionary genetics analysis (MEGA) (34). The DNA sequences of ospC were translated into amino acid sequences using Protein Machine software, and each deduced amino acid sequence then was aligned using Clustal W as described above. The phylogenetic trees were generated by using PAUP, Clustal W, PHYLIP, and MEGA.
Retrieved sequences.
To compare the relationship between our strains and the strains in different genospecies and to conduct phylogenetic analyses, ospC sequences of strains from the following genospecies (with strain names and database accession numbers) were used. They included B. burgdorferi sensu stricto [X69596 (B31T), L81131 (CA4), U91792 (HII), U91799 (IP1), L42887 (IP2), U91797 (IP3), U91798(L5), U91801(P1F), X69589 (PKa), X84783 (TXGW), X84778 (DUN), X84765 (BUR), L42868 (ZS7), X81522 (PBre), X73625 (DK7), U91795 (61BV3), U91794 (35B808), U04281 (HB19), L42894 (Z8691), L42893 (297), U91790 (212), U91802 (MIL), X81524 (T255), L42899 (21347), U01892 (2591), U04240 (N40), L42894 (28691), L42895 (28354), X84779 (MUL), X84782 (KIPP), X84785 (272), L42897 (26815), L81130 (SON188), L42896 (27579), X83555 (Borrelia pacificus), U91793 (Z136), U91796 (20006), U91800 (NE56), U91791 (ESP-1), L25413 (CA11.2), AF029860 (OC1), AF029861 (OC2), AF029862 (OC3), AF029863 (OC4), AF029864 (OC5), AF029866 (OC7), AF029865 (OC6), AF029867 (OC8), AF029868 (OC9), AF029869 (OC10), AF029870 (OC11), AF029871 (OC12), AF029872 (OC13), AF065143 (OC16), and AF065144 (OC17)], B. afzelii [L42871 (VS461T), X84777 (DK9), L42892 (ACAI), X84775 (DK5), L42874 (Orth), L42884 (J1), X81523 (P1J7), L42888 (H9), L42873 (Simon), L42890 (E61), X84768 (DK15), X84774 (DK4), X84776 (DK8), X73624 (DK26), X84767 (DK21), L42883 (JSB), X81521 (PBo), X84771 (DK3), X84766 (DK2), X73627 (DK1), X84769 (DK22), X83552 (PLud), X62162 (PKo), X69590 (PWudI), X80255 (PLe), D49501 (HT10), D49502 (HT25), and D49503 (HT61)], B. garinii [X69595 (PBi), X73626 (DK6), X83554 (PTrob), L42800 (KL11), L42869 (W), X83553 (PHei), X81526 (WABSou), X84770 (DK29), X73623 (DK27), X83556 (N34), L42879 (M57), X81525 (Tis1), L42889 (H13), L42900 (20047T), L42878 (NBS16), L42870 (VSDA), L42875 (NBS23), L42863 (153), X84773 (DK35), L42891 (BITS), L42882 (KL10), L42877 (NBSlab), L42886 (IP90), X69594 (pBr), X69593 (TN), X69592 (T25), D49499 (HT17), D49500 (JEM4), D49377 (HT57), and D49378 (HT64)], B. bissettii [U04280 (DN127T) and U04282 (25015)], B. andersonii [L42866 (21123T) and L42864 (19857)], B. valaisiana [L42872 (VS116T)], and Borrelia spp. [D49504 (HT22), D49505 (JEM1), D49506 (JEM2), D49381 (HT37), D49507 (HT19), D49508 (JEM3), and D49509 (HT55)].
Nucleotide sequence accession numbers.
The ospC nucleotide sequences of 19 B. burgdorferi sensu lato isolates were deposited in the GenBank database, and their accession numbers are AF278582 (MI-2), AF278583 (SCW-30h), AF278584 (MI-9), AF278585 (MOK-3a), AF278586 (MOD-5), AF278587 (BC-1), AF278588 (SCGT-8a), AF278589 (SCGT-10), AF278590 (MI-8), AF278591 (AI-1), AF278592 (FD-1), AF278593 (SI-1), AF278594 (SM-1), AF278595 (SCI-2), AF278596 (MI-6), AF278597 (MOD-1), AF278598 (MOS-1b), AF278599 (SI-10), and AF278600 (MI-5).
RESULTS
Amplification and sequencing of ospC genes.
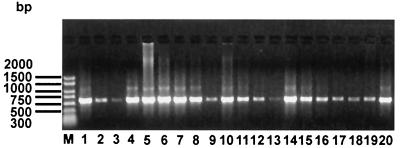
Excluding TXW-1, 606- to 624-bp coding regions of the ospC genes of the 19 southern B. burgdorferi sensu lato strains were amplified (Fig. 1) and sequenced. There was no suggestion of a relationship between the lengths of the ospC genes and the spirochete genospecies. However, the longest genes (624 bp) of ospC were found in SCGT-8a, SCGT-10, MI-6, and MOD-1 and the shortest one in BC-1 (606 bp); most southern strains have an average ospC gene length of 609 to 621 bp. Although various PCR conditions were used to amplify the ospC gene in strain TXW-1, failure suggests that TXW-1 may lack this gene or that the undetected gene in this isolate is truncated or otherwise altered at the primer sites. To confirm our sequencing results, the ospC gene of reference strain B31 was sequenced and compared to the B31 sequence retrieved from GenBank; identical results were obtained.
FIG. 1.
Amplified ospC genes of southern strains of B. burgdorferi sensu lato. The PCR products were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with a UV light transilluminator. Molecular size standards (in base pairs) are shown to the left of the gel. The 19 southern strains and the reference strain AI-1, FD-1, MI-6, MI-8, MI-9, MOD-1, SCW-30h, SCGT-8a, SCGT-10, SI-10, BC-1, MOS-1b, MOD-5, MOK-3a, SI-1, SM-1, MI-2, MI-5, SCI-2, and B31 are shown in lanes 1 to 20, respectively.
Analysis of ospC sequences of southern strains.
A similarity matrix of OspC amino acid sequences indicated a high degree of heterogeneity, and amino acid identities ranged from 59.9 to 100%; OspC from most strains ranged from 60 to 80% identity (Table 2). The lowest degree of similarity (59.9%) was found between SM-1 (B. burgdorferi sensu stricto) and 20047 (B. garinii). The highest degrees of similarity were shown between strains of B. burgdorferi sensu stricto, i.e., SM-1 and SI-1 or SCI-2 (99.5 and 100%), MI-2 and B31 (99.5%), B. andersonii strains MOS-1b and SI-10 (99.5%), B. bissettii strain MI-9, and B. burgdorferi sensu stricto strain MI-5 (99.5%), B. andersonii strain MOD-1 and B. bissettii strain MI-6 (99%), B. bissettii strains SCGT-8a and SCGT-10 (98.1%), MI-9 and FD-1 (95.6%), and B. burgdorferi sensu stricto strain MI-5 and B. bissettii strain FD-1 (95.1%). As shown above, high degrees of similarity among strains were usually found in those from the same genospecies but also sometimes among strains in different genospecies (Table 2), for example, between B. burgdorferi sensu stricto and B. bissettii or between B. andersonii and B. bissettii. Results indicated that B. burgdorferi sensu stricto, B. bissettii, and B. andersonii from North America are closely related.
TABLE 2.
Amino acid similarity between OspC sequences from different B. burgdorferi sensu lato isolates
| Isolate | % Amino acid similarity with OspC sequence from isolate:
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DN127 | VS116 | 21123 | 25015 | VS416 | 20047 | B31 | SI-1 | SI10 | SM1 | SCI2 | SCW30h | AI1 | FD-1 | MI-2 | MI-5 | MI-6 | MI-8 | MI-9 | SCGT-8a | SCGT-10 | MOD-1 | MOD-5 | BC-1 | MOK-3a | MOS-1b | |
| DN127 | 100.0 | 67.3 | 69.8 | 69.1 | 69.3 | 69.3 | 68.0 | 67.0 | 70.2 | 66.5 | 67.0 | 73.0 | 70.7 | 75.0 | 67.5 | 73.0 | 70.2 | 73.1 | 72.6 | 75.1 | 76.6 | 71.2 | 70.0 | 69.3 | 75.9 | 69.8 |
| VS116 | 100.0 | 66.8 | 66.3 | 69.8 | 63.4 | 68.8 | 63.9 | 64.9 | 63.3 | 63.9 | 63.3 | 68.3 | 69.3 | 68.3 | 67.3 | 70.3 | 69.3 | 66.8 | 66.8 | 68.8 | 70.8 | 62.4 | 62.9 | 65.3 | 64.4 | |
| 21123 | 100.0 | 67.6 | 63.9 | 62.9 | 66.5 | 70.0 | 81.6 | 66.5 | 70.0 | 65.0 | 71.2 | 69.6 | 66.5 | 70.6 | 73.9 | 70.7 | 70.1 | 70.5 | 71.0 | 74.4 | 67.0 | 72.3 | 68.5 | 82.1 | ||
| 25015 | 100.0 | 70.8 | 64.4 | 82.8 | 69.5 | 68.6 | 69.0 | 69.5 | 71.4 | 73.5 | 81.4 | 82.3 | 80.4 | 75.0 | 75.5 | 80.0 | 74.0 | 73.5 | 75.0 | 77.8 | 68.8 | 79.8 | 68.1 | |||
| VS461 | 100.0 | 74.8 | 73.8 | 67.8 | 63.9 | 67.3 | 67.8 | 70.8 | 70.8 | 70.8 | 73.3 | 69.8 | 69.3 | 70.3 | 69.3 | 69.8 | 69.8 | 69.3 | 67.8 | 65.3 | 68.3 | 63.4 | ||||
| 20047 | 100.0 | 64.4 | 60.4 | 63.9 | 59.9 | 60.4 | 66.8 | 63.4 | 66.8 | 64.9 | 64.9 | 63.9 | 68.3 | 64.4 | 69.8 | 68.8 | 63.9 | 62.9 | 60.4 | 63.9 | 63.4 | |||||
| B31 | 100.0 | 69.0 | 65.0 | 68.5 | 69.0 | 72.4 | 70.0 | 77.3 | 99.5 | 76.4 | 71.9 | 73.4 | 75.9 | 68.5 | 67.5 | 71.4 | 72.4 | 66.3 | 74.4 | 64.5 | ||||||
| SI-1 | 100.0 | 67.5 | 99.5 | 100.0 | 70.9 | 72.4 | 71.4 | 68.5 | 68.5 | 68.5 | 67.0 | 69.0 | 64.0 | 64.5 | 68.5 | 69.5 | 65.8 | 70.9 | 67.0 | |||||||
| SI10 | 100.0 | 67.0 | 67.5 | 66.5 | 70.2 | 69.6 | 65.0 | 69.9 | 72.9 | 70.7 | 69.1 | 69.6 | 70.5 | 73.4 | 65.5 | 71.3 | 69.5 | 99.5 | ||||||||
| SM1 | 100.0 | 99.5 | 70.4 | 71.9 | 70.9 | 68.0 | 68.0 | 68.0 | 66.5 | 68.5 | 63.5 | 64.0 | 68.0 | 69.0 | 65.3 | 70.4 | 66.5 | |||||||||
| SCI2 | 100.0 | 70.9 | 72.4 | 71.4 | 68.5 | 68.5 | 68.5 | 67.0 | 69.0 | 64.0 | 64.5 | 68.5 | 69.5 | 65.8 | 70.9 | 67.0 | ||||||||||
| SCW-30h | 100.0 | 77.3 | 72.4 | 71.9 | 71.4 | 69.0 | 73.4 | 70.9 | 70.0 | 69.5 | 68.5 | 68.0 | 64.9 | 71.9 | 66.0 | |||||||||||
| AI-1 | 100.0 | 76.5 | 69.5 | 73.5 | 75.1 | 75.1 | 73.0 | 68.8 | 70.2 | 74.6 | 71.4 | 68.3 | 76.4 | 69.8 | ||||||||||||
| FD-1 | 100.0 | 76.8 | 95.1 | 77.9 | 80.4 | 95.6 | 75.0 | 76.5 | 77.9 | 82.8 | 73.3 | 81.8 | 69.1 | |||||||||||||
| MI-2 | 100.0 | 75.9 | 71.4 | 72.9 | 75.4 | 68.0 | 67.0 | 70.9 | 71.9 | 66.3 | 74.0 | 64.5 | ||||||||||||||
| MI-5 | 100.0 | 76.5 | 79.4 | 99.5 | 72.1 | 73.5 | 76.0 | 81.8 | 74.3 | 80.3 | 69.1 | |||||||||||||||
| MI-6 | 100.0 | 75.6 | 76.0 | 73.6 | 74.5 | 99.0 | 70.4 | 73.3 | 73.4 | 72.5 | ||||||||||||||||
| MI-8 | 100.0 | 78.9 | 75.1 | 77.1 | 75.6 | 71.4 | 68.8 | 73.9 | 70.2 | |||||||||||||||||
| MI-9 | 100.0 | 72.5 | 74.0 | 75.5 | 82.3 | 73.8 | 79.8 | 68.6 | ||||||||||||||||||
| SCGT-8a | 100.0 | 98.1 | 73.6 | 68.5 | 67.8 | 70.0 | 69.6 | |||||||||||||||||||
| SCGT-10 | 100.0 | 74.5 | 70.4 | 67.8 | 70.9 | 70.5 | ||||||||||||||||||||
| MOD-1 | 100.0 | 70.0 | 73.3 | 74.4 | 72.9 | |||||||||||||||||||||
| MOD-5 | 100.0 | 69.8 | 78.8 | 65.5 | ||||||||||||||||||||||
| BC-1 | 100.0 | 71.3 | 70.8 | |||||||||||||||||||||||
| MOK-3a | 100.0 | 69.0 | ||||||||||||||||||||||||
| MOS-1b | 100.0 | |||||||||||||||||||||||||
The amino acid sequences of species-specific epitopes of southern strains varied between positions 28 and 31. Most southern B. bissettii and B. andersonii strains have an S at position 28, but southern B. burgdorferi sensu stricto strains either have an A or lack an amino acid at this position. At positions 29 and 30, most southern B. andersonii and B. bissettii strains have A and S, but southern B. burgdorferi sensu stricto strains have T and S or S and V. At position 31, southern B. burgdorferi sensu stricto strains have either an A or a gap; however, most southern B. bissettii and B. andersonii strains have T, and three B. andersonii strains have N.
Except for this region, each of the strains MOD-5, MOK-3a, SI-1, SM-1, SCI-2, SCW-30h, and AI-1 has a gap at position 70. Each of the strains SCGT-8a and SCGT-10 has an insertion of T at position 88. Strain BC-1 has a deletion at position 79, and strains MI-6 and MOD-1 have a 3-amino-acid insertion, G, L, and N. Strains BC-1, B31, MI-2, 25015, MI-5, MI-9, FD-1, MOD-5, and MOK-3a each have a deletion at position 115. Strains SCGT-8a and SCGT-10 have an insertion of N and G and strains SI-10 and MOS-1b have an insertion of D and G at positions 146 and 147. These differences in OspC sequences among southern strains reflect significant diversity at the genospecies level.
An alignment of the deduced amino acid sequences indicated that both the C-terminal and N-terminal ends of OspC appear to be conserved. The amino acid sequence variation was observed in the internal part of this protein; a variable region was located from amino acids 72 to 191 among these southern strains. Two hypervariable regions were located between amino acids 84 and 96 and between amino acids 146 and 176. In comparison with the previously reported hypervariable regions in other strains (30), the first hypervariable region among southern strains was located between amino acids 84 and 96 rather than between amino acids 83 and 95. The second hypervariable region among southern strains was located between amino acids 146 and 164 and extended to amino acid 176 rather than between amino acids 143 and 162. There were three identical amino acids in the second hypervariable region.
There are four domains in the OspC amino acid sequence according to a previous report (38): the conserved amino-terminal domain (C1), two variable domains (V1 and V2), and the semivariable domain (SV). For southern strains, C1 is located from amino acids 35 to 86, V1 from amino acids 86 to 99, SV from amino acids 99 to 118, and V2 from amino acids 118 to 191. The locations of these domains are shown in Table 3.
TABLE 3.
Amino acid sequences of OspC proteins of southern B. burgdorferi sensu lato and other strains deduced from the nucleotide sequences of ospCs aligned with Clustal Wa
| Isolate | Species specific sequencesC1V1SVV2
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ◂—————▸◂————————————————————————————————-▸◂——————--▸◂—————————-▸◂———————————————————————————————————————————————————————--—▸ | ||||||||
| SCGT-8a | SAILMTLFLFISCNNSGKDGNSAST-NPADESVKGPNLTEISKKIMDSNTVVLAVKEVEA | 59 | LLSSIDELATKAIGKKIDANGSLVADATDFNTSLLAGAYAISSLITEKLN---KLKNSEG | 116 | LKEKIAEAKKCSDDFTKKLKDSHQELGVANGAATADNAKKAILKTNATKDKGAEELEKLF | 176 | KAVESLSKAAQDMLTNSVKELTSPVVGKSPKX | 208 |
| SCGT-10 | SAILMTLFLFISCNNSGKDGNSAST-NPADESAKGPNLTEISKKITDSNTVVLAVKEVET | 59 | LLASIDELATKAIGKKIDANGSLVADATDFNTSLLAGAYAISSLITEKLN---KLKNSEG | 116 | LKEKIAEAKKCSDDFTKKLKDSHQELGVANGAATADNAKKAILKTNATKDKGAEELEKLF | 176 | KAVESLSKAAQDMLTNSVKELTSPVVGKSPKX | 208 |
| DN127 | SAILMTLFLFISCNNSGKDGNSAST-NPADESAKGPNLTEISKKITDSNAIVLAVKEVET | 59 | LLLSIDELA-KAIGKKINNNG--LDVLQNFNASLLGGAHTISKLITEKLS---KLNGSEE | 113 | LKEKIEAAKKCSDDFTKKLQSSHAELGVAGGATTDENAKKAILKSNADKTKGADELGKLF | 173 | ESVESLAKAAKEMLANSVKELTSPVVAETPKX | 205 |
| SI-10 | SAILMTLFLFISCNNSGKDGNSAST-NPVDESVKGPNLIEISKKITDSNAVVLAVQEVET | 59 | LLASIDEIAKKAIGQRIAPGNTLTADA-SRNGSLIVGVYTISTLITGKLN---GLKDLED | 115 | LKEKIEKAKKCSTEFTKKLLDSHAELGQADGAVTDDNAKRAILKTHNNTDKGAKELKDLS | 175 | ESVESLAKAAKEMLNNAVKELTKPVVAEAPKX | 207 |
| MOS-1b | SAILMTLFLFISCNNSGKDGNSAST-NPVDESVKGPNLIEISKKITDSNAVVLAVQEVET | 59 | LLASIDEIAKKAIGQRIAPGNTLTADA-SRNGSLIVGVYTISTLITGKLN---GLKDLED | 115 | LKEKIEKAKKCSTEFTKKLLDSHAELGQADGAVTDDNAKRAILKTHNNTDKGAKELKDLS | 175 | ESVESLAKAAKEMLNNAVKELTKPVVTEAPKX | 207 |
| 21123 | SAILMTLFLFISCNNSGKDGNSASN-NSADESVKGPNLVEISKKITDSNAVVLAVQEVET | 59 | LLASIDELAKKAIGQRIDQGNKLAVDA-DHNGSLLAGVYTISTLITEKLN---SLKISEE | 115 | LKEKIEKAKKYSTEFTKKLSDSHGVLGQVGGNATDVNAKAAILKTNQTTDKGAKELKELS | 175 | ESVESLAKAAKEMLNDSIKELTKPVVTEAPKX | 207 |
| BC-1 | SAILMTLFLFISCNNSGKDGNSASN-NSADESVKGPNLIEISKKITDSNVVVLAVKEIET | 59 | LLSSIDELGNKAIGKLIS-ANGLNVQA-GQNGSLLAGAHAISSLITQKLS---ALN-SEE | 113 | LKEKIKDAKDCSEKFTKKLSESHADLGQN---ATDDNAKRAILKTHTDTDKGAKELKELS | 170 | ASVESLAKAAKELLNNAVKELTKPIVAESPKX | 202 |
| MI-6 | SAILMTLFLFISCNNSGKDGNSAST-NPVDESVKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LLASIDQLATKAIGKKIDQNNALGTQA-GQNGSLLAGAYTISTLITEKLSGLNALKNAEE | 118 | LKEKIKDAKKFSEDFTKKLSDNHADLSPA--AATDDNAKAAILKTNNTKDKGAKELKELS | 176 | ESVEALSKAAQAMLTNSVKELTSPVVAESPKX | 208 |
| MOD-1 | SAILMTLFLFISCNNSGKDGNSAST-NPVDESVKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LLASIDQLATKAIGKKIDQNNALGTQA-GQNGSLLAGAYTISTLITEKLSGLNALKNAEE | 118 | LKEKIKDAKKFSEDFTKKLSDSHADLSPA--AATDDNAKAAILKTNNTKDKGAKELKELS | 176 | ESVEALSKAAQAMLTNSVKELTSPVVAETPKX | 208 |
| 831 | SAILMTLFLFISCNNSGKDGN-TSA-NSADESVKGPNLTEISKKITDSNAVLLAVKEVEA | 58 | LLSSIDEIAAKAIGKKIHQNNGLDTEN-NHNGSLLAGAYAISTLIKQKLD---GLK-NEG | 113 | LKEKIDAAKKCSETFTNKLKEKHTDLGKE--GVTDADAKEAILKTNGTKTKGAEELGKLF | 171 | ESVEVLSKAAKEMLANSVKELTSPVVAESPKX | 203 |
| MI-2 | SAILMTLFLFISCNNSGKDGN-TSA-NSADESVKGPNLTEISKKITDSNAVLLAVKEVEA | 58 | LLSSIDEIAAKAIGKKIHQNNGLDTEN-NHNGSLLAGAYAISTLIKQKLD---GLK-NEG | 113 | LKEKIDAAKKCSETFTNKLKEKHTDLGKE--GVTDADAKEAILKTNGTKTKGAEELGKLF | 171 | ESVEVLSKAAKEMLANSVKELTNPVVAESPKX | 203 |
| 25015 | SAILMTLFLFISCNNSGKDGNAAST-NPADESVKGPNLTEISKKITDSNAVVLAVKEVGA | 59 | LLTSIDELATKAIGKKIHQNNGLDTEN-NHNGSLLAGAYAISTLITQKLG---GLK-NEE | 114 | LKEKIAAVKKCSEEFTNKLKSSHTELGKQ--DAQDDDAKKAILRTHNTKDKGAEELDKLF | 172 | KAVENLSKAAKEMLSNSVKELTSPVVAESPKX | 204 |
| MI-5 | SAILMTLFLFISCNNSGKDGNSAST-NPADESTKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LIASIDELADKAIGKKIQQNNGLGTEN-NHNGSLLAGVYAISTLITQKLS---SLN-SEE | 114 | LKEKIKEAKDCSEKFTKKLETGHAELGKE--DATDDNAKKAVLKTNADKSKGAEELEKLF | 172 | KSVESLAKAAKESLNKSVKELTSPVVAESPKX | 204 |
| MI-9 | SAILMTLFLFISCNNSGKDGNSAST-NPADESTKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LIASIDELADKAIGKKIQQNNGLGTEN-NHNGSLLAGVYAISTLITQKLS---SLN-SEE | 114 | LKEKIKEAKDCSEKFTKKLETGHAELGKE--DATDDNAKKAVLKTNADKSKGAEELEKLF | 172 | KSVESLAKAAKESLNKSVKELTSPVVAKSPKX | 204 |
| FD-1 | SAILMTLFLFISCNNSGKVGNSAST-NPADESTKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LIASIDELADKAIGKKIQQNNGLGTEN-NHNGSLLAGAYAISTLITQKLS---KLN-SEE | 114 | LKEKIKEAKKCSEKFTKKLETGHAELGKE--DATDDNAKKAILKTNADKSKGAEELEKLF | 172 | KSVESLAKAAKEMLTNSVKELTSPVVAKNPKX | 204 |
| MOD-5 | SAILMTLFLFISCNNSGKDGNSASN-NSADESAKGPNLIEISKKITDSNAVVLAVKEVET | 59 | LIASIDELA-KAIGKKIQQNNGLGTEN-NQNGSLLAGVHAISTLITEKLN---ALN-SET | 113 | LKEKIKEVKKCSENFTNTLKQNHTELGKK--DANDDDAKKAILRTNGDKTKGAKELEDLF | 171 | KAVESLAKAAKEILNNSVKELTSPVVGKNPKX | 203 |
| MOK-3a | SAILMTLFLFISCNNSGKDGNSAST-NPADESVKGPNLTEISKKITDSNAVVLAVKEVET | 59 | LIASIDELA-KAIGKKIEPTGGLGTDN-NHNGSLLSGAHAISTLITQKLS---ALN-SEE | 113 | LKGKIEDVKKCSQEFTNQLKNSHTELGKQ--DATDENAKKAILKTNAAKDKGAEELGELF | 171 | ESVESLSKAAKEMLANAVKELTSPVVAETPKX | 203 |
| SI-1 | SAILMTLFLFISCNNSGKDGNASV--NSADESVKGPNLVEISKKITDSNAVVIAVKEVET | 58 | LLVSIDELA-KAIGKKIEAGGTLGSDG-AHNGSLLAGAYKIATEITANLS---KLKASED | 113 | LKEKITKAKECSEKFTDKLKSENAALGKQ--DASDDDAKKAILKTHNDITKGAKELKELS | 171 | ESVETLLKAAKEMLANSVKELTSPVVAKNPKX | 203 |
| SM-1 | SAILMTLFLLISCNNSGKDGNASV--NSADESVKGPNLVEISKKITDSNAVVIAVKEVET | 58 | LLVSIDELA-KAIGKKIEAGGTLGSDG-AHNGSLLAGAYKIATEITANLS---KLKASED | 113 | LKEKITKAKECSEKFTDKLKSENAALGKQ--DASDDDAKKAILKTHNDITKGAKELKELS | 171 | ESVETLLKAAKEMLANSVKELTSPVVAKNPKX | 203 |
| SCI-2 | SAILMTLFLFISCNNSGKDGNASV--NSADESVKGPNLVEISKKITDSNAVVIAVKEVET | 58 | LLVSIDELA-KAIGKKIEAGGTLGSDG-AHNGSLLAGAYKIATEITANLS---KLKASED | 113 | LKEKITKAKECSEKFTDLKSENAALGKQ--DASDDDAKKAILKTHNDITKGAKELKELS | 171 | ESVETLLKAAKEMLANSVKELTSPVVAKNPKX | 203 |
| SCW-30h | SAILMTLFLFISCNNSGKDGNSAS--NTADESVKGPNLTEISKKIADSNAVVLAVKEVEA | 58 | LLASIDELA-KAIGKKIKNDGSLEAEA-NRNESLVAGAYTISALITQKLG---KLKNSEE | 113 | LKEKISNAKDCAEAFTKKLKDNQAELGVQ--GVTDENSKKAILKTHGDKTKGAEELGKLF | 171 | ESVGNLSKAAKELLDNSVKELTSPVVAENPKX | 203 |
| AI-1 | SAILMTLFLFISCNNSGKDGNSASTNNPADESTKGPNLTEISKKITDSNAVVLAVKEVVT | 60 | LIASIDELS-KAIGKKFNQNGGLDAEA-DHNGSLLSGAYTISTLITQKLG---KLKNSEE | 115 | LKEKIENAKKCSEEFTKKLKDNQADLGKK--DATDENAKKAILKTHGTTDKGAKELKELS | 173 | ESVEILIKAAKDMLTNSVKALTSPVVAENPKX | 205 |
| MI-8 | SAILMTLFLFISCNNSGKDGNSAST-NPADESAKGPNLTEISKKITDSNTVVLAVKEVET | 59 | LLASIDELATKAIGKKIQQNGGLAAEA-GHNGTLLAGAYTISKLITQKLD---GLKNTEK | 115 | LKEIENAKKCSEDFTKKLEGEHVVLGIE--NVTDENAKKAILITDAAKDKGAAELEKLF | 173 | KSVESLAKAAQESLANSVKELTSPVVAENPKX | 205 |
| VS461 | SAILMTLFLFISCNNSGKGGDIAST-NP-DESAKGPNLTEISKKITDSNAVVLAVKEVEA | 58 | LLSSIDELA-KTIGKKIEANG-LGNEA-DKNGSLLAGAYAISTLIKQKLD---GLKGLEG | 112 | LNKEIAEAKKCSEAFTKKLQDSNADLGKHN--ATDADSKEAILKTNGTKTKGAKELEELF | 170 | KSVESLSKAAKEALSNSVKELTSPVVAESPKX | 202 |
| 20047 | SAILMTLFLFISCNNS--GGDTAST-NP-DESVKGPNLTEISKKITDSNAFVLAVKEVEA | 56 | LISSIDELA-KAIGQRIQQNG-LVADA-GHNSALLAGAHEISILITQKLD---GLKGLEG | 110 | LKAEIAEAKKYSEAFTKKLKDNHAQLGIQNGASLDDEAKKAILKTNVDKTKGAEELEKLF | 170 | KSVESLSKAAQEALTNSVKELTNPVVAETPKX | 202 |
| VS116 | SAILMTLFLFISCNNS--GGDTAST-NPVDESAKGPNLTEISKKITDSNAIVLAVKEVET | 57 | LLASINEIANKGIGKKINQNG-LDNLT-DHNGSLIAGAYVISTLITEKLN---NLKNSEG | 112 | LKEKIKKVKECSDKFTKKLTTSNGDLGKEN--VTDAHAQAAILKTNPTNDKGAKELGELF | 170 | ESVEILSKAAQEALTNSIAELTSPVVAENPKX | 202 |
| *************** * * ******** ****** ****** | * ** * ** * * * * * * * * | * * * ** * * ***** ** * | * * *** * **** *** | |||||
Dashes indicate gaps. The conserved amino-terminal (C1), variable (V1 and V2), and semivariable (SV) domains and species-specific motif of the OspC proteins are indicated. Identical amino acid sequences (indicated by asterisks) of strains suggest a lateral genetic transfer of part of the ospC genes within and between genospecies.
Lateral transfer and internal gene recombination of ospC gene and OspC sequence diversity.
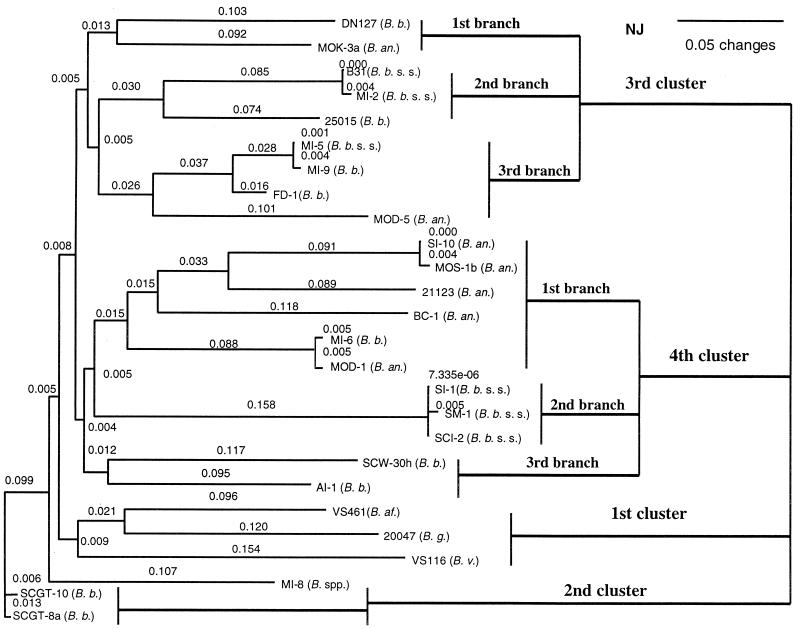
A close identity was found between B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii (Tables 2 and 3). The close identity (indicated in Table 3) among some amino acid sequences of southern strains and other strains belonging to different B. burgdorferi genospecies suggests that a lateral transfer and internal gene recombination of the ospC gene may have occurred among the three genospecies. Both phylogenetic trees also suggest that lateral transfer and internal gene recombination of the ospC gene occurred among these three genospecies (Fig. 2 and 3). Based on the result in which MI-6 (B. bissettii) was misplaced in the SI-10 cluster (B. andersonii) in the phylogenetic tree (Fig. 2), B. andersonii appears to be the donor and B. bissettii the recipient. However, B. andersonii might have obtained part of the ospC sequence from B. bissettii based on the grouping in which MOD-5 (B. andersonii) was misplaced in the FD-1 group (B. bissettii). Genetic transfer could happen in both directions. Moreover, B31 and MI-2 were misplaced in the 25015 group (B. bissettii), suggesting that B. burgdorferi sensu stricto might have obtained part of the ospC gene from B. bissettii.
FIG. 2.
Phylogenetic tree derived from the deduced amino acid sequences of southern and reference strains of Borrelia burgdorferi sensu lato. The tree was constructed with PAUP software and was based on a comparison of 208 OspC amino acids. B. b. ss, Borrelia burgdorferi sensu stricto; B. b., Borrelia bissettii; B. an., Borrelia andersonii; B. g., Borrelia garinii; B. af., Borrelia afzelii; B. v., Borrelia valaisiana.
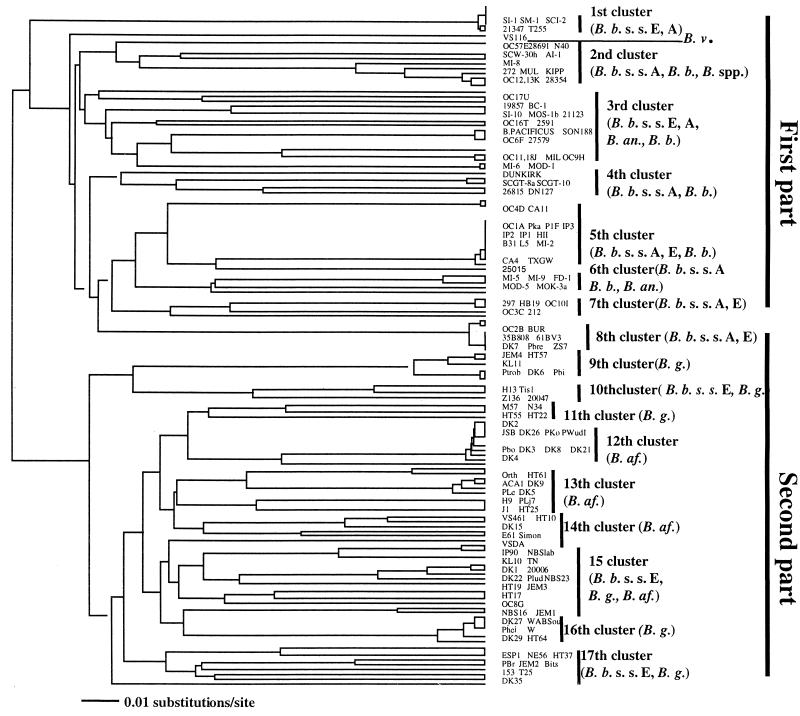
FIG. 3.
Phylogenetic tree determined from the DNA sequence of the ospC genes of southern and other genospecies strains of B. burgdorferi sensu lato. The UPGMA tree was constructed with PAUP software and is based on a comparison of 344 bp of ospC genes. The tree was compared with the trees produced by neighbor-joining, maximum-likelihood, and parsimony methods with PAUP software. The four methods produced similar results. Scale bar represents calculated distance value. The abbreviations are defined in the legend to Fig. 2. A and E, America and Europe, respectively (these letters indicate the geographic origins of the isolates).
Clinical and epidemiological implication of ospC groups.
Each ospC type is a group of strains in which divergence is greater than 8% between groups and less than 1% within groups (27, 57, 64). A branch length of 0.04 corresponds to 8% divergence between two sequences and is the cutoff that defines the ospC group (9, 21). Based on this definition, the 19 southern strains can be divided into 12 ospC groups (except for the MI-5, MI-9, and FD-1 group, whose branch length is 0.037 and is close to the cutoff) (Fig. 2, Table 4). Of the 12 southern strain ospC groups, two groups belong to the B. burgdorferi sensu stricto genospecies. One is the A group (Table 4) and contains MI-2 along with the IP1 group, all of which belong to a so-called invasive cluster (9, 57). MI-2 most closely aligns with a group that possesses proven human invasive strains.
TABLE 4.
Relationships between ospC major groups and their biological sources and geographic origins
| Major ospC group | Strains | Biological sourcesa | Origins |
|---|---|---|---|
| A | HII, IP1, IP2, IP3, P1F, L5, TXGW | Patients (blood, CSF, synovial fluid, skin) | USA (New York, California, Texas, Florida) |
| MI2 | Animal (P. gossypinus) | Italy, France, Austria, Germany | |
| B31, CA4, Pka | Ticks (I. scapularis, I. pacificus, I. ricinus) | ||
| B | 61BV3, BUR, DK7, Pbre | Patients (skin) | USA (New York) |
| 35B808, ZS7, OC2 | Ticks (I. scapularis, I. ricinus) | Germany, Denmark | |
| C | OC3 | Tick (I. scapularis) | USA (New York) |
| D | CA11.2A, OC4 | Ticks (I. scapularis, I. pacificus) | USA (New York, California) |
| E | 28691, N40, OC5, OC7 | Tick (I. scapularis) | USA (New York, Pennsylvania) |
| F | 27579, OC6, B. pacificus, Son188 | Ticks (I. scapularis, I. pacificus) | USA (New York, Connecticut, California) |
| G | OC8 | Tick (I. scapularis) | USA (New York) |
| H | OC9 | Tick (I. scapularis) | USA (New York) |
| I | 297, HB19, OC10 | Patients (blood, CSF) | USA (New York, Connecticut) |
| Tick (I. scapularis) | |||
| J | MIL, OC11 | Ticks (I. scapularis, I. ricinus) | USA (New York), Slovakia |
| K | 272, 297, KIPP, MUL, 28354, OC12, OC13 | Patients (CSF, skin) | USA (New York, Connecticut, Maryland) |
| Tick (I. scapularis) | |||
| L | 21347, T255 | Animal (P. leucopus) | USA (Wisconsin), Germany |
| Tick (I. ricinus) | |||
| M | 2591 | Animal (P. leucopus) | USA (Connecticut) |
| N | 26815 | Chipmunk | USA (Connecticut) |
| O | DUNKIRK | Patients (skin) | USA (New York) |
| P | 20006 | Tick (I. ricinus) | France |
| Q | 212 | Tick (I. ricinus) | France |
| R | ESP-1, NE56 | Tick (I. ricinus) | Spain, Switzerland |
| S | Z136 | Tick (I. ricinus) | Germany |
| T | OC16 | Patient (EM) | USA (New York) |
| U | OC17 | Patient (EM) | USA (New York) |
| V | S-1, SM-1, SC-12 | Animal (P. gossypinus) | USA (Georgia) |
| W | DN127 | Tick (I. pacificus) | USA (California) |
| X | 25015 | Tick (I. scapularis) | USA (New York) |
| Y | MI-5, MI-9, FD-1 | Animal (P. gossypinus, S. hispidus) | USA (Florida) |
| Z | MI-8 | Animal (S. hispidus) | USA (Florida) |
| A1 | AI1 | Animal (S. hispidus) | USA (Georgia) |
| B1 | SCW-30h | Tick (I. minor) | USA (South Carolina) |
| C1 | SCGT-8a, SCGT-10 | Animal (N. floridana) | USA (South Carolina) |
| Tick (I. minor) | |||
| D1 | MI6, MOD1 | Animal (S. hispidus) | USA (Georgia) |
| Tick (I. dentatus) | |||
| E1 | 21123 | Tick (I. dentatus) | USA (New York) |
| F1 | BC1 | Tick (I. dentatus) | USA (Georgia) |
| G1 | MOS1b, SI-10 | Ticks (I. dentatus, I. scapularis) | USA (Georgia, Missouri) |
| H1 | MOD5 | Tick (I. dentatus) | USA (Missouri) |
| I1 | MOK3a | Tick (I. dentatus) | USA (Missouri) |
| J1 | VS461 | Tick (I. ricinus) | Switzerland |
| K1 | 20047 | Tick (I. ricinus) | France |
| L1 | VS116 | Tick (I. ricinus) | Switzerland |
CSF, cerebrospinal fluid; EM, erythema migrans.
Transmission experiments show that this group of southern spirochetes are able to infect mice and hamsters and that the typical vector of Lyme disease, I. scapularis, can acquire the spirochetes from infected mammals (J. H. Oliver et al., unpublished data). Another B. burgdorferi sensu stricto group (V group) contains three southern strains, SI-1, SM-1, and SCI-2, and is a typical southern B. burgdorferi sensu stricto group. MI-5 (B. burgdorferi sensu stricto) and MI-9 and FD-1 (B. bissettii) comprise a mixed group, Y. MI-8, the undescribed new genospecies, constitutes the Z group. There are three B. bissettii groups, AI-1 (A), SCW-30h (B1), and the SCGT-8a-SCGT-10 group (C1), and two mixed groups, the D1 group (MI-6 and MOD-1) and the Y group (FD-1, MI-9, and MI-5). The B. andersonii genospecies includes BC-1 (F1), the MOS-1b-SI-10 group (G1), MOD-5 (H1), and MOK-3a (I1). Except for the mixed groups, which comprise different genospecies, the neighbor-joining tree grouped MOK-3a with DN127, and the MI-2 group clustered with 25015 (Fig. 2). It appears that horizontal genetic transfer and internal gene recombination of the ospC gene occurred between B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii.
Except for the MI-2 strain, which is included in the invasive cluster (ospC group A), most southern strains formed separate groups (Fig. 3, Table 4). We identified them as major ospC groups including V to I1. Of the 12 southern major ospC groups, two were B. burgdorferi sensu stricto groups, three were B. bissettii groups, four were B. andersonii groups, one was an undescribed genospecies group, and two were mixed groups (formed by B. burgdorferi sensu stricto and B. bissettii and by B. bissettii and B. andersonii) (Fig. 3, Table 4).
Phylogenetic analysis of ospC gene.
To compare and further investigate the phylogenetic relationships within and among southern strains and other genospecies available in GenBank, phylogenetic trees were constructed based on the almost complete amino acid sequences of OspC and partial nucleotide sequences of ospC. Comparison of phylogenetic trees based on these results indicated that all trees were consistent, with minor variation (Fig. 2 and 3).
Phylogenetic trees inferred from 606 to 624 bp of nucleotide sequences of ospC (from nucleotides 18 to 643) and 202 to 208 corresponding amino acids of OspC (from amino acids 7 to 218) (Table 3) of southern strains and reference strains of B. burgdorferi sensu stricto, B. andersonii, B. bissettii, B. garinii, B. afzelii, and B. valaisiana were exactly the same. There were four major clusters in the neighbor-joining tree (Fig. 2). Clearly, genetic diversity exists among southern strains, especially B. bissettii and B. andersonii, compared to United States B. burgdorferi sensu stricto strains. B. bissettii strains were present in six branches; in five of them, B. bissettii combined with B. andersonii, B. burgdorferi sensu stricto, and the undescribed genospecies MI-8. Also, diverse B. andersonii strains were present in three branches.
The deduced OspC amino acid sequences of southern strains were aligned and compared with previously published OspC sequences of other genospecies (Table 3). Significant variability was observed among the OspC sequences present in the 19 southern B. burgdorferi sensu lato strains and other genospecies. According to a previous study of the OspC sequence and its polymorphism in B. burgdorferi (69) and the N-terminal sequences of Vmp3 and Vmp33 of B. hermsii (12, 16), a high degree of variation exists between the Vmps of B. hermsii and OspCs of B. burgdorferi sensu lato. Species-specific epitopes were found between amino acids 20 and 33, which is highly conserved among B. afzelii and B. garinii strains but is significantly different compared with that of B. burgdorferi sensu stricto strains. The highest degree of genetic diversity among them was observed in the two variable domains (V1 and V2), the semivariable domain (SV), and the species-specific epitopes (between amino acids 28 and 31). Except for these regions, amino acid insertion, deletion, and replacement of OspC among the three genospecies occurred at many positions in the variable regions. These differences in OspC sequences among southern strains reflect the diversity at the strain and genospecies levels.
Phylogenetic trees were also derived from 344 bp of nucleotide sequences (from nucleotides 108 to 552) of ospC genes and 146 deduced amino acid sequences (from amino acids 36 to 184) of OspCs. This fragment included the variable regions of ospC. The southern strains were compared with some other strains available in GenBank, and two trees indicated similar results; one is presented in Fig. 3. There were 17 clusters in the UPGMA tree which could be divided into two parts. The first part consisted of B. burgdorferi sensu stricto (including both North American and some European strains), B. andersonii, and B. bissettii strains. The second part consisted of Eurasian B. garinii, B. afzelii, and some European B. burgdorferi sensu stricto strains. The B. burgdorferi sensu stricto strains separated into 11 clusters, 8 of them located in the first part of the tree; in fact, they were the major strains in that part. Three of them shared clusters with B. garinii and B. afzelii in the second part of the tree. Three strains in three clusters of the second part were European B. burgdorferi sensu stricto strains (20006, Z136, ESP1, and NE56); no North American strains were found in that part.
Compared to European B. burgdorferi sensu stricto strains, North American B. burgdorferi sensu stricto strains appear to have greater ospC heterogeneity. North American B. burgdorferi sensu stricto strains were dispersed in all eight clusters in the B. burgdorferi sensu stricto part of the tree, and three clusters were exclusively North American strains, but European B. burgdorferi sensu stricto strains were included in only five clusters (Fig. 3). Several of the European B. burgdorferi sensu stricto strains were closely related to North American B. burgdorferi sensu stricto strains, inasmuch as they shared five clusters. Nevertheless, they were different. European B. burgdorferi sensu stricto strains appeared genetically closer to each other and grouped into two major clusters. For example, PKa, P1F, IP1, IP2, IP3, and HII were located in the same branch as strains 35B808, 61BV3, DK7, PBre, and ZS7. Interestingly, only European B. burgdorferi sensu stricto strains were found in the European genospecies B. garinii and B. afzelii part of the tree (Fig. 3).
Greater genetic differences were observed among southern strains than northern strains in North America. Nineteen southern strains separated into all six clusters in the B. burgdorferi sensu stricto part of the tree and were dispersed into nine branches (Fig. 3). Southern B. burgdorferi sensu stricto strains occupied three branches. The first branch consisted of SI-1, SM-1, and SCI-2, which shared the cluster with North American and European B. burgdorferi sensu stricto strains. The second branch contained southern strains MI-2 and North American strain B31 as well as several European strains that have the same ospC sequence. The third branch was shared by southern B. burgdorferi sensu stricto strain MI-5, southern B. bissettii strains MI-9 and FD-1, and southern B. andersonii strains MOD-5 and MOK-3a. Southern B. andersonii strains BC-1, SI-10, and MOS-1b and southern B. bissettii strains SCW-30h and AI-1 occupy separate branches. However, southern B. bissettii strain MI-6 shared a branch with southern B. andersonii strain MOD-1. Interestingly, but not surprisingly, MI-8, as noted in our previous reports (35; Lin et al., Proc. VIII Int. Conf. Lyme Borreliosis Other Emerg. Tick-Borne Dis.), is present but distinct from B. burgdorferi sensu stricto strains. As noted earlier, its taxonomic status remains uncertain, but it appears to be an undescribed genospecies.
DISCUSSION
Usually, the greater the genetic diversity of a given species, the longer the time that was available for its evolution. When several phylogenetic trees derived from different genes or molecular markers are compared, if the topologies of the trees are different, this may suggest that genetic transfer or recombination has occurred. The phylogenetic trees generated from the ospC gene of southern strains show greater differences than phylogenetic trees derived from the rrf-rrl intergenetic spacers (35), pulsed-field gel electrophoresis (39; Lin et al., Proc. VIII Int. Conf. Lyme Borreliosis Other Emerg. Tick-Borne Dis.), and randomly amplified polymorphic DNA fingerprint (Lin et al., Proc. VIII Int. Conf. Lyme Borreliosis Other Emerg. Tick-Borne Dis.).
Similarity matrixes, amino acid sequences, phylogenetic trees, and major ospC groups all suggest that lateral transfer and internal gene recombination of the ospC gene occurred between B. burgdorferi sensu stricto and B. bissettii or between B. bissettii and B. andersonii. OspC diversity may be promoted by several mechanisms, including host-stimulated immunological selection, gene transfer, intragenomic gene recombination, effects of environmental constraints, or other factors. Of these factors, frequency-dependent selection might be the major force maintaining the variation in ospC (21, 64).
The great diversity of OspC compared to OspA (a single type in B. burgdorferi sensu stricto and B. afzelii) (30, 70) is strong evidence that immunological selection and gene transfer happened frequently in B. burgdorferi sensu stricto and B. afzelii. This frequency-dependent selection is created by the host immune system, because OspC is a major protective antigen and is expressed on the surface of spirochetes during tick feeding (54). It can cause a strong immunoglobulin M response at an early stage of disease (19). Interestingly, OspC can protect animals against subsequent infection by the same strain (26) but not by heterologous strains (49). Also, injection with polyclonal and monoclonal antibodies to OspC can cure chronically infected mice and prevent reinfection with the same strain. It can also cure current infection (72). To escape selective immunological pressure caused by hosts, spirochetes can change their ospC sequences and thus cause variation of the ospC gene.
In addition to immunological selection, there is another strategy for bacteria to escape immunological attack. The spirochetes which produce stable OspA are kept in the tick, and spirochetes which produce OspC or nothing are able to enter a host. Recent studies show that before the blood meal, spirochetes in the tick are a homogeneous population that mainly produce OspA. During the blood meal, the population becomes more heterogeneous. Many spirochetes produce both OspA and OspC, whereas others produce only a single Osp, and a few produce neither Osp. Most of the spirochetes in the salivary glands and host dermis produce neither OspA nor OspC, and a significant minority produce only OspC. OspC may play a role in allowing the spirochetes to escape from the gut into the hemocoele. OspA serves to bind spirochetes in the tick midgut, and OspC facilitates transfer from the gut lumen to the hemocoele (41). By downregulating OspA, the spirochetes might free themselves to migrate through the midgut epithelium and out of the midgut (26, 46, 59, 60). Spirochetes change their expression pattern of Osps in ticks when they move from the gut to the salivary gland and host dermis (41).
Isolate MI-8 was considered an undescribed genospecies in our previous report (35), and the current ospC analysis supports this conclusion. It occupies a separate branch in both of the phylogenetic trees (Fig. 2 and 3) derived from amino acid sequences and partial nucleotide sequences of ospC. It shares a branch with southern B. bissettii strains SCGT-8a and SCGT-10 in the tree derived from amino acid sequences (Fig. 2). In the tree generated from partial nucleotide sequences, it is related but distinct from B. burgdorferi sensu stricto strains 272, 28354, MUL, KIPP, 28691, and N40 and southern B. bissettii strains AI-1 and SCW-30h (Fig. 3). Whole DNA-DNA hybridization analysis with a group of strains containing MI-8 did not allow a clear conclusion because the hybridization levels were in borderline positions (Guy Baranton, Pasteur Institute, Paris, France, personal communication).
Since the North American B. burgdorferi sensu stricto strains in the clusters noted above were isolated from human skin, the major tick vector I. scapularis, or the common animal reservoir, P. gossypinus, perhaps the MI-8 strain and the southern B. bissettii strains SCW-30h and AI-1 in this cluster might be infective to humans (Fig. 3). Also needing further evaluation is another undescribed southern spirochete isolate, TXW-1. As noted in the Results, we were unable to amplify the ospC gene under various PCR conditions. Whether it lacks the ospC gene or the undetected gene in this isolate is truncated or otherwise altered at the primer sites remains to be determined. Although this strain appeared to be close to B. garinii in our previous analysis (35) using PCR-restriction fragment length polymorphism of the rrf-rrl intergenic spacer, data in the present study suggest that it belongs to an undescribed species among relapsing fever borreliae.
OspC might play a direct role in determining the potential for a given isolate to cause disseminated disease. The major ospC groups of B. burgdorferi sensu stricto found in human erythema migrans lesions are only a subset of the groups found in ticks based on single-strand conformation polymorphism analysis (27, 64). Moreover, of the 58 defined ospC groups in B. burgdorferi sensu stricto, B. garinii, and B. afzelii, only 10 groups belong to known invasive Borrelia strains that are able to cause disseminated infection in humans (9, 21).
Infectivity and pathogenicity of most southern strains remain to be determined. Thus far, only B. burgdorferi sensu stricto has been confirmed to be pathogenic for humans in the United States. B. andersonii occurs in the eastern half of the United States and appears to exist primarily in an enzootic cycle involving cottontail rabbits and I. dentatus. Therefore, B. andersonii is thought not to infect humans. B. bissettii has been isolated only from ticks and nonhuman animals to date in the United States. Whether it is infectious or pathogenic for humans is unknown; however, several isolates identified as B. bissettii have been isolated from Slovenian patients (46). This suggests that B. bissettii might cause human Lyme disease in the United States. Earlier, strain 25015 from New York, the reference strain for this genospecies, was thought to be infectious but nonpathogenic in mice (3). However, a later study reported it to cause mild arthritis (23).
Typical and atypical B. burgdorferi sensu stricto strains are found among southern isolates (Fig. 2 and 3). Southern B. burgdorferi sensu stricto isolates appear in almost all of the branches in the North American part of the tree. This indicates great genetic diversity among southern B. burgdorferi sensu stricto. Typical southern B. burgdorferi sensu stricto strains, such as MI-2, are quite similar to the B31 reference strain and are located in the same branch (Fig. 2 and 3). Previous analyses based on rrf-rrl intergenic spacer sequences indicate the same association (35). Except for B31 which was isolated from the tick I. scapularis, all the strains in this branch of the tree were isolated from cerebrospinal fluid, blood, synovial fluid, and skin of patients in Europe (Fig. 2 and 3). MI-2 most closely aligns with a group that possesses many proven human-invasive strains (9, 57). TXGW and MI-2 are currently the only two known southern isolates that belong to this group.
The ospC genotype is associated with invasiveness and infectivity of B. burgdorferi and might determine the difference in pathogenicity between clones (9, 57). A given strain is defined as invasive if it is isolated from a secondary site in the body, for example, blood, joints, or the central nervous system. Although OspC is not expressed as a major protein in B. burgdorferi in culture, it is an immunodominant protein of the early humoral immune response in humans (1, 19, 65, 67; B. Wilske, V. Preac-Mursic, G. Schierz, G. Liegl, and W. Gueye, Proc. Lyme Borreliosis Update Europe, p. 299-309, 1987) and is a specific and sensitive marker for the early stage of Lyme disease (43, 68, 69). The expression pattern of ospC plays a role in the infection cycle of the spirochete (54, 59-61). ospC is expressed and ospA is repressed when the ticks begin feeding on mice and humans (24, 54). Factors that regulate the switch from expression of OspA to OspC are likely varied and complex. The switch is in part due to the change in temperature; ospC is induced at 32 to 37°C, but not at 24°C (54, 59-61). Other factors, such as cell density (18), growth phase (50), or a change in pH (14, 15), involve the expression regulation of OspC and OspA. OspC expression was shown to increase in cultures treated with hemolymph (31). Evidence suggests coregulation of the ospA and ospC genes (33).
The major ospC groups found in human erythema migrans lesions are only a subset of the ospC groups found in ticks (57). Moreover, of the 22 major ospC groups in B. burgdorferi sensu stricto, only four groups (A, B, I, and K) had higher frequencies in the primary infected sites than in ticks and were the only groups found in secondary sites. The frequency distribution of ospC groups from ticks is significantly different from that from primary sites (erythema migrans lesion or skin rash), which in turn is significantly different from that in secondary sites (blood, joints, or the central nervous system) (57). Similarly, of the 14 ospC major groups in B. afzelii, only 2 groups contain the putative invasive isolates, and of 22 ospC groups in B. garinii, 4 groups contain all the putative invasive isolates (9, 57). Thus, there are only a minority (10 of 58) of ospC groups in the three genospecies (B. burgdorferi sensu stricto, B. garinii, and B. afzelii) that are invasive and able to colonize and persist in organs other than skin. Other groups may cause erythema migrans but do not appear to be invasive and disseminate to other organs.
Twelve major ospC groups are recognized among our 19 southern strains analyzed here. Likewise, of the 12 ospC groups, only one is considered to be in the invasive group (A). Southern B. burgdorferi sensu stricto strain MI-2, isolated from a cotton mouse from Florida, most closely aligns with a group that possesses many proven human invasive strains. Transmission experiments showed that this group of southern spirochetes is able to infect mice and hamsters and that I. scapularis can transmit it (J. H. Oliver et al., unpublished data). This suggests that typical B. burgdorferi sensu stricto strains with strong virulence and invasiveness exist in certain enzootic cycles in the southern United States and that MI-2 might be able to cause invasive human Lyme disease. Other southern B. burgdorferi sensu stricto strains, such as SI-1, SM-1, and SCI-2, are genetically closely related to North American B. burgdorferi sensu stricto strain 21347 (L) and European B. burgdorferi sensu stricto strain T255 (ospC group L), from the white-footed mouse and I. ricinus, respectively.
The diversity of ospC in one local population in New York was almost as great as the diversity worldwide (64). However, our results suggest that the distribution of ospC major groups might be associated with geographic sites. Of the three ospC groups containing B. burgdorferi sensu stricto isolates, two of them (SI-1 group and MI-5 mixture group) separated into different branches in the tree and formed different ospC groups (Fig. 2, Table 4).
B. andersonii and B. bissettii strains were distributed throughout the clusters in the B. burgdorferi sensu stricto part of the tree, suggesting that these three genospecies are closely related (Fig. 3). Perhaps genetic transfer or intragenomic gene recombination occurred among southern strains and also between southern and northern strains. Several methods can be used to determine genetic transfer or recombination based on sequence data (21). The first method is to compare several gene trees generated from the same set of strains but different molecular markers; a difference in branching order is evidence for recombination. Comparison of the phylogenetic trees generated from rrf-rrl intergenic spacer, fla, rrs, pulsed-field gel electrophoresis, and random amplified polymorphic DNA shows that the phylogenetic trees generated from the ospC data are different from the trees derived from others. This suggests that lateral transfer of the ospC gene or intragenomic gene recombination occurred between B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii, as mentioned in the Results.
Another method is to find significant similarities in sequences among different genospecies. Close identity was observed between the ospC sequences of B. burgdorferi sensu stricto and B. bissettii and between B. bissettii and B. andersonii. All the differences between the phylogenetic trees generated from ospC data and the trees generated from other genes and the close identity of ospC sequences among genospecies are caused by the introduction of a small piece of DNA (about 500 bp) into the central variable region of the ospC gene (Table 3).
Southern B. burgdorferi sensu stricto strain MI-5 shared a branch with southern B. bissettii strains MI-9 and FD-1, and this branch combined with southern B. andersonii strains MOD-5 and MOK-3a (Fig. 3). They were located in the fourth cluster of the tree, which contained most of the typical North American and European B. burgdorferi sensu stricto strains. This distribution adds additional evidence suggesting that the three southern B. burgdorferi genospecies are closely related. The evolutionary process continues, and the different levels of infectivity and virulence among the strains may be due to selection pressures in the tick vectors or animal hosts or to environmental conditions.
It is not strange that the southern B. andersonii strains MOD-1, BC-1, SI-10, and MOS-1b shared the same cluster as B. andersonii strains 21123 and 19857 from the north. All of them belong to the B. andersonii genospecies and were isolated from I. dentatus and/or cottontail rabbits except for SI-10, which was isolated from I. scapularis. Unexpectedly, however, they were grouped into the same cluster as the southern B. bissettii strain MI-6 and the North American B. burgdorferi sensu stricto strains 27579, B. pacificus (strain name, not genospecies name), SON188, and the European B. burgdorferi sensu stricto strain MIL (Fig. 3). The strains in the major ospC groups F, H, J, T, and U also grouped into this cluster (Fig. 3). Except for MI-6, which was isolated from a cotton rat in Florida, all these strains were isolated from ticks in the I. ricinus species complex. B. pacificus and SON188 were isolated from I. pacificus ticks in California, 27579 was isolated from I. scapularis in Connecticut, MIL was isolated from I. ricinus in Slovakia, and strains in ospC groups F, H, J, T, and U were isolated from I. scapularis from New York. The data suggest that the tick vector plays a role in the evolutionary process of the ospC gene. This interaction is probably influenced by geographic location and the genospecies distribution of Borrelia strains. That is probably why Borrelia strains from different genospecies and different geographic locations occupy distinct branches in the same cluster in phylogenetic trees (Fig. 2 and 3).
Acknowledgments
This research was supported in part by grant R37 AI-24899 from the National Institutes of Health and cooperative agreement U50/CCU410282 from the Centers for Disease Control and Prevention.
We thank G. Baranton (Pasteur Institute, Paris, France), B. Wilske (Max von Pettenkofer Institut fur Hygiene und Medizinische Mikrobiologie, University of Munich, Munich, Germany), and M. Fukunaga (Fukuyama University, Fukuyama, Japan) for reviewing the manuscript and G. Teltow (Texas Department of Health) for sending us the Dermacentor variabilis tick from which we isolated the TXW-1 spirochete strain. We thank two anonymous reviewers for suggestions to improve the manuscript.
REFERENCES
- 1.Aguero-Rosenfeld, M. E., J. Nowakowski, D. F. McKenna, C. A. Carbonaro, and G. P. Wormser. 1993. Serodiagnosis in early Lyme disease. J. Clin. Microbiol. 31:3090-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. F., L. A. Magnarelli, R. B. LeFebvre, T. G. Andreadis, J. B. McAninch, G. C. Perng, and R. C. Johnson. 1989. Antigenically variable Borrelia burgdorferi isolated from cottontail rabbits and Ixodes dentatus in rural and urban areas. J. Clin. Microbiol. 27:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. F., S. W. Barthold, and L. A. Magnarelli. 1990. Infectious but nonpathogenic isolate of Borrelia burgdorferi. J. Clin. Microbiol. 28:2693-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. F., R. A. Flavell, L. A. Magnarelli, S. W. Barthold, F. S. Kantor, R. Wallich, D. H. Persing, D. Matheisen, and E. Fikrig. 1996. Novel Borrelia burgdorferi isolates from Ixodes scapularis and Ixodes dentatus ticks feeding on humans. J. Clin. Microbiol. 34:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assous, M. V., D. Postic, G. Paul, P. Nevot, and G. Baranton. 1993. Western blot analysis of sera from Lyme borreliosis patients according to the genomic species of the Borrelia strains used as antigens. Eur. J. Clin. Microbiol. Infect. Dis. 12:261-268. [DOI] [PubMed] [Google Scholar]
- 6.Assous, M. V., D. Postic, G. Paul, P. Nevot, and G. Baranton. 1994. Individualisation of two new genomic groups among American Borrelia burgdorferi sensu lato strains. FEMS Microbiol. Lett. 121:93-98. [DOI] [PubMed] [Google Scholar]
- 7.Balmelli, T., and J. C. Piffaretti. 1996. Analysis of the genetic polymorphism of Borrelia burgdorferi sensu lato by multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 46:167-172. [DOI] [PubMed] [Google Scholar]
- 8.Baranton, G., D. Postic, I. Saint Girons, P. Boerlin, J. C. Piffaretti, M. Assous, and P. A. Grimont. 1992. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS 461 associated with Lyme borreliosis. Int. J. Syst. Bacteriol. 42:378-383. [DOI] [PubMed] [Google Scholar]
- 9.Baranton, G., G. Seinost, G. Theodore, D. Postic, and D. Dykhuizen. 2001. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res. Microbiol. 152:149-156. [DOI] [PubMed] [Google Scholar]
- 10.Brown, R. N., and R. S. Lane. 1992. Lyme disease in California: a novel enzootic transmission cycle of Borrelia burgdorferi. Science 256:1439-1442. [DOI] [PubMed] [Google Scholar]
- 11.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 12.Cadavid, D., D. D. Thomas, R. Crawley, and A. G. Barbour. 1994. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J. Exp. Med. 179:631-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canica, M. M., F. Nato, L. du Merle, J. C. Mazie, G. Baranton, and D. Postic. 1993. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand. J. Infect. Dis. 25:441-448. [DOI] [PubMed] [Google Scholar]
- 14.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter, C. J., S. Bergstrom, S. Norris, and A. G. Barbour. 1994. A family of surface-exposed proteins of 20 kDa in the genus Borrelia. Infect. Immun. 62:2792-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. 2001. Lyme disease—United States, 1999. Morb. Mortal. Wkly. Rep. 50:181-185. [PubMed] [Google Scholar]
- 18.De Silva, A. M., N. S. Zeidner, Y. Zhang, M. C. Dolan, J. Piesman, and E. Fikrig. 1999. Influence of outer surface protein A antibody on Borrelia burgdorferi within feeding ticks. Infect. Immun. 67:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dressler, F., J. A. Whalen, B. N. Reinhardt, and A. C. Steere. 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167:392-400. [DOI] [PubMed] [Google Scholar]
- 20.Dykhuizen, D. E., D. S. Polin, J. J. Dunn, B. Wilske, V. Preac-Mursic, R. J. Dattwyler, and B. J. Luft. 1993. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. USA 90:10163-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykhuizen, D. E., and G. Baranton. 2001. Implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 23.Fikrig, E., S. W. Barthold, D. H. Persing, X. Sun, F. S. Kantor, and R. A. Flavell. 1992. Borrelia burgdorferi strain 25015: characterization of outer surface protein A and vaccination against infection. J. Immunol. 148:2256-2260. [PubMed] [Google Scholar]
- 24.Fingerle, V., G. Liegl, U. Munderloh, and B. Wilske. 1998. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus ticks removed from humans. Med. Microbiol. Immunol. 187:121-126. [DOI] [PubMed] [Google Scholar]
- 25.Fingerle, V., H. Laux, U. G. Munderloh, U. Schulte-Spechtel, and B. Wilske. 2000. Differential expression of outer surface proteins A and C by individual Borrelia burgdorferi in different genospecies. Med. Microbiol. Immunol. 189:59-66. [DOI] [PubMed] [Google Scholar]
- 26.Gilmore, R. D., K. J. Kappel, M. C. Dolan, T. R. Burkot, and B. J. B. Johnson. 1996. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect. Immun. 64:2234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman, D. S., P. W. Wang, I. N. Wang, E. M. Bosler, B. J. Luft, and D. E. Dykhuizen. 1996. Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand conformation polymorphism analysis. J. Clin. Microbiol. 34:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins, D. G., and R. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biol. Sci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 29.Jauris-Heipke, S., R. Fuchs, M. Motz, V. Preac-Mursic, E. Schwab, E. Soutschk, G. Will, and B. Wilske. 1993. Genetic heterogeneity of the genes coding for the outer surface protein C (ospC) and the flagellin of Borrelia burgdorferi. Med. Microbiol. Immunol. 182:37-50. [DOI] [PubMed] [Google Scholar]
- 30.Jauris-Heipke, S., G. Liegl, V. Preac-Mursic, D. Rössler, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1995. Molecular analysis of genes encoding outer surface protein C (ospC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J. Clin. Microbiol. 33:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johns, R. H., D. E. Sonenshine, and W. L. Hynes. 2000. Enhancement of OspC expression by Borrelia burgdorferi in the presence of tick hemolymph. FEMS Microbiol. Lett. 193:137-141. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, R. C., G. P. Schmid, F. W. Hyde, G. Steigerwalt, and D. J. Brenner. 1984. Borrelia burgdorferi sp. nov.: etiological agent of Lyme disease. Int. J. Syst. Bacteriol. 34:496-497. [Google Scholar]
- 33.Jonsson, M., and S. Bergstrom. 1995. Transcriptional and translational regulation of the expression of the major outer surface proteins in Lyme disease Borrelia strains. Microbiology 141:1321-1329. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetic analysis, 1.01 ed. Pennsylvania State University, University Park, Pa.
- 35.Lin, T., J. H. Oliver, Jr., L. Gao, T. M. Kollars, Jr., and K. L. Clark. 2001. Genetic heterogeneity of Borrelia burgdorferi sensu lato in the southern United States based on restriction fragment length polymorphism and sequence analysis. J. Clin. Microbiol. 39:2500-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liveris, D., A. Gazumyan, and I. Schwartz. 1995. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marconi, R. T., D. Liveris, and I. Schwartz. 1995. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J. Clin. Microbiol. 33:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marti Ras, N., D. Postic, M. Foretz, and G. Baranton. 1997. Borrelia burgdorferi sensu stricto, a bacterial species made in the USA? J. Syst. Bacteriol. 47:1112-1117. [DOI] [PubMed] [Google Scholar]
- 39.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 40.O'Connell, S., M. Granstrom, J. S. Gray, and G. Stanek. 1998. Epidemiology of European Lyme borreliosis. Zentbl. Bakteriol. 287:229-240. [DOI] [PubMed] [Google Scholar]
- 41.Ohnishi, J., J. Piesman, and A. M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA 98:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver, J. H., Jr. 1996. Lyme borreliosis in the southern United States: a review. J. Parasitol. 82:926-935. [PubMed] [Google Scholar]
- 43.Padula, S. J., F. Dias, A. Sampieri, R. B. Craven, and R. W. Ryan. 1994. Use of recombinant OspC from Borrelia burgdorferi for serodiagnosis of early Lyme disease. J. Clin. Microbiol. 32:1733-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, and J. F. Anderson. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picken, R. N., Y. Cheng, F. Strle, J. Cimperman, V. Maraspin, S. Lotric-Furlan, E. Ruzic-Sabljic, D. Han, J. A. Nelson, M. M. Picken, and G. M. Trenholme. 1996. Molecular characterization of Borrelia burgdorferi sensu lato from Slovenia revealing significant differences between tick and human isolates. Eur. J. Clin. Microbiol. Infect. Dis. 15:313-323. [DOI] [PubMed] [Google Scholar]
- 46.Picken, R. N., Y. Cheng, F. Strle, and M. M. Picken. 1996. Patient isolates of Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities of strain 25015. J. Infect. Dis. 174:1112-1115. [DOI] [PubMed] [Google Scholar]
- 47.Postic, D., M. V. Assous, P. A. D. Grimont, and G. Baranton. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf(5S)-rrl(23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743-752. [DOI] [PubMed] [Google Scholar]
- 48.Postic, D., N. M. Ras, R. S. Lane, M. Hendson, and G. Baranton. 1998. Expanded diversity among California Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127). J. Clin. Microbiol. 36:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Probert, W. S., M. Crawford, R. B. Cadiz, and R. B. LeFebvre. 1997. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis. 175:400-405. [DOI] [PubMed] [Google Scholar]
- 50.Ramamoorthy, R., and M. T. Philipp. 1998. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect. Immun. 66:5119-5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadziene, A., B. Wilske, M. S. Ferdows, and A. G. Barbour. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, B. S., N. S. Zeidner, T. R. Burkot, G. O. Maupin, and J. Piesman. 2000. Borrelia isolates in northern Colorado identified as Borrelia bissettii. J. Clin. Microbiol. 38:3103-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete. Borrelia burgdorferi during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwan, T. G., and J. Piesman. 2002. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg. Infect. Dis. 8:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seinost, G., D. E. Dykhuizen, R. J. Dattwyler, W. T. Golde, J. J. Dunn, I. N. Wang, G. P. Wormser, M. E. Schriefer, and B. J. Luft. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67:3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, Calif.
- 59.Stevenson, B., L. K. Bogkenstedt, and S. W. Barthold. 1994. Expression and gene sequence of outer surface protein C of Borrelia burgdorferi reisolated from chronically infected mice. Infect. Immun. 62:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevenson, B., and S. W. Barthold. 1994. Expression and sequence of outer surface protein C among North American isolates of Borrelia burgdorferi. FEMS Microbiol. Lett. 124:367-372. [DOI] [PubMed] [Google Scholar]
- 61.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swofford, D. L. 1992. Paup: phylogenetic analysis using parsimony, version 3.0. Illinois Natural History Survey, Champaign, Ill.
- 63.Wang, G., A. P. van Dam, and J. Dankert. 1999. Evidence for frequent ospC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol. Lett. 177:289-296. [DOI] [PubMed] [Google Scholar]
- 64.Wang, I. N., D. E. Dykhuizen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilske, B., V. Preac-Mursic, G. Schierz, and K. von Busch. 1986.. Immunochemical and immunological analysis of European Borrelia burgdorferi strains. Zentbl. Bakteriol. Mikrobiol. Hyg. Ser. A 263:92-102. [DOI] [PubMed]
- 66.Wilske, B., V. Preac-Mursic, U. B. Gobel, B. Graf, S. Jauris-Heipke, E. Soutschek, E. Schwab, and G. Zumstein. 1993. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J. Clin. Microbiol. 31:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilske, B., V. Fingerle, P. Herzer, A. Hofmann, G. Lehnert, H. Peters, H. W. Pfister, V. Preac-Mursic, E. Soutschek, and K. Weber. 1993. Recombinant immunoblot in the serodiagnosis of Lyme borreliosis. Med. Microbiol. Immunol. 182:255-270. [DOI] [PubMed] [Google Scholar]
- 68.Wilske, B., V. Preac-Mursic, S. Jauris, A. Hofmann, I. Pradel, E. Soutschek, E. Schwab, G. Will, and G. Wanner. 1993. Immunological and molecular polymorphisms of ospC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect. Immun. 61:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilske, B., V. Fingerle, V. Preac-Mursic, S. Jauris-Heipke, A. Hofmann, H. Loy, H. W. Pfister, D. Rossler, and E. Soutschek. 1994. Immunoblot using recombinant antigens derived from different genospecies of Borrelia burgdorferi sensu lato. Med. Microbiol. Immunol. 183:43-59. [DOI] [PubMed] [Google Scholar]
- 70.Wilske, B., S. Jauris-Heipke, R. Lobentanzer, I. Pradel, V. Preac-Mursic, D. Rössler, E. Soutschek, and R. C. Johnson. 1995. Phenotypic analysis of outer surface protein C (ospC) of Borrelia burgdorferi sensu lato by monoclonal antibodies: relationship to genospecies and OspA serotype. J. Clin. Microbiol. 33:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, and K. E. Hagman. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 72.Zhong, W., T. Stehle, C. Museteanu, A. Siebers, and L. Gern. 1997. Therapeutic passive vaccination against chronic Lyme disease in mice. Proc. Natl. Acad. Sci. USA 94:12533-12538. [DOI] [PMC free article] [PubMed] [Google Scholar]