Abstract
Liquid biopsy offers a noninvasive method to identify and monitor tumor-derived biomarkers, including circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), exosomes, microRNAs, and tumor-educated platelets, that provide real-time insights into the biological behavior of gynecological cancers. The detection of these markers has the potential to revolutionize cancer management by enabling earlier detection, providing novel data to personalize treatments, and predicting disease recurrence before clinical imaging can confirm progression, thereby also guiding complex clinical decision-making. However, because this new “omics” layer introduces additional complexity, it must be fully understood, from its biological rationale to technical development and clinical integration, to prevent confusion or misapplication. That is why, focusing on 14 critical fields of inquiry, our goal is to map the current state of liquid biopsy from bench to bedside while highlighting practical considerations for clinical integration. Each topic integrates recent advances in assay sensitivity, biomarker variability, and data interpretation, underscoring how standardized protocols and robust analytical methods are pivotal for reliable results. We then translate these findings into disease-specific insights, examining how liquid biopsy could refine early detection, minimal residual disease assessment, and therapy guidance in endometrial, cervical, and ovarian cancers. Although several FDA-approved assays and promising commercial tests illustrate the field’s rapid evolution, many translational hurdles remain, including the need for harmonized protocols, larger prospective clinical trials, and cost-effectiveness analyses. Crucially, our synthesis clarifies the pivotal role of interdisciplinary collaboration. Oncologists, laboratory scientists, and industry partners must align on standardized procedures and clinically relevant endpoints. Without such coordination, promising biomarkers may remain confined to research settings, limiting their practical benefit. Taken together, our review offers a translational view designed to contextualize liquid biopsy in gynecological oncology.
Keywords: Liquid biopsy, Gynecological cancer, Translational medicine, Clinical implementation, Precision oncology, Narrative review
Introduction
The era of precision oncology mandates a paradigm shift in diagnosing, monitoring, and treating gynecological malignancies. Over 1.3 million women in the United States, encompassing 862,875 endometrial, 295,748 cervical, and 238,484 ovarian cancer cases, highlight the urgent need to translate molecular insights into clinical practice [1]. In this context, liquid biopsy has emerged as a non-invasive diagnostic tool with significant promise, facilitating the detection of tumor-derived biomarkers [2–7]. Despite this promise, translating emerging molecular findings into routine gynecological oncology care remains challenging. Each step, from in vitro experiments to clinical validation, introduces distinct hurdles, as illustrated by Curry’s conceptual framework on translational science [8]. Moreover, the complexity of oncology requires approaches that do not overwhelm practitioners, making standardized, domain-agnostic strategies critical for practical implementation [9]. In gynecological oncology specifically, protocols must consider the molecular heterogeneity seen across endometrial, cervical, and ovarian cancers, all while remaining feasible in real-world settings. A truly translational perspective recognizes the bidirectional nature of research and clinical practice: clinicians require clear guidance on interpreting and applying molecular data, while scientists need insights from clinical realities to refine diagnostics and treatments [10, 11]. Understanding this interplay is paramount in gynecological oncology, where the nuances of tumor biology intersect with patient-centered care needs. This narrative review thus spotlights liquid biopsy as both a scientific breakthrough and a case study in effective translation. By examining its technical advances alongside real-world considerations, we aim to clarify how liquid biopsy can be harnessed to enhance gynecological cancer care.
Methods
We adopted a narrative, translational approach to address 14 critical issues in liquid biopsy for gynecological oncology, guided by the PRISMA flowchart (Supplementary Document 1). Each issue was explored either via focused search strategies or an experience-based review (Table 1):
Narrative Exploration: Foundational topics were identified through landmark studies and the authors’ expertise.
Targeted Searches: Where specific data were needed, we used predefined keywords in PubMed, Embase, and Cochrane (2019–2024), limiting to English-language human studies of clear relevance.
Screening: Retrieved articles were manually screened to exclude non-gynecological malignancies, case reports, editorials, or non-original data.
Table 1.
Summary of approaches, search strategies, and outcomes for 14 critical issue related to liquid biopsy in gynecological cancers
Overall, our goal was to integrate molecular rationale, technical considerations, and real-world clinical implications for each query.
The targets of liquid biopsy
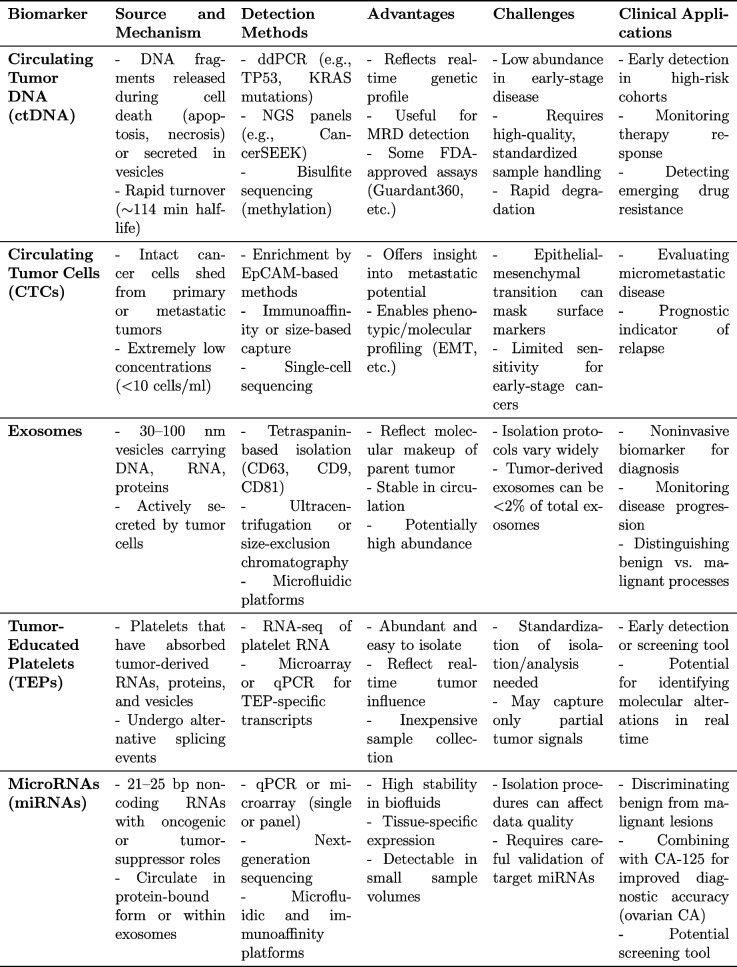
Liquid biopsy represents a significant advancement in cancer diagnostics by enabling noninvasive sampling of tumor-derived materials from blood, urine, ascitic fluid, pleural fluid, cerebrospinal fluid, sputum, saliva, and feces [2–7] (Table 2 and Fig. 1). The principal analytes include:
Table 2.
Summary of liquid biopsy biomarkers in gynecological cancers
Fig. 1.
Graphic representation of liquid biopsy markers and sources for cancer detection. Liquid biopsy markers encompass circulating tumor cells, extracellular vesicles, tumor-derived microRNAs, and microbial cell-free DNA derived from the microbiota, which can be utilized to extrapolate cancer-associated microbiota signatures. These markers are released and detectable in various bodily fluids, including blood, urine, saliva, or stool (created with BioRender.com)
- Cell-free DNA (cfDNA) and Circulating Tumor DNA (ctDNA)
- Cell-free DNA (cfDNA) refers to DNA fragments found in circulation, and its tumor-derived fraction is termed ctDNA. In healthy individuals, plasma cfDNA levels range from 65 to 877 ng/ml, while those in cancer patients often exceed 1000 ng/ml [12]. This tumor-derived fraction possesses genetic and epigenetic alterations and typically has a brief half-life of around 114 minutes [13]. Clearance primarily occurs via the reticuloendothelial system, with the liver accounting for 70–90%, spleen ~3%, kidneys ~4%, and the remainder undergoing enzymatic degradation [14, 15].
- Circulating Tumor Cells (CTCs)
- CTCs are intact cancer cells shed from primary or metastatic sites into the bloodstream, generally at very low concentrations (<10 cells/ml) [8]. Their half-life ranges from 1 to 2.4 hours, and they are commonly identified using epithelial markers such as epithelial cell adhesion molecule (EpCAM) or via distinct cellular traits [16].
- Exosomes
- Exosomes are 30–100 nm vesicles formed via endocytic pathways [21–25]. They carry DNA, RNA, miRNAs, and proteins, and can be isolated using tetraspanins like CD63, CD9, and CD81 [26]. Because they reflect the molecular makeup of their cells of origin, exosomes play crucial roles in tumor communication and metastasis.
- MicroRNAs (miRNAs)
How cancer relates to liquid biopsy targets in gynecologic oncology
Although each liquid biopsy component ultimately reflects tumor presence, each biomarker enters circulation through distinct biological mechanisms:
CfDNA enters the circulation via apoptosis, necrosis, and NETosis [29]. During apoptosis, DNA is fragmented by enzymes such as DNA fragmentation factor B (DFFB), DNASE1, and DNASE1L3 [30, 31], and may also be released under conditions of cellular stress or injury [32]. Furthermore, their altered chromatin structure, which is more “open” due to intensified transcription, renders DNA susceptible to nuclease-mediated fragmentation [33]. These unique fragmentation patterns mirror profound changes in nuclear organization and gene expression seen in malignant transformation [34–36], a field referred to as “fragmentomics.” In circulation, it is protected by binding to nucleosomes, argonauts, and lipoproteins (HDL, LDL), or by encapsulation in vesicles [37, 38]. Cancer cells can also actively secrete DNA through extracellular vesicles, reflecting their high metabolic activity [39].
CTCs enter the bloodstream when cells detach from the primary tumor or metastases, reflecting key molecular alterations essential for malignancy. These include modified cell adhesion, heightened survival mechanisms, and resistance to anoikis (programmed cell death caused by detachment from the extracellular matrix). Their mere presence in peripheral blood points to the tumor’s invasive potential [16, 40]
TEPs are platelets whose molecular profiles have been reshaped by close interaction with the tumor microenvironment. Cancer cells release soluble factors, such as RNA and proteins, which platelets absorb, triggering alternative splicing events. These modifications indicate both direct tumor influence and the body’s wider response to malignancy [17–20]. Although early-phase or “phase 0” studies in gynecological cancers have hinted at the strong potential of TEPs for cancer detection and monitoring, robust and up-to-date evidence specifically within gynecological malignancies remains limited [41, 42].
Exosomes in cancer undergo changes in both quantity and composition because malignant cells frequently boost exosome production as part of survival and growth strategies, promoting cell-to-cell communication, remodeling the tumor microenvironment, facilitating invasive behavior, and contributing to drug resistance. Moreover, their cargo, mutated DNA, regulatory RNAs, and proteins, mirrors the parental tumor’s molecular signature [22–25].
Abnormal miRNA profiles in cancer represent a breakdown of crucial regulatory pathways. Elevated or depleted miRNA expression can bolster the malignant phenotype. These small RNAs reach circulation via active vesicular secretion or cell death; they remain stable either by binding protective proteins or by encapsulation in vesicles. Their distinct expression signatures in patient plasma often correlate with tumor presence and progression [25, 27, 28].
Techniques for biomarker detection
From a practical standpoint, having the option to analyze multiple biomarkers makes liquid biopsy especially appealing to clinicians. Nonetheless, standardizing protocols, managing costs, and accurately interpreting complex genomic data remain significant barriers to routine adoption. The principal methods include:
DNA methylation represents one of the earliest and most stable cancer-associated alterations. It is typically assessed via bisulfite conversion, which distinguishes methylated from unmethylated cytosines [43]. Panels targeting genes such as RASSF1A, OPCML, and BRCA1 can achieve sensitivity and specificity as high as 91%, thereby increasing confidence in early malignancy detection [44]. More advanced methods, including high-resolution melting analysis (HRMA) and next-generation sequencing (NGS), offer deeper insight into methylation profiles, though they come with higher costs and longer turnaround times [44]. One of the most famous multi-cancer early detection test focusing on ctDNA methylation signatures to detect multiple cancers is the Grail Galleri. With one large study suggesting the potential to identify over 50 tumor types from a single blood draw [45], its performance specifically in endometrial, cervical, or ovarian cancer still requires further validation and FDA approval is yet to come.
Because ctDNA reflects both genetic and epigenetic aspects of a tumor, measuring it can guide targeted therapy or detect minimal residual disease [46]. Techniques like droplet digital PCR (ddPCR) can reliably identify mutations (e.g., TP53, KRAS) with over 99% specificity. NGS-based panels also allow for simultaneous evaluation of multiple alterations. Importantly, multi-analyte tests now integrate ctDNA with additional biomarkers, such as selected proteins, combined with machine-learning algorithms to achieve early multi-cancer detection. For instance, CancerSEEK includes 16 gene mutations and 8 circulating proteins, and has reported sensitivities exceeding 70–90% for certain tumor types [46, 47]. In gynecologic oncology, ctDNA monitoring has shown practical clinical applications; for instance, Toboni et al. demonstrated in advanced or recurrent endometrial cancer that ctDNA status, measured via the Signatera™ multiplex PCR assay, correlates with clinical outcomes [48]. Personalized NGS approaches have likewise been used to measure responses to PARP inhibitors in ovarian cancer. Large-scale sequencing of ctDNA has further delineated actionable molecular profiles in advanced endometrial cancer [49]. Despite these advances, ctDNA test sensitivity can decrease when ctDNA levels are extremely low, emphasizing the necessity of high-quality plasma samples and rigorous laboratory protocols [50]. Several ctDNA-based assays have received FDA approval, including Guardant360 and FoundationOne® Liquid CDx [44]. FoundationOne® Liquid has also been applied to endometrial and other advanced tumors (lung, colon, ovarian, breast), where ctDNA fraction guides real-time management [51]. Moreover, low-coverage Illumina HiSeq2500 sequencing studies indicate that cfDNA can provide relevant molecular insights even without direct tumor references, further extending its diagnostic and monitoring utility [51].
These small, stable, tissue-specific RNA molecules continue to generate interest in clinical oncology. They can be measured by qPCR or microarray for targeted screening, while NGS allows a more comprehensive miRNA survey [52]. Exosomal miRNAs in particular reflect ongoing tumor activity, but isolation protocols significantly affect yield and data quality. For example, ultracentrifugation can retrieve plentiful exosomes but risks vesicle damage, whereas size-exclusion chromatography (SEC) better preserves vesicle integrity but may be labor-intensive [52]. Microfluidic platforms that integrate both size-based and immunoaffinity-based isolation are emerging as a reliable alternative with promising specificity and recovery [53]. Furthermore, new techniques like ELSA-seq (Enhanced Linear-Splinter Amplification Sequencing) have shown predictive accuracies up to 97% in ovarian cancer, underscoring the role of ctDNA methylation profiling [54].
Biomarkers could reveal the story of gynecological cancers
Each biomarker type, ctDNA, CTCs, cfRNA, TEPs, and exosomes, uniquely reflects a tumor’s genetic profile, proliferative behavior, and metastatic potential.
- ctDNA: Tracing Tumor Genetic Footprints.
- Because ctDNA captures the active genetic alterations that drive a patient’s cancer, it can detect minimal residual disease (MRD) even before clinical or imaging signs emerge [55]. In ovarian cancer, clinicians often track ctDNA levels to forecast outcomes and tailor therapies, whereas in endometrial carcinoma, fluctuating ctDNA can suggest a heightened recurrence risk, prompting closer monitoring or additional intervention.
- CTCs: Migratory Messengers of Tumors.
- Unlike ctDNA, which reflects genetic mutations, CTCs are intact cancer cells that escape from primary or metastatic lesions [46, 56, 57]. Detecting even a small number can expose a latent potential for spread. In ovarian cancer, CTC enumeration and molecular profiling reveal micrometastatic disease, while in endometrial cancer, the presence of CTCs, sometimes detectable in early stages, may signal impending recurrence [58–61]. A notable hurdle is that CTCs can undergo epithelial-to-mesenchymal transition (EMT), losing conventional surface markers and thereby requiring more advanced technologies for reliable capture.
- cfRNA and TEPs: Unveiling Subtler Layers of Tumor Biology.
- Not all tumor-related signals come from DNA fragments or whole cells. cfRNA (including mRNA, miRNA, circRNA, and lncRNA) reflects active gene expression in tumors and can remain surprisingly stable in circulation [55]. In ovarian cancer, certain miRNAs and circRNAs appear promising for diagnosing early-stage disease, and preliminary research on mRNA within utero-tubal lavage fluid hints at additional detection approaches. Beyond cfRNA, TEPs offer another vantage point; platelets “educated” by cancer cells incorporate malignant RNA signatures that can distinguish tumors from benign states, though more validation is needed before such tests become routine.
- Exosomes: Tiny Couriers of Tumor Cargo.
- Exosomes encapsulate a tumor’s molecular fingerprint, making them promising candidates for noninvasive diagnosis, prognosis, and follow-up [55]. For example, in cervical cancer, exosomal circSLC26A4 correlates with advanced FIGO stages and lymph node metastases, suggesting its utility in gauging disease severity [62]. Exosome-based assays also show potential in differentiating benign from malignant ovarian masses, though further clinical trials are required to standardize their application.
Does chemotherapy have an impact on liquid biopsy detection rate?
Chemotherapy can alter both the release and detectability of ctDNA and CTCs. Early in treatment, tumor cell apoptosis and necrosis often lead to short-term spikes in ctDNA as dying cells shed DNA fragments [63]. Although this increase may seem concerning, it typically signifies a positive treatment response rather than disease progression. Over time, stable declines in ctDNA or CTC counts indicate meaningful tumor regression [64], whereas persistently elevated or resurgent biomarker levels suggest drug resistance or minimal residual disease [65]. For instance, platinum-resistant ovarian tumors generally exhibit higher CTC counts [65], while increasing ctDNA during PARP inhibitor therapy can signal emerging treatment failure [66, 67]. Epigenetic modifications, like HOXA9 promoter methylation, further refine therapeutic choices and prognostication [68]. By serially monitoring these biomarkers, clinicians can distinguish between chemotherapy-induced cell death and emerging resistance [69].
Ideal Scenarios for Liquid Biopsy: What are the clear-cut situations or clinical indications in gynecological cancers where liquid biopsy is particularly advantageous?
Liquid biopsy is especially beneficial in gynecological cancers for early detection, often identifying malignancies before symptoms appear, particularly in high-risk groups [70]. It also facilitates real-time monitoring of therapeutic responses, enabling treatment modifications if ctDNA levels rise or remain elevated. Post-treatment detection of minimal residual disease (MRD) via ctDNA can guide follow-up and additional interventions. In addition, liquid biopsy pinpoints actionable mutations for personalized therapy and reveals tumor heterogeneity to refine treatment plan. Finally, routine ctDNA assessments can determine recurrence risk, ensuring targeted surveillance for high risk patients [71].
Warnings on liquid biopsy
Liquid biopsy can be unreliable under certain biological and technical conditions. In early-stage (I or II) disease, ctDNA concentrations can be more than tenfold lower than in advanced disease, greatly increasing the likelihood of false negatives [72]. Additionally, ctDNA’s short half-life, ranging from 15 minutes to 2 hours, makes it susceptible to degradation, while cell lysis can further contaminate samples unless handled swiftly under stringent protocols [73, 74]. Exosomes pose similar hurdles: tumor-derived vesicles often account for less than 2% of circulating exosomes and are rapidly cleared, requiring high-throughput, highly sensitive methods for accurate analysis [75, 76]. Although TEPs are abundant and simple to isolate, they may capture only a fraction of a tumor’s genetic profile [72]. Combined, these issues can lead to suboptimal or false-negative outcomes, particularly in heterogeneous tumors or when specific histopathological detail is essential for treatment decisions [77]. Understanding and compensating for such biological complexities is crucial to preserving the accuracy and clinical utility of liquid biopsy results.
False positives and negatives in liquid biopsy
Biological complexity and technical challenges often lead to false results in liquid biopsies for gynecological cancers. For instance, Fourier transform infrared (FTIR) spectroscopy can reach nearly 99% sensitivity, but at the cost of lowered specificity, causing benign conditions to appear malignant [78] . Conversely, boosting specificity risks missing early tumors that release minimal ctDNA [78]. In fluorescence liquid biopsy protocols, benign ovarian cysts may mimic malignant fluorescence signals, raising false positives, while some ovarian tumors lack a distinct fluorescence shift, increasing false negatives [79]. Similarly, in microvesicle proteomics of uterine fluid, benign conditions can mimic malignant signatures, leading to over-diagnosis, while low microvesicle counts in early-stage cancer may go undetected [77].
Emerging biotechnological techniques under investigation for liquid biopsy
A variety of next-generation laboratory methods promise to bolster biomarker detection and revolutionize current diagnostic workflows. Digital PCR partitions samples to precisely count nucleic acids, obviating reference standards and enhancing sensitivity for rare ctDNA mutations [80]. NGS provides deep genomic snapshots, capturing tumor heterogeneity and enabling real-time monitoring of emerging subclones [81]. Nanoparticle-based assays detect signals from low-abundance targets and can be integrated into portable, point-of-care devices [82]. Meanwhile, CRISPR-Cas systems leverage genome-editing principles for rapid, targeted nucleic-acid detection, cutting turnaround times and facilitating flexible assay design [83]. Single-cell sequencing dissects individual CTCs at the genomic and epigenomic levels, identifying subtle subclones that fuel resistance or relapse [84]. Finally, electrochemical and optical biosensors transform biological interactions into quantifiable signals, delivering cost-effective, sensitive detection of biomarkers such as miRNAs, ctDNA, and exosomes [85].
AI Integration in Liquid Biopsy
Artificial intelligence can substantially enhance liquid biopsy applications by synthesizing multiple layers of cfDNA fragmentomics, methylomics, and epigenetic data, thereby boosting both sensitivity and specificity. Combining fragmentation-focused approaches like DELFI with three-dimensional genome mapping tools pinpoints tumor-specific changes in cfDNA length and distribution while determining tissue of origin via chromatin conformation and nucleosome positioning [33, 86]. Further metrics, such as promoter fragmentation entropy and windowed protection scores, illuminate gene activation and transcription factor binding [87, 88]. Orientation-aware fragmentation refines tissue-of-origin analyses by mapping nucleosome placement in open chromatin areas [89]. Methylation-based methods classify cfDNA fragments based on inferred methylation patterns, attributing them to specific tissues [90, 91]. Different databases like consolidate broad omics data with cfDNA sequencing, offering a holistic perspective on gene regulation and fragmentomics [92].
Liquid biopsy in ovarian cancer
- Screening
- Liquid biopsy screening is particularly relevant for BRCA1/2 mutation carriers, whose ovarian cancer risk is two to four times higher [93]. Various biomarkers have shown promise. Plasma protein signatures (e.g., SPARC, THBS1) have been proposed [94], and uterine lavage analyses identified a seven-protein panel with over 99% negative predictive value [95]. Pap test–based methods demonstrated 52% overall accuracy and 26% sensitivity for eight-gene panels [96], while p53 variants were found in archival Pap smears up to six years before diagnosis [97]. Methylation profiling of tumor suppressor genes ranges from 41% to 100% sensitivity, with OPCML emerging as a particularly robust biomarker [98].
- Early Diagnosis
- Meta-analysis of circulating cell-free DNA demonstrated 70% sensitivity and 90% specificity, with a diagnostic odds ratio of 26.05 and negative likelihood ratio of 0.34 [99]. Updated meta-analyses incorporating 22 studies confirmed these findings with slightly improved pooled sensitivity of 73% while maintaining 90% specificity [100]. miRNA analysis showed strong diagnostic potential, with meta-analyses revealing 89% sensitivity and 64% specificity. Multiple miRNA panels demonstrate superior performance compared to single markers, with diagnostic odds ratios of 30.06 versus 13.21 [101]. Recent research has identified nine upregulated miRNAs in ovarian cancer patients, with MiR-145 and MiR-205 showing the highest fold change exceeding 2-fold [102]. Studies have consistently demonstrated improved accuracy when combining CA125 with investigated miRNAs compared to either marker alone.
- Diagnosis
- ctDNA correlates well with tumor DNA, though heterogeneity may affect accuracy [103]. Extracellular vesicles containing miRNAs also display notable differences in expression between ovarian cancer patients and controls [104]. Diagnostic performance varies according to plasma vs. serum collection, extraction protocols, hormonal factors, and menstrual status [105, 106].
- Staging
- Several liquid biopsy biomarkers help distinguish disease stages. For instance, CD117 expression on cells and on extracellular vesicles is significantly higher in recurrent disease, particularly in high-grade serous carcinoma [107]. Advanced-stage ovarian cancer typically features elevated cfDNA levels [108] and higher ctDNA detection rates [109]. Low-coverage whole-genome sequencing of plasma cfDNA can differentiate early- from late-stage cancers [110]. Exosomal miR-205 and certain metabolic markers (phenylpyruvic acid, 4-hydroxyphenylpyruvic acid) are also linked to advanced disease [55, 111].
- Evaluation for Treatment Response
- Liquid biopsy enables real-time monitoring of chemotherapy and PARP inhibitor responses [55]. ctDNA tracks primary and acquired resistance; for instance, in BRCA-mutated cases, increased HOXA9 methylation during treatment correlates with poor PARP inhibitor efficacy [68]. Serum soluble PD-L2 levels can predict platinum response since high levels demonstrate association with platinum therapy response and low levels indicate resistance and poorer prognosis [112]. For high-grade serous ovarian cancer patients, ctDNA analysis detects chemotherapy response earlier than CA-125, with TP53 mutant allele fraction serving as a predictor of poor outcomes and rapid progression [113]. Extracellular vesicle markers CD24 and EpCAM in plasma show elevated levels in non-responding versus responding patients [114]. Additionally, cfDNA analysis through next-generation sequencing before and during treatment can track tumor progression and genetic evolution during chemotherapy [115].
- Follow-up
- For post-treatment surveillance, ctDNA shows high prognostic utility. In high-grade serous ovarian cancer, elevated ctDNA (≥0.2 copies/μL) at three months post-chemotherapy correlates with a 58.3% recurrence risk compared to patients with low levels (<0.2 copies/μL), who demonstrate only 6.7% recurrence risk [63]. Liquid biopsy can detect relapse up to seven months earlier than CT imaging [116] and outperforms CA125 in predicting progression [117]. Elevated ctDNA levels are also associated with worse survival, and HOXA9 methylation positivity raises the relapse risk more than threefold [118]. Additional studies have confirmed that ctDNA quantification can indicate recurrence months before conventional clinical methods, providing an objective definition of complete cytoreduction [119].
Liquid biopsy in endometrial cancer
- Screening
- Endometrial cancer (EC) often presents asymptomatically, prompting interest in minimally invasive screening. A major systematic review reported 56 blood-based biomarker studies and only one employing urine [120]. Recent methods emphasize site-specific sampling to improve sensitivity and specificity. A tampon-based test evaluating methylated DNA markers in vaginal fluid reached 76% sensitivity and 96% specificity (AUC=0.88) in 192 participants, and further refinements raised sensitivity to 82% [121]. Another technique, endometrial fluid sampling akin to saline infusion sonohysterography, revealed distinct microRNAs (miR-183-5p, miR-429, miR-146a-5p) differentiating malignant from benign cases in both an exploratory and validation cohort [122]. Although these strategies reduce nonspecific signals by sampling near the tumor site, large-scale validation remains necessary [120–122].
- Diagnosis
- While standard endometrial biopsy remains the gold standard, liquid biopsy may aid patients who cannot undergo invasive sampling or who yield inadequate tissue. Exosomal, proteomic, and metabolomic markers, along with ctDNA, cfDNA, and survivin-expressing cells, have all shown promise but have not yet matched the accuracy of tissue biopsy [120]. Nonetheless, certain exosomal microRNAs (e.g., miR-21, miR-27a, miR-223) and proteomic markers (YKL-40, DJ-1) demonstrated AUC values approaching 0.925 [120]. In a pilot study, CTCs were detectable in 80% of ovarian venous blood but absent in peripheral blood of patients with early-stage EC, suggesting a localized dissemination route [58]. Meanwhile, cfDNA profiling in 61 advanced EC patients had 87.5% concordance with tissue-based molecular classification and uncovered actionable alterations in 65% [123]. Another methodological study combined suction curettage without anesthesia, liquid-based cytology, and micro-histology to achieve 92.3% sensitivity and 100% specificity in 100 patients [122]. Collectively, these findings indicate that blood, uterine aspirates, and other minimally invasive samples, together with molecular profiling, may guide targeted EC therapies [58, 120, 122, 123].
- Follow-up and Prognostic Assessment
- Liquid biopsy also aids in tracking disease progression and refining prognosis. In a prospective study of 198 patients, 29.38% had detectable ctDNA at surgery; this correlated with higher tumor grade and advanced stage [124]. Elevated cfDNA (>25 ng/mL) was linked to shorter DFS and DSS, and ctDNA positivity detected relapse around 4.7 months before clinical or radiologic confirmation [124]. Another pilot comparing ctDNA and tumor DNA in 21 patients found shared mutations in two-thirds of cases, although the detection of DNMT3A and TET2 mutations in older individuals emphasized the confounding issue of clonal hematopoiesis [59]. Microsatellite instability (MSI) monitored via ddPCR in 90 uterine aspirates showed 96.67% concordance with mismatch repair protein status; in one case, MSI markers identified recurrence before clinical detection [124]. Despite these advances, about 20% of relapse cases were ctDNA-negative, highlighting the need for assay improvements or multipronged biomarker approaches [124, 125].
- Perspectives and Emerging Techniques
- Ongoing refinements aim to enhance liquid biopsy’s sensitivity, specificity, and practicality. An exosome metabolic fingerprinting study that used a Fe3O4@COF@Au-Apt nanoplatform evaluated 105 plasma samples (51 EC, 54 controls) and attained an AUC of 0.924 in blind testing [126]. Machine learning identified four metabolites, hydroxychalcone, L-acetylcarnitine, elaidic acid, and glutathione, that produced 94.9% classification accuracy [126]. Similar AI-assisted analytics, along with advanced sampling (e.g., suction curettage, tampon-based fluid collection) and molecular assays (NGS, ddPCR-MSI), could further transform EC detection and monitoring [120, 121, 126].
Liquid Biopsy in Cervical Cancer
- Early Diagnosis and Screening
- Human papillomavirus (HPV) is the primary etiologic agent in cervical cancer, with subtypes 16 and 18 accounting for about 70% of cases [127]. Digital droplet PCR of HPV-specific genes E7 and L1 in plasma samples from 138 Hong Kong Chinese women with cervical cancer revealed that higher viral loads correlate with increased five-year recurrence and mortality risk, highlighting circulating HPV DNA as a critical surveillance marker [128].
- Diagnosis and Disease Monitoring
- Several serum protein markers offer clinical utility in monitoring cervical cancer. For example, VCAM-1 and ICAM-1 levels can predict radiotherapy or chemoradiotherapy response in a cohort of 189 patients [129]. Regulatory proteins Rspo1 and Slit2 correlate positively with radiotherapy tolerance and negatively with hematologic and cardiac toxicity [130]. Pre-treatment hemoglobin below 11 g/dl is linked to treatment resistance, whereas hemoglobin above 12.7 g/dl is significantly associated with complete radiotherapy response (p<0.001), as well as improved overall and disease-free survival [131].
- Staging and Treatment Response Evaluation
- Multiple studies have established threshold values for Squamous cell carcinoma antigen (SCC-Ag) across clinical contexts. Pre-treatment levels above 2 ng/ml predict distant recurrence within five years [132], while levels exceeding 6.5 ng/ml suggest a benefit from adjuvant chemotherapy, reducing systemic recurrence [133]. Post-treatment thresholds above 1.15 ng/ml correlate with decreased three-year overall survival (84%) in chemoradiotherapy patients, and levels exceeding 1.20 ng/ml predict worse survival (95%) in radiotherapy-only cohorts. Additionally, post-treatment SCC-Ag above 1.0 ng/ml is linked to higher recurrence risk in stage IB–IIIB disease [134, 135]. Analyzing cfDNA in 93 plasma samples from 57 patients demonstrated a marked decrease in allele fraction deviation (AFD) after treatment (p=0.029), correlating cfDNA reduction with tumor shrinkage and confirming the predictive value of AFD for disease progression and relapse [136].
- Follow-up and Surveillance
- In a study of 99 patients with locally advanced cervical cancer (FIGO stage IIB–IVA), elevated CTCs and SCC-Ag levels emerged as independent prognostic factors for disease-free survival. A combined CTC/SCC-Ag risk model outperformed individual markers, and multivariate analysis confirmed serum CTC count, FIGO stage, and serum SCC-Ag as independent predictors for two-year disease-free survival [137]. Monitoring treatment response through VEGF1 reduction, alongside tissue HIF-1a levels, correlated strongly with complete chemoradiotherapy response [138]. Implementing cfDNA surveillance enabled early detection of treatment response and progression, while serial plasma sampling facilitated longitudinal genomic tracking. Systematic examination of circulating HPV DNA similarly reflected disease status, with shifts in viral load providing an early sign of therapy response and recurrence risk [139]. Integrating multiple biomarkers yields superior clinical utility over single-marker approaches, particularly in evaluating early response, anticipating recurrence, adjusting treatment strategies, and ensuring long-term surveillance.
Analogistic practical considerations: liquid biopsy versus pathological analysis
Empirical data from a broad range of tumor types underscore how liquid biopsy has become powerful, minimally invasive tool that complements traditional tissue biopsy, expanding the horizons of precision oncology (Table 3). Although many of these studies center on non-gynecological cancers, they provide instructive analogies for potential applications in endometrial, cervical, and ovarian tumors. Capturing tumor heterogeneity in real time provides vital insights into mutational burdens, emerging resistance, and minimal residual disease [140]. However, important challenges persist. Early-stage malignancies may release scant amounts of ctDNA, limiting sensitivity and increasing the risk of false negatives, and clonal hematopoiesis can obscure ctDNA findings in older patients [141, 142]. Further, while liquid biopsy excels at providing a dynamic portrait of disease evolution, it lacks the exhaustive morphological data gleaned from tissue specimens, which remain the gold standard for grading, staging, and comprehensive immunohistochemical assessments [143, 144]. Despite these constraints, the real-time feedback offered by liquid biopsy is invaluable for guiding therapy modifications, detecting emerging driver mutations, and flagging early relapse, often before radiographic imaging can confirm disease progression [145, 146]. Moreover, it can be particularly advantageous when tumor sites are anatomically difficult to biopsy or when patients’ overall health precludes surgical intervention [147]. Current investigative trends include refining molecular assays for higher sensitivity in early-stage tumors and exploring additional biofluids, such as ascites and cerebrospinal fluid, to expand diagnostic reach [148]. In moving forward, the synergy between liquid biopsy and traditional pathological analysis promises the most comprehensive patient stratification. Tissue biopsy remains indispensable for initial diagnosis, detailed morphological evaluation, and robust molecular classification; meanwhile, liquid biopsy offers a dynamic and repeatable snapshot of tumor evolution that optimizes personalized treatment and surveillance strategies [141, 142]. By integrating both modalities, clinicians can capitalize on real-time genetic insights while retaining the diagnostic depth essential for accurate staging and targeted therapy. Ultimately, ongoing technological advances and interdisciplinary collaboration will further refine these approaches, facilitating earlier detection, improved outcomes, and a more patient-centric model of cancer care.
Table 3.
Clinical and practical overview of liquid biopsy use across gynecological oncology
Conclusion
Liquid biopsy has emerged as a pivotal, noninvasive tool for the detection and monitoring of gynecological cancers, offering real-time insights into tumor biology that complement standard tissue-based approaches, but it has not yet been entered into routine clinical practice in gynecological oncology. This review aims to clarify the fundamental biological rationale and technical foundations of various liquid biopsy biomarkers in gynecological oncology, which is essential for both researchers and clinicians seeking to contextualize current evidence. Indeed, while many studies report encouraging performance in controlled or advanced disease settings, true clinical utility remains unproven in larger, prospective trials. A further consideration is the growing interest in multi-cancer early detection tests that are still awaiting FDA approval [149, 150]. Although these platforms have generated considerable excitement, real-world data has highlighted potential shortcomings. Sensitivity for early stage disease can be modest, raising questions about whether early detection of a tumor will ultimately translate into improved survival or merely reflect lead-time bias. Likewise, even a small false-positive rate, when projected to a population-wide screening program, could lead to unnecessary, invasive diagnostic procedures and substantial costs. For gynecological malignancies, specifically, data remain sparse, underscoring the need for robust clinical trials that assess actual reductions in cancer-specific mortality rather than rely solely on stage-shift endpoints. By contrasting robust technical advances with these variables, and sometimes limited, clinical outcomes, we aim to underscore that liquid biopsy, though innovative, is not a one-size-fits-all solution; its utility must be assessed in a disease- and context-specific manner. By outlining the mechanistic basis, we illustrate where current approaches might fail under practical conditions and how targeted technical improvements could address these pitfalls. Ultimately, ongoing interdisciplinary efforts, larger prospective trials, cost-effectiveness analyses, and meticulous follow-up will be essential for resolving these gaps and ensuring that promising laboratory data translate into meaningful, patient-centered outcomes in gynecologic oncology.
Acknowledgements
The authors acknowledge the contributions of their respective institutions for providing the necessary infrastructure and support to carry out this work.
Authors’ contributions
CM contributed to conceptualization, methodology, investigation, original and final draft writing, supervision, and project administration. AE was involved in supervision and validation. GV provided resources, supervision, and validation. SRB engaged in investigation, writing review/editing, and visualization. MC participated in investigation, data curation, and writing review/editing. VS was responsible for investigation, data curation, and writing review/editing. HK conducted formal analysis, software development, and visualization. SB was involved in investigation. GM contributed to investigation and data curation. IM participated in investigation. MNT engaged in investigation and writing review/editing. SR and MA provided resources and supervision. AP was involved in investigation and writing review/editing. VT participated in investigation and writing review/editing. SiPa contributed to investigation and resources. StPe provided supervision and resources. GI was involved in resources, validation, and writing review/editing. CCN, CP, and LP participated in investigation and writing review/editing. AI contributed to validation and resources. SC conducted formal analysis, validation, and writing review/editing. AG was responsible for conceptualization, funding acquisition, and supervision. All authors have read and approved the manuscript.
Funding
This study did not receive any funding or financial support.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not required as this manuscript doesn’t include details, images or videos related to the participants.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: the authors identified spelling, grammar, and typo errors in the body of the article.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/30/2025
The original online version of this article was revised: the authors identified spelling, grammar, and typo errors in the body of the article
Change history
5/30/2025
A Correction to this paper has been published: 10.1186/s13046-025-03429-0
Contributor Information
Salvatore Cortellino, Email: s.cortellino@ssmeridionale.it.
Antonio Giordano, Email: giordano@temple.edu, Email: giordano12@unisi.it.
References
- 1.SEER*Explorer: An interactive website for SEER cancer statistics [Internet]. Surveillance Research Program, National Cancer Institute; 2024 Apr 17. [updated: 2024 Nov 5; cited 2024 Dec 30]. Available from: https://seer.cancer.gov/statistics-network/explorer/. Data source(s): U.S. 2021 cancer prevalence estimates are based on 2021 cancer prevalence proportions from the SEER 12 Areas (excluding the Alaska Native Tumor Registry) and 1/1/2021 U.S. population estimates based on the average of 2020 and 2021 population estimates from the U.S. Bureau of the Census.
- 2.Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Cancer Res. 2017Aug 15;23(16):4716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villatoro S, Mayo-de-las-Casas C, Jordana-Ariza N, Viteri-Ramírez S, Garzón-Ibañez M, Moya-Horno I, et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol Oncol. 2019Dec;13(12):2633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han MR, Lee SH, Park JY, Hong H, Ho JY, Hur SY, et al. Clinical Implications of Circulating Tumor DNA from Ascites and Serial Plasma in Ovarian Cancer. Cancer Res Treat. 2020Jul 15;52(3):779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019Jan;565(7741):654–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nonaka T, Wong DTW. Saliva diagnostics. J Am Dent Assoc. 2023Aug;154(8):696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022Mar 18;21(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry SH. Translational Science: Past, Present, and Future. Biotechniques. 2008Feb;44(sup2):1. [DOI] [PubMed] [Google Scholar]
- 9.Smith EMR, Molldrem S, Farroni JS, Tumilty E. Articulating the social responsibilities of translational science. Humanit Soc Sci Commun. 2024Jan 9;11(1):85. [Google Scholar]
- 10.Tong L, Ding N, Tong X, Li J, Zhang Y, Wang X, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9(19):5532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobillo S, Crespo M, Escudero L, Mayor R, Raheja P, Carpio C, et al. Cell free circulating tumor DNA in cerebrospinal fluid detects and monitors central nervous system involvement of B-cell lymphomas. Haematologica. 2020Feb 20;106(2):513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldshtein H, Hausmann MJ, Douvdevani A. A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann Clin Biochem Int J Lab Med. 2009Nov;46(6):488–94. [DOI] [PubMed] [Google Scholar]
- 13.Millholland JM, Li S, Fernandez CA, Shuber AP. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res Rep Urol. 2012;4:33-40. 10.2147/RRU.S32736. [DOI] [PMC free article] [PubMed]
- 14.Martin M, Leffler J, Smoląg KI, Mytych J, Björk A, Chaves LD, et al. Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 2016May;23(5):903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephan F, Marsman G, Bakker LM, Bulder I, Stavenuiter F, Aarden LA, et al. Cooperation of Factor VII–Activating Protease and Serum DNase I in the Release of Nucleosomes From Necrotic Cells. Arthritis Rheumatol. 2014Mar;66(3):686–93. [DOI] [PubMed] [Google Scholar]
- 16.Alix-Panabières C, Pantel K. Technologies for detection of circulating tumor cells: facts and vision. Lab Chip. 2014;14(1):57–62. [DOI] [PubMed] [Google Scholar]
- 17.Lakka Klement G, Yip TT, Cassiola F, Kikuchi L, Cervi D, Podust V, et al. Platelets actively sequester angiogenesis regulators. Blood. 2009Mar 19;113(12):2835–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calverley DC, Phang TL, Choudhury QG, Gao B, Oton AB, Weyant MJ, et al. Significant Downregulation of Platelet Gene Expression in Metastatic Lung Cancer. Clin Transl Sci. 2010Oct;3(5):227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson RJA, Balaj L, Hulleman E, Van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011Sep 29;118(13):3680–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015Nov;28(5):666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016Dec;30(6):836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidle UH, Birzele F, Kollmorgen G, Rüger R. The Multiple Roles of Exosomes in Metastasis. Cancer Genomics Proteomics. 2017Jan 1;14(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell. 2014Apr;25(4):501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bach D, Hong J, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017Jul 15;141(2):220–30. [DOI] [PubMed] [Google Scholar]
- 25.Bhome R, Del Vecchio F, Lee GH, Bullock MD, Primrose JN, Sayan AE, et al. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018Apr;420:228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17(21):3558–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016Jan 28;1(1):15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J Cell Physiol. 2018Sep;233(9):6370–80. [DOI] [PubMed] [Google Scholar]
- 29.Vorobjeva NV, Chernyak BV. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochem Mosc. 2020Oct;85(10):1178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Widlak P, Li P, Wang X, Garrard WT. Cleavage Preferences of the Apoptotic Endonuclease DFF40 (Caspase-activated DNase or Nuclease) on Naked DNA and Chromatin Substrates. J Biol Chem. 2000Mar;275(11):8226–32. [DOI] [PubMed] [Google Scholar]
- 31.Han DSC, Ni M, Chan RWY, Chan VWH, Lui KO, Chiu RWK, et al. The Biology of Cell-free DNA Fragmentation and the Roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. 2020Feb;106(2):202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018Nov 29;9(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019Jun;570(7761):385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov M, Baranova A, Butler T, Spellman P, Mileyko V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics. 2015Dec;16(S13):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez C, Snyder MW, Tanos R, Shendure J, Thierry AR. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. Npj Genomic Med. 2018Nov 23;3(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment Length of Circulating Tumor DNA. Kwiatkowski DJ, editor. PLOS Genet. 2016;12(7):e1006162. [DOI] [PMC free article] [PubMed]
- 37.Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018Dec;7(1):1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuchs TA, Kremer Hovinga JA, Schatzberg D, Wagner DD, Lämmle B. Circulating DNA and myeloperoxidase indicate disease activity in patients with thrombotic microangiopathies. Blood. 2012Aug 9;120(6):1157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019Aug 3;20(8):1057–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Bode AM, Dong Z. Circulating Tumor Cells: Moving Biological Insights into Detection. Theranostics. 2017;7(10):2606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Łukasiewicz M, Pastuszak K, Łapińska-Szumczyk S, Różański R, Veld SGJGI ‘t, Bieńkowski M, et al. Diagnostic Accuracy of Liquid Biopsy in Endometrial Cancer. Cancers. 2021;13(22):5731. [DOI] [PMC free article] [PubMed]
- 42.Patel D, Thankachan S, Sreeram S, Kavitha KP, Suresh PS. The role of tumor-educated platelets in ovarian cancer: A comprehensive review and update. Pathol - Res Pract. 2023Jan;1(241): 154267. [DOI] [PubMed] [Google Scholar]
- 43.Li P, Liu S, Du L, Mohseni G, Zhang Y, Wang C. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin Epigenetics. 2022Dec;14(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo XM, Miller H, Matsuo K, Roman LD, Salhia B. Circulating Cell-Free DNA Methylation Profiles in the Early Detection of Ovarian Cancer: A Scoping Review of the Literature. Cancers. 2021Feb 17;13(4):838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neal RD, Johnson P, Clarke CA, Hamilton SA, Zhang N, Kumar H, et al. Cell-Free DNA-Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers. 2022Oct 1;14(19):4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, Tang J, Zhao Z, Zhao C, Xiang Y. Circulating tumor DNA: a noninvasive biomarker for tracking ovarian cancer. Reprod Biol Endocrinol. 2021Dec 3;19(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018Feb 23;359(6378):926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toboni M, Maxwell L, Scalise CB, Berman T, Hardesty M, Ketch P, et al. Personalized circulating tumor DNA analysis used for ctDNA detection and response monitoring among patients with advanced or recurrent endometrial cancer. Gynecol Oncol. 2024Nov;190:S208–9. [Google Scholar]
- 49.Soberanis Pina P, Clemens K, Bubie A, Grant B, Haynes G, Zhang N, et al. Genomic Landscape of ctDNA and Real-World Outcomes in Advanced Endometrial Cancer. Clin Cancer Res. 2024Dec 16;30(24):5657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castellano T, Bae-Jump V, Secord AA, Danziger N, Graf R, Quintanilha JCF, et al. Liquid biopsy for advanced endometrial cancer: Prognostic value of plasma circulating DNA fraction in real-world settings (1320). Gynecol Oncol. 2023Sep;176:S194–5. [Google Scholar]
- 51.Pinet S, Durand S, Perani A, Darnaud L, Amadjikpe F, Yon M, et al. Clinical management of molecular alterations identified by high throughput sequencing in patients with advanced solid tumors in treatment failure: Real-world data from a French hospital. Front Oncol. 2023Feb;27(13):1104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galvão-Lima LJ, Morais AHF, Valentim RAM, Barreto EJSS. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. Biomed Eng OnLine. 2021Dec;20(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramnauth N, Neubarth E, Makler-Disatham A, Sher M, Soini S, Merk V, et al. Development of a Microfluidic Device for Exosome Isolation in Point-of-Care Settings. Sensors. 2023Oct 7;23(19):8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang N, Li B, Jia Z, Wang C, Wu P, Zheng T, et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat Biomed Eng. 2021Jun 15;5(6):586–99. [DOI] [PubMed] [Google Scholar]
- 55.Zhu JW, Charkhchi P, Akbari MR. Potential clinical utility of liquid biopsies in ovarian cancer. Mol Cancer. 2022Dec;21(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med CCLM. 2018Jan 26;56(2):186–97. [DOI] [PubMed] [Google Scholar]
- 57.Jou HJ, Ling PY, Hsu HT. Circulating tumor cells as a “real-time liquid biopsy”: Recent advances and the application in ovarian cancer. Taiwan J Obstet Gynecol. 2022Jan;61(1):34–9. [DOI] [PubMed] [Google Scholar]
- 58.Francini S, Duraes M, Rathat G, Macioce V, Mollevi C, Pages L, et al. Circulating Tumor Cell Detection by Liquid Biopsy during Early-Stage Endometrial Cancer Surgery: A Pilot Study. Biomolecules. 2023Feb 24;13(3):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kodada D, Hyblova M, Krumpolec P, Janostiakova N, Barath P, Grendar M, et al. The Potential of Liquid Biopsy in Detection of Endometrial Cancer Biomarkers: A Pilot Study. Int J Mol Sci. 2023Apr 25;24(9):7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y, Shi R, Zhao R, Wang H. Clinical application of liquid biopsy in endometrial carcinoma. Med Oncol. 2023Feb 9;40(3):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vizza E, Corrado G, De Angeli M, Carosi M, Mancini E, Baiocco E, et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J Exp Clin Cancer Res. 2018Dec;37(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong Y, Jia L, Li M, Li H, Wang S. Identification of exosomal circSLC26A4 as a liquid biopsy marker for cervical cancer. Cobucci RNO, editor. PLOS ONE. 2024;19(6):e0305050. [DOI] [PMC free article] [PubMed]
- 63.Kim YM, Lee SW, Lee YJ, Lee HY, Lee JE, Choi EK. Prospective study of the efficacy and utility of TP53 mutations in circulating tumor DNA as a non-invasive biomarker of treatment response monitoring in patients with high-grade serous ovarian carcinoma. J Gynecol Oncol. 2019;30(3): e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sánchez-Herrero E, Serna-Blasco R, Robado De Lope L, González-Rumayor V, Romero A, Provencio M. Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That may Impact on ctDNA Analysis. Front Oncol. 2022;12:943253. [DOI] [PMC free article] [PubMed]
- 65.Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020Jan;468:59–71. [DOI] [PubMed] [Google Scholar]
- 66.Senturk Kirmizitas T, Van Den Berg C, Boers R, Helmijr J, Makrodimitris S, Dag HH, et al. Epigenetic and Genomic Hallmarks of PARP-Inhibitor Resistance in Ovarian Cancer Patients. Genes. 2024Jun 7;15(6):750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lheureux S, Prokopec SD, Oldfield LE, Gonzalez-Ochoa E, Bruce JP, Wong D, et al. Identifying Mechanisms of Resistance by Circulating Tumor DNA in EVOLVE, a Phase II Trial of Cediranib Plus Olaparib for Ovarian Cancer at Time of PARP Inhibitor Progression. Clin Cancer Res. 2023Sep 15;29(18):3706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rusan M, Andersen RF, Jakobsen A, Steffensen KD. Circulating HOXA9-methylated tumour DNA: A novel biomarker of response to poly (ADP-ribose) polymerase inhibition in BRCA-mutated epithelial ovarian cancer. Eur J Cancer. 2020Jan;125:121–9. [DOI] [PubMed] [Google Scholar]
- 69.Murtaza M, Dawson SJ, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013May;497(7447):108–12. [DOI] [PubMed] [Google Scholar]
- 70.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med [Internet]. 2014 Feb 19 [cited 2025 Jan 4];6(224). Available from: https://www.science.org/doi/10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed]
- 71.Golara A, Kozłowski M, Cymbaluk-Płoska A. The Role of Circulating Tumor DNA in Ovarian Cancer. Cancers. 2024Sep 10;16(18):3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karimi F, Azadbakht O, Veisi A, Sabaghan M, Owjfard M, Kharazinejad E, et al. Liquid biopsy in ovarian cancer: advantages and limitations for prognosis and diagnosis. Med Oncol. 2023Aug 10;40(9):265. [DOI] [PubMed] [Google Scholar]
- 73.Ntzifa A, Lianidou E. Pre-analytical conditions and implementation of quality control steps in liquid biopsy analysis. Crit Rev Clin Lab Sci. 2023Nov 17;60(8):573–94. [DOI] [PubMed] [Google Scholar]
- 74.Gao Y, Zhou N, Liu J. Ovarian Cancer Diagnosis and Prognosis Based on Cell-Free DNA Methylation. Cancer Control. 2024Jan;31:10732748241255548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadeghi S, Tehrani FR, Tahmasebi S, Shafiee A, Hashemi SM. Exosome engineering in cell therapy and drug delivery. Inflammopharmacology. 2023Feb;31(1):145–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022Dec;21(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anitha K, Posinasetty B, Naveen Kumari K, Chenchula S, Padmavathi R, Prakash S, et al. Liquid biopsy for precision diagnostics and therapeutics. Clin Chim Acta. 2024Feb;554: 117746. [DOI] [PubMed] [Google Scholar]
- 78.Cameron JM, Sala A, Antoniou G, Brennan PM, Butler HJ, Conn JJA, et al. A spectroscopic liquid biopsy for the earlier detection of multiple cancer types. Br J Cancer. 2023Nov 9;129(10):1658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morcuende-Ventura V, Hermoso-Durán S, Abian-Franco N, Pazo-Cid R, Ojeda JL, Vega S, et al. Fluorescence Liquid Biopsy for Cancer Detection Is Improved by Using Cationic Dendronized Hyperbranched Polymer. Int J Mol Sci. 2021Jun 17;22(12):6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn. 2015May;17(3):265–72. [DOI] [PubMed] [Google Scholar]
- 81.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017Apr;17(4):223–38. [DOI] [PubMed] [Google Scholar]
- 82.Martín-Gracia B, Martín-Barreiro A, Cuestas-Ayllón C, Grazú V, Line A, Llorente A, et al. Nanoparticle-based biosensors for detection of extracellular vesicles in liquid biopsies. J Mater Chem B. 2020;8(31):6710–38. [DOI] [PubMed] [Google Scholar]
- 83.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018Apr 27;360(6387):436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015Oct;25(10):1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Labib M, Sargent EH, Kelley SO. Electrochemical Methods for the Analysis of Clinically Relevant Biomolecules. Chem Rev. 2016Aug 24;116(16):9001–90. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y, Liu TY, Weinberg DE, White BW, De La Torre CJ, Tan CL, et al. Spatial co-fragmentation pattern of cell-free DNA recapitulates in vivo chromatin organization and identifies tissues-of-origin [Internet]. Genomics; 2019 [cited 2025 Jan 4]. Available from: http://biorxiv.org/lookup/doi/10.1101/564773
- 87.Esfahani MS, Hamilton EG, Mehrmohamadi M, Nabet BY, Alig SK, King DA, et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat Biotechnol. 2022Apr;40(4):585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanley KE, Jatsenko T, Tuveri S, Sudhakaran D, Lannoo L, Van Calsteren K, et al. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat Commun. 2024Mar 12;15(1):2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun K, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019Mar;29(3):418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng H, Zhu MS, Liu Y. FinaleDB: a browser and database of cell-free DNA fragmentation patterns. Peter R, editor. Bioinformatics. 2021;37(16):2502–3. [DOI] [PMC free article] [PubMed]
- 91.Lu H, Liu Y, Wang J, Fu S, Wang L, Huang C, et al. Detection of ovarian cancer using plasma cell-free DNA methylomes. Clin Epigenetics. 2022Dec;14(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, Reed SC, Lo C, Choudhury AD, Parsons HA, Stover DG, et al. FinaleMe: Predicting DNA methylation by the fragmentation patterns of plasma cell-free DNA [Internet]. Bioinformatics; 2024 [cited 2025 Jan 4]. Available from: http://biorxiv.org/lookup/doi/10.1101/2024.01.02.573710 [DOI] [PMC free article] [PubMed]
- 93.Sessa C, Balmaña J, Bober SL, Cardoso MJ, Colombo N, Curigliano G, et al. Risk reduction and screening of cancer in hereditary breast-ovarian cancer syndromes: ESMO Clinical Practice Guideline. Ann Oncol. 2023Jan;34(1):33–47. [DOI] [PubMed] [Google Scholar]
- 94.Ahn HS, Ho JY, Yu J, Yeom J, Lee S, Hur SY, et al. Plasma Protein Biomarkers Associated with Higher Ovarian Cancer Risk in BRCA1/2 Carriers. Cancers. 2021May 11;13(10):2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bahar-Shany K, Barnabas GD, Deutsch L, Deutsch N, Glick-Saar E, Dominissini D, et al. Proteomic signature for detection of high-grade ovarian cancer in germline BRCA mutation carriers. Int J Cancer. 2023Feb 15;152(4):781–93. [DOI] [PubMed] [Google Scholar]
- 96.Van Bommel MHD, Pijnenborg JMA, Van Der Putten LJM, Bulten J, Snijders MPLM, Küsters-Vandevelde HVN, et al. Diagnostic accuracy of mutational analysis along the Müllerian tract to detect ovarian cancer. Int J Gynecol Cancer. 2022Dec;32(12):1568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paracchini L, Pesenti C, Delle Marchette M, Beltrame L, Bianchi T, Grassi T, et al. Detection of TP53 Clonal Variants in Papanicolaou Test Samples Collected up to 6 Years Prior to High-Grade Serous Epithelial Ovarian Cancer Diagnosis. JAMA Netw Open. 2020Jul 1;3(7): e207566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Terp SK, Stoico MP, Dybkær K, Pedersen IS. Early diagnosis of ovarian cancer based on methylation profiles in peripheral blood cell-free DNA: a systematic review. Clin Epigenetics. 2023Feb 14;15(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Q, Li W, Leng B, Zheng W, He Z, Zuo M, et al. Circulating Cell Free DNA as the Diagnostic Marker for Ovarian Cancer: A Systematic Review and Meta-Analysis. Chalmers J, editor. PLOS ONE. 2016;11(6):e0155495. [DOI] [PMC free article] [PubMed]
- 100.Li B, Pu K, Ge L, Wu X. Diagnostic significance assessment of the circulating cell-free DNA in ovarian cancer: An updated meta-analysis. Gene. 2019Sep;714: 143993. [DOI] [PubMed] [Google Scholar]
- 101.Wang X, Kong D, Wang C, Ding X, Zhang L, Zhao M, et al. Circulating microRNAs as novel potential diagnostic biomarkers for ovarian cancer: a systematic review and updated meta-analysis. J Ovarian Res. 2019Dec;12(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frisk NLS, Sørensen AE, Pedersen OBV, Dalgaard LT. Circulating microRNAs for Early Diagnosis of Ovarian Cancer: A Systematic Review and Meta-Analysis. Biomolecules. 2023May 22;13(5):871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thusgaard CF, Korsholm M, Koldby KM, Kruse TA, Thomassen M, Jochumsen KM. Epithelial ovarian cancer and the use of circulating tumor DNA: A systematic review. Gynecol Oncol. 2021Jun;161(3):884–95. [DOI] [PubMed] [Google Scholar]
- 104.Lucidi A, Buca D, Ronsini C, Tinari S, Bologna G, Buca D, et al. Role of Extracellular Vesicles in Epithelial Ovarian Cancer: A Systematic Review. Int J Mol Sci. 2020Nov 19;21(22):8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao Q, Lin YP, Li BS, Wang GQ, Dong LQ, Shen BY, et al. Unintrusive multi-cancer detection by circulating cell-free DNA methylation sequencing (THUNDER): development and independent validation studies. Ann Oncol. 2023May;34(5):486–95. [DOI] [PubMed] [Google Scholar]
- 106.Marinelli LM, Kisiel JB, Slettedahl SW, Mahoney DW, Lemens MA, Shridhar V, et al. Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility. Gynecol Oncol. 2022Jun;165(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shnaider PV, Petrushanko IYu, Aleshikova OI, Babaeva NA, Ashrafyan LA, Borovkova EI, et al. Expression level of CD117 (KIT) on ovarian cancer extracellular vesicles correlates with tumor aggressiveness. Front Cell Dev Biol. 2023;11:1057484. [DOI] [PMC free article] [PubMed]
- 108.Shao X, He Y, Ji M, Chen X, Qi J, Shi W, et al. Quantitative analysis of cell-free DNA in ovarian cancer. Oncol Lett. 2015Dec;10(6):3478–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ogasawara A, Hihara T, Shintani D, Yabuno A, Ikeda Y, Tai K, et al. Evaluation of Circulating Tumor DNA in Patients with Ovarian Cancer Harboring Somatic PIK3CA or KRAS Mutations. Cancer Res Treat [Internet]. 2020 May 6 [cited 2025 Jan 4]; Available from: http://www.e-crt.org/journal/view.php?doi=10.4143/crt.2019.688 [DOI] [PMC free article] [PubMed]
- 110.Chen L, Ma R, Luo C, Xie Q, Ning X, Sun K, et al. Noninvasive early differential diagnosis and progression monitoring of ovarian cancer using the copy number alterations of plasma cell-free DNA. Transl Res. 2023Dec;262:12–24. [DOI] [PubMed] [Google Scholar]
- 111.Grobben Y, Den Ouden JE, Aguado C, Van Altena AM, Kraneveld AD, Zaman GJR. Amino Acid-Metabolizing Enzymes in Advanced High-Grade Serous Ovarian Cancer Patients: Value of Ascites as Biomarker Source and Role for IL4I1 and IDO1. Cancers. 2023Jan 31;15(3):893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Buderath P, Schwich E, Jensen C, Horn PA, Kimmig R, Kasimir-Bauer S, et al. Soluble Programmed Death Receptor Ligands sPD-L1 and sPD-L2 as Liquid Biopsy Markers for Prognosis and Platinum Response in Epithelial Ovarian Cancer. Front Oncol. 2019Oct;15(9):1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. Mardis ER, editor. PLOS Med. 2016;13(12):e1002198. [DOI] [PMC free article] [PubMed]
- 114.Chang L, Ni J, Zhu Y, Pang B, Graham P, Zhang H, et al. Liquid biopsy in ovarian cancer: recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics. 2019;9(14):4130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arend RC, Londoño AI, Montgomery AM, Smith HJ, Dobbin ZC, Katre AA, et al. Molecular Response to Neoadjuvant Chemotherapy in High-Grade Serous Ovarian Carcinoma. Mol Cancer Res. 2018May 1;16(5):813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, et al. Personalized Circulating Tumor DNA Biomarkers Dynamically Predict Treatment Response and Survival In Gynecologic Cancers. Samimi G, editor. PLOS ONE. 2015;10(12):e0145754. [DOI] [PMC free article] [PubMed]
- 117.Paracchini L, D’Incalci M, Marchini S. Liquid Biopsy in the Clinical Management of High-Grade Serous Epithelial Ovarian Cancer—Current Use and Future Opportunities. Cancers. 2021May 14;13(10):2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taliento C, Morciano G, Nero C, Froyman W, Vizzielli G, Pavone M, et al. Circulating tumor DNA as a biomarker for predicting progression-free survival and overall survival in patients with epithelial ovarian cancer: a systematic review and meta-analysis. Int J Gynecol Cancer. 2024Jun;34(6):906–18. [DOI] [PubMed] [Google Scholar]
- 119.Llueca A, Canete-Mota S, Jaureguí A, Barneo M, Ibañez MV, Neef A, et al. The Impact of Liquid Biopsy in Advanced Ovarian Cancer Care. Diagnostics. 2024Aug 26;14(17):1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karkia R, Wali S, Payne A, Karteris E, Chatterjee J. Diagnostic Accuracy of Liquid Biomarkers for the Non-Invasive Diagnosis of Endometrial Cancer: A Systematic Review and Meta-Analysis. Cancers. 2022Sep 25;14(19):4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bakkum-Gamez JN, Sherman ME, Slettedahl SW, Mahoney DW, Lemens MA, Laughlin-Tommaso SK, et al. Detection of endometrial cancer using tampon-based collection and methylated DNA markers. Gynecol Oncol. 2023Jul;174:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Barkley JE, Bhattarai B, Firouzi K, Monk BJ, Coonrod DV, et al. Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid. Int J Mol Sci. 2023May 12;24(10):8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Blanc-Durand F, Camilleri GM, Bayle A, Aldea M, Vasseur D, Ouali K, et al. Clinical utility of comprehensive liquid molecular profiling in patients with advanced endometrial cancer. Cancer. 2024Oct;130(19):3311–20. [DOI] [PubMed] [Google Scholar]
- 124.Casas-Arozamena C, Vilar A, Cueva J, Arias E, Sampayo V, Diaz E, et al. Role of cfDNA and ctDNA to improve the risk stratification and the disease follow-up in patients with endometrial cancer: towards the clinical application. J Exp Clin Cancer Res. 2024Sep 20;43(1):264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Casas-Arozamena C, Moiola CP, Vilar A, Bouso M, Cueva J, Cabrera S, et al. Noninvasive detection of microsatellite instability in patients with endometrial cancer. Int J Cancer. 2023May 15;152(10):2206–17. [DOI] [PubMed] [Google Scholar]
- 126.Yang H, Wu P, Li B, Huang X, Shi Q, Qiao L, et al. Diagnosis and Biomarker Screening of Endometrial Cancer Enabled by a Versatile Exosome Metabolic Fingerprint Platform. Anal Chem. 2024Nov 5;96(44):17679–88. [DOI] [PubMed] [Google Scholar]
- 127.Burd EM. Human Papillomavirus and Cervical Cancer. Clin Microbiol Rev. 2003Jan;16(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheung TH, Yim SF, Yu MY, Worley MJ, Fiascone SJ, Chiu RWK, et al. Liquid biopsy of HPV DNA in cervical cancer. J Clin Virol. 2019May;114:32–6. [DOI] [PubMed] [Google Scholar]
- 129.Choi KH, Lee SW, Yu M, Jeong S, Lee JW, Lee JH. Significance of elevated SCC-Ag level on tumor recurrence and patient survival in patients with squamous-cell carcinoma of uterine cervix following definitive chemoradiotherapy: a multi-institutional analysis. J Gynecol Oncol. 2019;30(1): e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nie J, Shao J, Guo SW, Liu X. The relevance of plasma R-spondin 1 and Slit2 as predictive biomarkers in cervical cancer chemotherapy and radiotherapy. Ann Transl Med. 2021May;9(10):837–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moreno-Acosta P, Carrillo S, Gamboa O, Romero-Rojas A, Acosta J, Molano M, et al. Novel predictive biomarkers for cervical cancer prognosis. Mol Clin Oncol. 2016Dec;5(6):792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kang S, Nam BH, Park JY, Seo SS, Ryu SY, Kim JW, et al. Risk Assessment Tool for Distant Recurrence After Platinum-Based Concurrent Chemoradiation in Patients With Locally Advanced Cervical Cancer: A Korean Gynecologic Oncology Group Study. J Clin Oncol. 2012Jul 1;30(19):2369–74. [DOI] [PubMed] [Google Scholar]
- 133.Zhang G, Miao L, Wu H, Zhang Y, Fu C. Pretreatment Squamous Cell Carcinoma Antigen (SCC-Ag) as a Predictive Factor for the Use of Consolidation Chemotherapy in Cervical Cancer Patients After Postoperative Extended-Field Concurrent Chemoradiotherapy. Technol Cancer Res Treat. 2021Jan;20:15330338211044626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kawaguchi R, Furukawa N, Kobayashi H, Asakawa I. Posttreatment cut-off levels of squamous cell carcinoma antigen as a prognostic factor in patients with locally advanced cervical cancer treated with radiotherapy. J Gynecol Oncol. 2013;24(4):313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ryu HK, Baek JS, Kang WD, Kim SM. The prognostic value of squamous cell carcinoma antigen for predicting tumor recurrence in cervical squamous cell carcinoma patients. Obstet Gynecol Sci. 2015;58(5):368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tian J, Geng Y, Lv D, Li P, Cordova M, Liao Y, et al. Using plasma cell-free DNA to monitor the chemoradiotherapy course of cervical cancer. Int J Cancer. 2019Nov;145(9):2547–57. [DOI] [PubMed] [Google Scholar]
- 137.Wen YF, Cheng TT, Chen XL, Huang WJ, Peng HH, Zhou TC, et al. Elevated circulating tumor cells and squamous cell carcinoma antigen levels predict poor survival for patients with locally advanced cervical cancer treated with radiotherapy. Katoh M, editor. PLOS ONE. 2018;13(10):e0204334. [DOI] [PMC free article] [PubMed]
- 138.Zhao K, Hu M, Yang R, Liu J, Zeng P, Zhao T. Decreasing expression of HIF-1α, VEGF-A, and Ki67 with efficacy of neoadjuvant therapy in locally advanced cervical cancer. Medicine (Baltimore). 2023May 19;102(20): e33820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dong SM, Pai SI, Rha SH, Hildesheim A, Kurman RJ, Schwartz PE, et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2002Jan;11(1):3–6. [PubMed] [Google Scholar]
- 140.Bai Y, Zhao H. Liquid biopsy in tumors: opportunities and challenges. Ann Transl Med. 2018Nov;6(S1):S89–S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin LH, Allison DHR, Feng Y, Jour G, Park K, Zhou F, et al. Comparison of solid tissue sequencing and liquid biopsy accuracy in identification of clinically relevant gene mutations and rearrangements in lung adenocarcinomas. Mod Pathol. 2021Dec;34(12):2168–74. [DOI] [PubMed] [Google Scholar]
- 142.Raez LE, Brice K, Dumais K, Lopez-Cohen A, Wietecha D, Izquierdo PA, et al. Liquid Biopsy Versus Tissue Biopsy to Determine Front Line Therapy in Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin Lung Cancer. 2023Mar;24(2):120–9. [DOI] [PubMed] [Google Scholar]
- 143.Xie S, Wang Y, Gong Z, Li Y, Yang W, Liu G, et al. Liquid Biopsy and Tissue Biopsy Comparison with Digital PCR and IHC/FISH for HER2 Amplification Detection in Breast Cancer Patients. J Cancer. 2022;13(3):744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mazzitelli C, Santini D, Corradini AG, Zamagni C, Trerè D, Montanaro L, et al. Liquid Biopsy in the Management of Breast Cancer Patients: Where Are We Now and Where Are We Going. Diagnostics. 2023Mar 25;13(7):1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arriola E, Paredes-Lario A, García-Gomez R, Diz-Tain P, Constenla M, García-Girón C, et al. Comparison of plasma ctDNA and tissue/cytology-based techniques for the detection of EGFR mutation status in advanced NSCLC: Spanish data subset from ASSESS. Clin Transl Oncol. 2018Oct;20(10):1261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019Sep;25(9):1415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ionescu F, Zhang J, Wang L. Clinical Applications of Liquid Biopsy in Prostate Cancer: From Screening to Predictive Biomarker. Cancers. 2022Mar 29;14(7):1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol OncolJ Hematol Oncol. 2022Sep 12;15(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Turnbull C, Wald N, Sullivan R, Pharoah P, Houlston RS, Aggarwal A, et al. GRAIL-Galleri: why the special treatment? The Lancet. 2024Feb 3;403(10425):431–2. [DOI] [PubMed] [Google Scholar]
- 150.Multi-cancer Early Detection Tests | MCED | GRAIL Galleri Test [Internet]. [cited 2025 Mar 8]. Available from: https://www.cancer.org/cancer/screening/multi-cancer-early-detection-tests.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.