Abstract
The emergence of proteomics, the large-scale analysis of proteins, has been inspired by the realization that the final product of a gene is inherently more complex and closer to function than the gene itself. Shortfalls in the ability of bioinformatics to predict both the existence and function of genes have also illustrated the need for protein analysis. Moreover, only through the study of proteins can posttranslational modifications be determined, which can profoundly affect protein function. Proteomics has been enabled by the accumulation of both DNA and protein sequence databases, improvements in mass spectrometry, and the development of computer algorithms for database searching. In this review, we describe why proteomics is important, how it is conducted, and how it can be applied to complement other existing technologies. We conclude that currently, the most practical application of proteomics is the analysis of target proteins as opposed to entire proteomes. This type of proteomics, referred to as functional proteomics, is always driven by a specific biological question. In this way, protein identification and characterization has a meaningful outcome. We discuss some of the advantages of a functional proteomics approach and provide examples of how different methodologies can be utilized to address a wide variety of biological problems.
INTRODUCTION
This review is intended to give the molecular biologist a rudimentary understanding of the technologies behind proteomics and their application to address biological questions. Entry of our laboratory into proteomics 5 years ago was driven by a need to define a complex mixture of proteins (∼36 proteins) we had affinity isolated that bound specifically to the catalytic subunit of protein phosphatase 1 (PP-1, a serine/threonine protein phosphatase that regulates multiple dephosphorylation events in cells) (26). We were faced with the task of trying to understand the significance of these proteins, and the only obvious way to begin to do this was to identify them by sequencing. We then bought an Applied Biosystems automated Edman sequencer (not having the budget for a mass spectrometer at the time). Since the majority of intact eukaryotic proteins are not immediately accessible to Edman sequencing due to posttranslational N-terminal modifications, we invented mixed-peptide sequencing (38). This method, described in detail later, essentially enables internal peptide sequence information to be derived from proteins electroblotted onto hydrophobic membranes. Using the mixed-peptide sequencing strategy, we identified all 36 proteins in about a week. The mixture contained at least two known PP-1 regulatory subunits, but most were identified in the expressed sequence tag or unannotated DNA databases and were novel proteins of unknown function. Since that time, we have been using various molecular biological approaches to determine the functions of some of these proteins. Herein lies the lesson of proteomics. Identifying long lists of potentially interesting proteins often generates more questions than it seeks to answer.
Despite learning this obvious lesson, our early sequencing experiences were an epiphany that has subsequently altered our whole scientific strategy for probing protein function in cells. The sequencing of the 36 proteins has opened new avenues to further explore the functions of PP-1 in intact cells. Because of increased sensitivity, our approaches now routinely use state-of-the-art mass spectrometry (MS) techniques. However, rather than using proteomics to simply characterize large numbers of proteins in complex mixtures, we see the real application of this technology as a tool to enhance the power of existing approaches currently used by the modern molecular biologist such as classical yeast and mouse genetics, tissue culture, protein expression systems, and site-directed mutagenesis. Importantly, the one message we would want the reader to take away from reading this review is that one should always let the biological question in mind drive the application of proteomics rather than simply engaging in an orgy of protein sequencing. From our experiences, we believe that if the appropriate controls are performed, proteomics is an extremely powerful approach for addressing important physiological questions. One should always design experiments to define a selected number of relevant proteins in the mixture of interest. Examples of such experiments that we routinely perform include defining early phosphorylation events in complex protein mixtures after hormone treatment of intact cells or comparing patterns of protein derived from a stimulated versus nonstimulated cell in an affinity pull-down experiment. Only the proteins that were specifically phosphorylated or bound in response to the stimulus are sequenced in the complex mixtures. Sequencing proteins that are regulated then has a meaningful outcome and directs all subsequent biological investigation.
Definitions
The term “proteomics” was first coined in 1995 and was defined as the large-scale characterization of the entire protein complement of a cell line, tissue, or organism (13, 163, 167). Today, two definitions of proteomics are encountered. The first is the more classical definition, restricting the large-scale analysis of gene products to studies involving only proteins. The second and more inclusive definition combines protein studies with analyses that have a genetic readout such as mRNA analysis, genomics, and the yeast two-hybrid analysis (123). However, the goal of proteomics remains the same, i.e., to obtain a more global and integrated view of biology by studying all the proteins of a cell rather than each one individually.
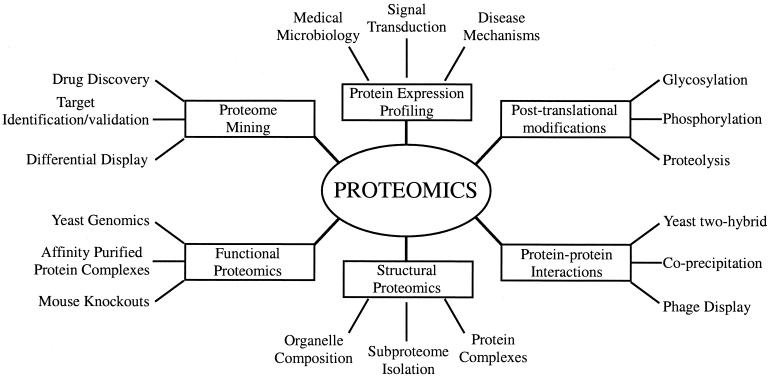
Using the more inclusive definition of proteomics, many different areas of study are now grouped under the rubric of proteomics (Fig. 1). These include protein-protein interaction studies, protein modifications, protein function, and protein localization studies to name a few. The aim of proteomics is not only to identify all the proteins in a cell but also to create a complete three-dimensional (3-D) map of the cell indicating where proteins are located. These ambitious goals will certainly require the involvement of a large number of different disciplines such as molecular biology, biochemistry, and bioinformatics. It is likely that in bioinformatics alone, more powerful computers will have to be devised to organize the immense amount of information generated from these endeavors.
FIG. 1.
Types of proteomics and their applications to biology.
In the quest to characterize the proteome of a given cell or organism, it should be remembered that the proteome is dynamic. The proteome of a cell will reflect the immediate environment in which it is studied. In response to internal or external cues, proteins can be modified by posttranslational modifications, undergo translocations within the cell, or be synthesized or degraded. Thus, examination of the proteome of a cell is like taking a “snapshot” of the protein environment at any given time. Considering all the possibilities, it is likely that any given genome can potentially give rise to an infinite number of proteomes.
Proteomics Origins
The first protein studies that can be called proteomics began in 1975 with the introduction of the two-dimensional gel by O'Farrell (119), Klose (87), and Scheele (140), who began mapping proteins from Escherichia coli, mouse, and guinea pig, respectively. Although many proteins could be separated and visualized, they could not be identified. Despite these limitations, shortly thereafter a large-scale analysis of all human proteins was proposed. The goal of this project, termed the human protein index, was to use two-dimensional protein electrophoresis (2-DE) and other methods to catalog all human proteins (14). However, lack of funding and technical limitations prevented this project from continuing.
Although the development of 2-DE was a major step forward, the science of proteomics would have to wait until the proteins displayed by 2-DE could be identified. One problem that had to be overcome was the lack of sensitive protein-sequencing technology. Improving sensitivity was critical for success because biological samples are often limiting and both one-dimensional (1-D) and two-dimensional (2-D) gels have limits in protein loading capacity. The first major technology to emerge for the identification of proteins was the sequencing of proteins by Edman degradation (45). A major breakthrough was the development of microsequencing techniques for electroblotted proteins (6-8). This technique was used for the identification of proteins from 2-D gels to create the first 2-D databases (31). Improvements in microsequencing technology resulted in increased sensitivity of Edman sequencing in the 1990s to high-picomole amounts (6).
One of the most important developments in protein identification has been the development of MS technology (11). In the last decade, the sensitivity of analysis and accuracy of results for protein identification by MS have increased by several orders of magnitude (11, 123). It is now estimated that proteins in the femtomolar range can be identified in gels. Because MS is more sensitive, can tolerate protein mixtures, and is amenable to high-throughput operations, it has essentially replaced Edman sequencing as the protein identification tool of choice.
Genome Information
The growth of proteomics is a direct result of advances made in large-scale nucleotide sequencing of expressed sequence tags and genomic DNA. Without this information, proteins could not be identified even with the improvements made in MS. Protein identification (by MS or Edman sequencing) relies on the presence of some form of database for the given organism (122, 146). The majority of DNA and protein sequence information has accumulated within the last 5 to 10 years (23). In 1995, the first complete genome of an organism was sequenced, that of Haemophilus influenzae (56). At the time of this writing, the sequencing of the genomes of 45 microorganisms has been completed and that of 170 more is under way (http://www.tiger.org/tdb/mdb/mdbcomplete.html). To date, five eukaryotic genomes have been completed: Arabidopsis thaliana (154), Saccharomyces cerevisiae (58), Schizosaccharomyces pombe (128), Caenorhabditis elegans (1), and Drosophila melanogaster (3, 113, 138). In addition, the rice (105), mouse (178a), and human (93, 161) genomes are near completion.
Why Proteomics?
Many types of information cannot be obtained from the study of genes alone. For example, proteins, not genes, are responsible for the phenotypes of cells. It is impossible to elucidate mechanisms of disease, aging, and effects of the environment solely by studying the genome. Only through the study of proteins can protein modifications be characterized and the targets of drugs identified.
Annotation of the genome.
One of the first applications of proteomics will be to identify the total number of genes in a given genome. This “functional annotation” of a genome is necessary because it is still difficult to predict genes accurately from genomic data (46). One problem is that the exon-intron structure of most genes cannot be accurately predicted by bioinformatics (43). To achieve this goal, genomic information will have to be integrated with data obtained from protein studies to confirm the existence of a particular gene.
Protein expression studies.
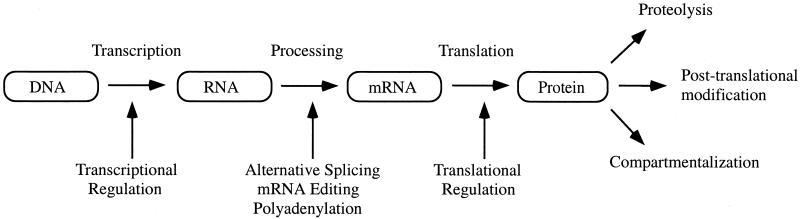
In recent years, the analysis of mRNA expression by various methods has become increasingly popular. These methods include serial analysis of gene expression (SAGE) (160) and DNA microarray technology (142, 143). However, the analysis of mRNA is not a direct reflection of the protein content in the cell. Consequently, many studies have now shown a poor correlation between mRNA and protein expression levels (2, 12, 67, 75). The formation of mRNA is only the first step in a long sequence of events resulting in the synthesis of a protein (Fig. 2). First, mRNA is subject to posttranscriptional control in the form of alternative splicing, polyadenylation, and mRNA editing (117). Many different protein isoforms can be generated from a single gene at this step. Second, mRNA then can be subject to regulation at the level of protein translation (78). Proteins, having been formed, are subject to posttranslational modification. It is estimated that up to 200 different types of posttranslational protein modification exist (89). Proteins can also be regulated by proteolysis (86) and compartmentalization (33). The average number of protein forms per gene was predicted to be one or two in bacteria, three in yeast, and three or more in humans (168). Therefore, it is clear that the tenet of “one gene, one protein” is an oversimplification. In addition, some bodily fluids such as serum or urine have no mRNA source and therefore cannot be studied by mRNA analysis.
FIG. 2.
Mechanisms by which a single gene can give rise to multiple gene products. Multiple protein isoforms can be generated by RNA processing when RNA is alternatively spliced or edited to form mature mRNA. mRNA, in turn, can be regulated by stability and efficiency of translation. Proteins can be regulated by additional mechanisms, including posttranslational modification, proteolysis, or compartmentalization.
Protein function.
According to one study, no function can be assigned to about one-third of the sequences in organisms for which the genomes have been sequenced (47). The complete identification of all proteins in a genome will aid the field of structural genomics in which the ultimate goal is to obtain 3-D structures for all proteins in a proteome. This is necessary because the functions of many proteins can only be inferred by examination of their 3-D structure (24).
Protein modifications.
One of the most important applications of proteomics will be the characterization of posttranslational protein modifications. Proteins are known to be modified posttranslationally in response to a variety of intracellular and extracellular signals (74). For example, protein phosphorylation is an important signaling mechanism and disregulation of protein kinases or phosphatases can result in oncogenesis (74). By using a proteomics approach, changes in the modifications of many proteins expressed by a cell can be analyzed simultaneously.
Protein localization and compartmentalization.
One of the most important regulatory mechanisms known is protein localization. The mislocalization of proteins is known to have profound effects on cellular function (e.g., cystic fibrosis) (42). Proteomics aims to identify the subcellular location of each protein. This information can be used to create a 3-D protein map of the cell, providing novel information about protein regulation.
Protein-protein interactions.
Of fundamental importance in biology is the understanding of protein-protein interactions. The process of cell growth, programmed cell death, and the decision to proceed through the cell cycle are all regulated by signal transduction through protein complexes (127). Proteomics aims to develop a complete 3-D map of all protein interactions in the cell. One step toward this goal was recently completed for the microorganism Helicobacter pylori (133). Using the yeast two-hybrid method to detect protein interactions, 1,200 connections were identified between H. pylori proteins covering 46.6% of the genome (133). A comprehensive two-hybrid analysis has also been performed on all the proteins from the yeast S. cerevisiae (157).
Types of Proteomics
Protein expression proteomics.
The quantitative study of protein expression between samples that differ by some variable is known as expression proteomics. In this approach, protein expression of the entire proteome or of subproteomes between samples can be compared. Information from this approach can identify novel proteins in signal transduction or identify disease-specific proteins.
Structural proteomics.
Proteomics studies whose goal is to map out the structure of protein complexes or the proteins present in a specific cellular organelle are known as “cell map” or structural proteomics (21). Structural proteomics attempts to identify all the proteins within a protein complex or organelle, determine where they are located, and characterize all protein-protein interactions. An example of structural proteomics was the recent analysis of the nuclear pore complex (137). Isolation of specific subcellular organelles or protein complexes by purification can greatly simplify the proteomic analysis (83). This information will help piece together the overall architecture of cells and explain how expression of certain proteins gives a cell its unique characteristics.
Functional proteomics.
“Functional proteomics” is a broad term for many specific, directed proteomics approaches. In some cases, specific subproteomes are isolated by affinity chromatography for further analysis. This could include the isolation of protein complexes or the use of protein ligands to isolate specific types of proteins. This approach allows a selected group of proteins to be studied and characterized and can provide important information about protein signaling, disease mechanisms or protein-drug interactions.
TECHNOLOGY OF PROTEOMICS
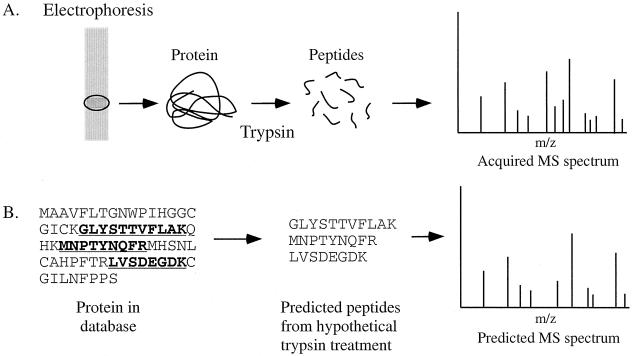
An integral part of the growth of proteomics has been in the advances made in protein technologies. Twenty-six years ago, when 2-DE was introduced, very few tools existed for proteomics. Since that time, new technologies have emerged and old ones have been improved in areas from protein separation to protein identification. However, it is also clear that it is still not feasible to conduct many types of proteomics because of limitations in technology. These problems will have to be solved and new technologies must be developed for proteomics to reach its full potential. A typical proteomics experiment (such as protein expression profiling) can be broken down into the following categories: (i) the separation and isolation of proteins from a cell line, tissue, or organism; (ii) the acquisition of protein structural information for the purposes of protein identification and characterization; and (iii) database utilization.
Separation and Isolation of Proteins
By the very definition of proteomics, it is inevitable that complex protein mixtures will be encountered. Therefore, methods must exist to resolve these protein mixtures into their individual components so that the proteins can be visualized, identified, and characterized. The predominant technology for protein separation and isolation is polyacrylamide gel electrophoresis. Unlike the breakthroughs in molecular biology that eventually enabled the sequencing of the human genome, some aspects of protein science have shown little progress over the years. Protein separation technology is one of them. Since its inception some 32 years ago (92), protein electrophoresis still remains the most effective way to resolve a complex mixture of proteins. In many applications, it is at this stage where the bottleneck occurs. This is because 1- or 2-DE is a slow, tedious procedure that is not easily automated. However, until something replaces this methodology, it will remain an essential component of proteomics.
One- and two-dimensional gel electrophoresis.
For many proteomics applications, 1-DE is the method of choice to resolve protein mixtures. In 1-DE, proteins are separated on the basis of molecular mass. Because proteins are solubilized in sodium dodecyl sulfate (SDS), protein solubility is rarely a problem. Moreover, 1-DE is simple to perform, is reproducible, and can be used to resolve proteins with molecular masses of 10 to 300 kDa. The most common application of 1-DE is the characterization of proteins after some form of protein purification. This is because of the limited resolving power of a 1-D gel. If a more complex protein mixture such as a crude cell lysate is encountered, then 2-DE can be used. In 2-DE, proteins are separated by two distinct properties. They are resolved according to their net charge in the first dimension and according to their molecular mass in the second dimension. The combination of these two techniques produces resolution far exceeding that obtained in 1-DE.
One of the greatest strengths of 2-DE is the ability to resolve proteins that have undergone some form of posttranslational modification. This resolution is possible in 2-DE because many types of protein modifications confer a difference in charge as well as a change in mass on the protein. One such example is protein phosphorylation. Frequently, the phosphorylated form of a protein can be resolved from the nonphosphorylated form by 2-DE. In this case, a single phosphoprotein will appear as multiple spots on a 2-D gel (94). In addition, 2-DE can detect different forms of proteins that arise from alternative mRNA splicing or proteolytic processing.
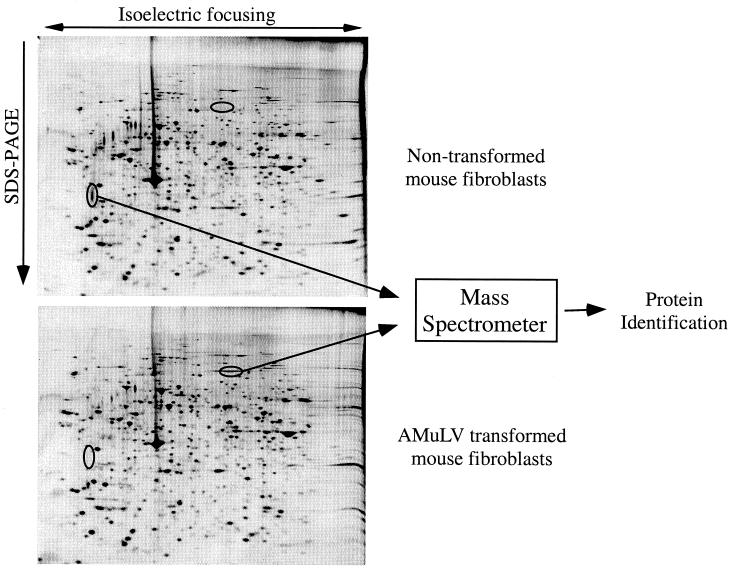
The primary application of 2-DE continues to be protein expression profiling. In this approach, the protein expression of any two samples can be qualitatively and quantitatively compared. The appearance or disappearance of spots can provide information about differential protein expression, while the intensity of those spots provides quantitative information about protein expression levels. Protein expression profiling can be used for samples from whole organisms, cell lines, tissues, or bodily fluids. Examples of this technique include the comparison of normal and diseased tissues (44) or of cells treated with various drugs or stimuli (30, 57, 69, 141, 144). An example of 2-DE used in protein profiling is shown in Fig. 3.
FIG. 3.
Protein expression profiling by 2-DE. Whole-cell lysates from nontransformed and Abelson murine leukemia virus (AMuLV)-transformed mouse fibroblasts were resolved by 2-DE, and proteins were visualized by silver staining. Differentially expressed proteins were excised from the gel and identified by MS.
Another application of 2-DE is in cell map proteomics. 2-DE is used to map proteins from microorganisms (28, 146), cellular organelles (83), and protein complexes (134). It can also be used to resolve and characterize proteins in subproteomes that have been created by some form of purification of a proteome (26, 35, 38, 83). Because a single 2-DE gel can resolve thousands of proteins (30, 44, 146), it remains a powerful tool for the cataloging of proteins. Many 2-DE databases have been constructed and are available on the World Wide Web (15).
A number of improvements have been made in 2-DE over the years (13, 29). One of the biggest improvements was the introduction of immobilized pH gradients, which greatly improved the reproducibility of 2-DE (20, 59). The use of fluorescent dyes has improved the sensitivity of protein detection (126), and specialized pH gradients are able to resolve more proteins (59). The speed of running 2-DE has been improved, and 2-D gels can now be run in the minigel format (139). In addition, there have been efforts to automate 2-DE. Hochstrasser's group has automated the process of 2-DE from gel running to image analysis and spot picking (156). The use of computers has aided the analysis of complex 2-D gel images (16). This is a critical aspect of 2-DE because a high degree of accuracy is required in spot detection and annotation if artifacts are to be avoided. Recently, a molecular scanner was developed to record 2-DE images (19). Software programs such as Melanie compare computer images of 2-D gels and facilitate both the identification and quantitation of protein spots between samples (171). A recent exciting advance in 2-DE was developed by Minden and coworkers (158). This technology is called difference gel electrophoresis (DIGE) and utilizes fluorescent tagging of two protein samples with two different dyes. The tagged proteins are run on the same 2-D gel, and postrun fluorescence imaging of the gel is used to create two images, which are superimposed to identify pattern differences. The dyes are amine reactive and are designed to ensure that proteins common to both samples have the same relative mobility regardless of the dye used to tag them. This technique circumvents the need to compare several 2-D gels. In their original paper, DIGE was used to detect differences between exogenous proteins in two D. melanogaster embryo extracts at nanogram levels (158). Moreover, an inducible protein from Escherichia coli was detected after 15 min of induction. This technology is now commercially available from Amersham/Pharmacia.
However, a number of problems with 2-DE still remain. Despite efforts to automate protein analysis by 2-DE, it is still a labor-intensive and time-consuming process. A typical 2-DE experiment can take two days, and only a single sample can be analyzed per gel. In addition, 2-DE is limited by both the number and type of proteins that can be resolved. For example, the protein mixture obtained from a eukaryotic cell lysate is too complex to be completely resolved on a single 2-D gel (29). Many large or hydrophobic proteins will not enter the gel during the first dimension, and proteins of extreme acidity or basicity (proteins with pIs below pH 3 and above pH 10) are not well represented (59). Some of these problems can be overcome with different solubilization conditions and pH gradients (59). Another limitation of 2-DE is the inability to detect low-copy proteins when a total-cell lysate is analyzed (67, 96, 146). In a crude cell extract, the most abundant proteins can dominate the gel, making the detection of low-copy proteins difficult. It was determined in the analysis of yeast proteins by 2-DE that no proteins defined as low-copy proteins were visible by 2-DE (67). Yet it is estimated that over half of the 6,000 genes in yeast may encode low-copy proteins (58). In mammalian cells, the dynamic range of protein expression is estimated to be between 7 and 9 orders of magnitude (36). This problem cannot be overcome by simply loading more protein on the gel, because the resolution will decrease and the comigration of proteins will increase (36). Because of these limitations, the largest application of 2-DE in the future will probably involve the analysis of protein complexes or subproteomes as opposed to whole proteomes.
Alternatives to electrophoresis.
The limitations of 2-DE have inspired a number of approaches to bypass protein gel electrophoresis. One approach is to convert an entire protein mixture to peptides (usually by digestion with trypsin) and then purify the peptides before subjecting them to analysis by MS. Various methods for peptide purification have been devised, including liquid chromatography (95, 106, 174), capillary electrophoresis (55, 155), and a combination of techniques such as multidimensional protein identification (95) or cation-exchange chromatography and reverse-phase (RP) chromatography (120). The advantage of these methods is that because a 2-D gel is avoided, a greater number of proteins in the mixture can be represented. The disadvantage is that it can require an immense amount of time and computing power to deconvolute the data obtained. In addition, considerable time and effort may be expended in the analysis of uninteresting proteins. One of the most exciting techniques to emerge as an alternative to protein electrophoresis is that of isotope-coded affinity tags (ICAT). This method allows the quantitative protein profiling between different samples without the use of electrophoresis (see “Proteomics applications” below).
Acquisition of Protein Structure Information
Edman sequencing.
One of the earliest methods used for protein identification was microsequencing by Edman chemistry to obtain N-terminal amino acid sequences. Little has changed in Edman chemistry since its introduction, but improvements in sequencing technology have increased the sensitivity and ease of Edman sequencing. Although the use of Edman sequencing is waning in the field of proteomics, it is still a very useful tool for several reasons. First, because Edman sequencing existed before MS as a sequencing tool, a considerable number of investigators continue to use Edman sequencing. Second, Edman sequencing of relatively abundant proteins is a viable alternative to MS if a mass spectrometer is in high demand for the identification of low-copy proteins or is not available. Finally, Edman sequencing is used to obtain the N-terminal sequence of a protein (if possible) to determine its true start.
The N-terminal sequencing of proteins was introduced by Edman in 1949 (45). Today, Edman sequencing is most often used to identify proteins after they are transferred to membranes. The development of membranes compatible with sequencing chemicals allowed Edman sequencing to become a more applicable sequencing method for the identification of proteins separated by SDS-polyacrylamide gel electrophoresis (8, 159). One of the biggest problems that has limited the success of Edman sequencing in the past is N-terminal modification of proteins. Since it is difficult to tell if a protein is N-terminally blocked before it is sequenced, precious samples were often lost in failed sequencing attempts. To overcome this problem, we developed a novel approach called mixed-peptide sequencing (38). In mixed-peptide sequencing, a protein is converted into peptides by cleavage with cyanogen bromide (CNBr) or skatole and the peptides are sequenced in an Edman sequencer simultaneously (9, 38, 99).
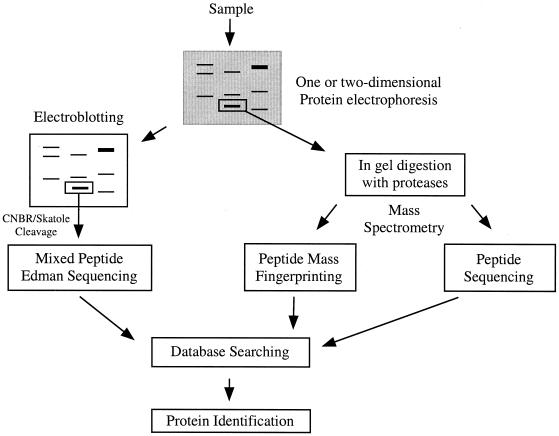
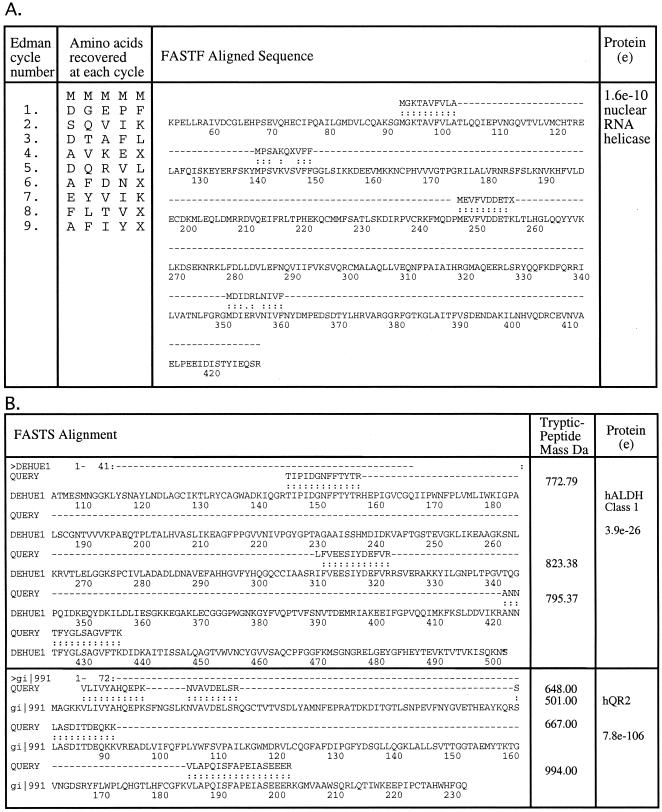
Briefly, the process of mixed-peptide sequencing involves separation of a complex protein mixture by polyacrylamide gel electrophoresis (1-D or 2-D) and then transfer of the proteins to an inert membrane by electroblotting (Fig. 4). The proteins of interest are visualized on the membrane surface, excised, and fragmented chemically at methionine (by CNBr) or tryptophan (by skatole) into several large peptide fragments. On average, three to five peptide fragments are generated, consistent with the frequency of occurrence of methionine and tryptophan in most proteins. The membrane piece is placed directly into an automated Edman sequencer without further manipulation. Between 6 and 12 automated Edman cycles are carried out (4 to 8 h), and the mixed-sequence data are fed into the FASTF or TFASTF algorithms, which sort and match the data against protein (FASTF) and DNA (TFASTF) databases to unambiguously identify the protein. The FASTF and TFASTF programs were written in collaboration with William Pearson (Department of Biochemistry, University of Virginia). Because minimal sample handling is involved, mixed-peptide sequencing can be a sensitive approach for identifying proteins in polyacrylamide gels at the 0.1- to 1-pmol level. An example of mixed-peptide sequencing is shown in Fig. 5A. The mixed-sequence approach has the advantage of enabling subsequent searches to be carried out against unannotated or non-species-specific DNA databases as well as annotated protein databases. This is because the T/FASTF algorithms utilize actual amino acid sequence and are therefore able to tolerate errors in the database as well as polymorphisms or conservative substitutions. A recent variation of T/FASTF has been devised for MS (101) (Fig. 5B). The T/FASTF/S programs are available at http://fasta.bioch.virginia.edu/ (Table 1).
FIG. 4.
Strategies for protein identification. The identification of proteins from a polyacrylamide gel by mixed-peptide sequencing or MS is depicted. For mixed-peptide sequencing, proteins are transferred to a membrane and cleaved with CNB or skatole, and the resulting peptides are sequenced simultaneously by Edman degradation. For MS, proteins are in-gel digested with proteases and the resulting peptides are mass fingerprinted or sequenced. Information from all these methods is used to search nucleotide and protein databases for protein identification.
FIG. 5.
The FASTF and FASTS search programs. (A) Example of a FASTF search where the amino acid sequence is obtained by Edman sequencing of a mixture of peptides. The information is then deconvoluted by a computer algorithm, and the results are given an expectation score (e). (B) With the FASTS program, a similar type of search is conducted except that peptide sequences obtained from MS are used.
TABLE 1.
World Wide Web tools for searching databases with protein information obtained either from mass spectrometry or from Edman degradation
| Site name | URL | Information available | Reference |
|---|---|---|---|
| MOWSE | http://srs.hgmp.mrc.ac.uk/cgi-bin/mowse | Peptide mass mapping and sequencing | 125 |
| ProFound | http://prowl.rockefeller.edu/cgi-bin/ProFound | Peptide mass mapping and sequencing | 176 |
| PeptIdent | http://www.expasy.ch/tools/peptident. | Peptide mass mapping and sequencing | 165 |
| PepSea | http://195.41.108.38/PepSeaIntro.html | Peptide mass mapping and sequencing | 102 |
| MASCOT | http://www.matrixscience.com/ | Peptide mass mapping and sequencing | 129 |
| PepFrag | http://www.proteometrics.com/ | Peptide mass mapping and sequencing | 54 |
| Protein Prospector | http://prospector.ucsf.edu/ | Peptide mass mapping and sequencing | 32 |
| FindMod | http://www.expasy.ch/tools/findmod/ | Posttranslational modification | 166 |
| SEAQUEST | http://fields.scripps.edu/sequest/ | Uninterpreted MS/MS searching | 49 |
| FASTA Search Programs | http://fasta.bioch.virginia.edu/ | Protein and nucleotide database searching | 101 |
| Cleaved Radioactivity of Phosphopeptides | http://fasta.bioch.virginia.edu/crp | Protein phosphorylation site mapping | MacDonald et al. submitted |
Mass spectrometry.
MS enables protein structural information, such as peptide masses or amino acid sequences, to be obtained. This information can be used to identify the protein by searching nucleotide and protein databases (Fig. 4). It also can be used to determine the type and location of protein modifications. The harvesting of protein information by MS can be divided into three stages: (i) sample preparation, (ii) sample ionization, and (iii) mass analysis.
(i) Sample preparation.
In most of proteomics, a protein is resolved from a mixture by using a 1- or 2-D polyacrylamide gel. The challenge is to extract the protein or its constituent peptides from the gel, purify the sample, and analyze it by MS. The extraction of whole proteins from gels is inefficient; however, if a protein is “in-gel” digested with a protease, many of the peptides can be extracted from the gel. A method for in-gel protein digestion was developed (149, 169) and is now commonly applied to both 1- and 2-D gels (136). In-gel digestion is more efficient at sample recovery than other common methods such as electroblotting (37). In addition, the conversion of a protein into its constituent peptides provides more information than can be obtained from the whole protein itself. For many applications, the peptides recovered following in-gel digestion need to be purified to remove gel contaminants. Common impurities from electrophoresis such as salts, buffers, and detergents can interfere with MS (172). In addition, peptide samples often require concentration before being analyzed by MS. One method of peptide purification commonly employed for this purpose is reverse-phase chromatography, which is available in a variety of formats. Peptides can be purified with ZipTips (Millipore) or Poros R2 perfusion material (PerSeptive Biosystems, Framingham, Mass.) (149, 169, 170) or by high-pressure liquid chromatography (HPLC).
(ii) Sample ionization.
For biological samples to be analyzed by MS, the molecules must be charged and dry. This is accomplished by converting them to desolvated ions. The two most common methods for this are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In both methods, peptides are converted to ions by the addition or loss of one or more protons. ESI and MALDI are “soft” ionization methods that allow the formation of ions without significant loss of sample integrity. This is important because it enables accurate mass information to be obtained about proteins and peptides in their native states.
(a) Electrospray ionization.
In ESI, a liquid sample flows from a microcapillary tube into the orifice of the mass spectrometer, where a potential difference between the capillary and the inlet to the mass spectrometer results in the generation of a fine mist of charged droplets (52, 72, 172). As the solvent evaporates, the sizes of the droplets decrease, resulting in the formation of desolvated ions (52). A significant improvement in ESI technology occurred with the development of nanospray ionization (169, 170). In nanospray ionization, the microcapillary tube has a spraying orifice of 1 to 2 μm and flow rates as low as 5 to 10 nl/min (170). The low flow rates possible with nanospray ionization reduce the amount of sample consumed and increase the time available for analysis (148, 149). For ESI, there are several ways to deliver the sample to the mass spectrometer. The simplest method is to load individual microcapillary tubes with sample. Because a new microcapillary tube is used for each sample, cross-contamination is avoided. In ESI, peptides require some form of purification after in-gel digestion, and this can be accomplished directly in the microcapillary tubes. The drawback to both the purification and manual loading of microcapillary tubes is that it is tedious and slow. As an alternative, electrospray sources have been connected in line with liquid chromatography (LC) systems that automatically purify and deliver the sample to the mass spectrometer. Examples of this method are LC (39, 55, 95, 106), reverse-phase LC (RP-LC) (64) and reverse-phase microcapillary LC (RP-μLC) (41).
(b) Matrix-assisted laser desorption/ionization.
In MALDI, the sample is incorporated into matrix molecules and then subjected to irradiation by a laser. The laser promotes the formation of molecular ions (84). The matrix is typically a small energy-absorbing molecule such as 2,5-dihydroxybenzoic acid or α-cyano-4-hydroxycinnamic acid. The analyte is spotted, along with the matrix, on a metal plate and allowed to evaporate, resulting in the formation of crystals. The plate, which can be 96-well format, is then placed in the mass spectrometer, and the laser is automatically targeted to specific places on the plate. Since sample application can be performed by a robot, the entire process including data collection and analysis can be automated. This is the single biggest advantage of MALDI. Another advantage of MALDI over ESI is that samples can often be used directly without any purification after in-gel digestion (131).
(iii) Mass analysis.
Mass analysis follows the conversion of proteins or peptides to molecular ions. This is accomplished by the mass analyzers in a mass spectrometer, which resolve the molecular ions on the basis of their mass and charge in a vacuum.
(a) Quadrupole mass analyzers.
One of the most common mass analyzers is the quadrupole mass analyzer. Here, ions are transmitted through an electric field created by an array of four parallel metal rods, the quadrupole (172). A quadrupole can act to transmit all ions or as a mass filter to allow the transmission of ions of a certain mass-to-charge (m/z) ratio. If multiple quadrupoles are combined, they can be used to obtain information about the amino acid sequence of a peptide. For a more detailed review of the operating principles of a quadrupole mass analyzer, the reader is directed to several excellent reviews (25, 109, 172).
(b) Time of flight.
A time-of-flight (TOF) instrument is one of the simplest mass analyzers. It measures the m/z ratio of an ion by determining the time required for it to traverse the length of a flight tube. Some TOF mass analyzers include an ion mirror at the end of the flight tube, which reflects ions back through the flight tube to a detector. In this way, the ion mirror serves to increase the length of the flight tube. The ion mirror also corrects for small energy differences among ions (172). Both of these factors contribute to an increase in mass resolution.
(c) Ion trap.
Ion trap mass analyzers function to trap molecular ions in a 3-D electric field. In contrast to a quadrupole mass analyzer, in which ions are discarded before the analysis begins, the main advantage of an ion trap mass analyzer is the ability to allow ions to be “stored” and then selectively ejected from the ion trap, increasing sensitivity (172). For a review of the operating principles of an ion trap mass spectrometer, see reference 34.
(iv) Types of mass spectrometers.
Most mass spectrometers consist of four basic elements: (i) an ionization source, (ii) one or more mass analyzers, (iii) an ion mirror, and (iv) a detector. The names of the various instruments are derived from the name of their ionization source and the mass analyzer. Some of the most common mass spectrometers are discussed; for a more comprehensive review of mass spectrometers, the reader is directed to references (76 and 172). The analysis of proteins or peptides by MS can be divided into two general categories: (i) peptide mass analysis and (ii) amino acid sequencing. In peptide mass analysis or peptide mass fingerprinting, the masses of individual peptides in a mixture are measured and used to create a mass spectrum (70). In amino acid sequencing, a procedure known as tandem mass spectrometry, or MS/MS, is used to fragment a specific peptide into smaller peptides, which can then be used to deduce the amino acid sequence.
(a) Triple quadrupole.
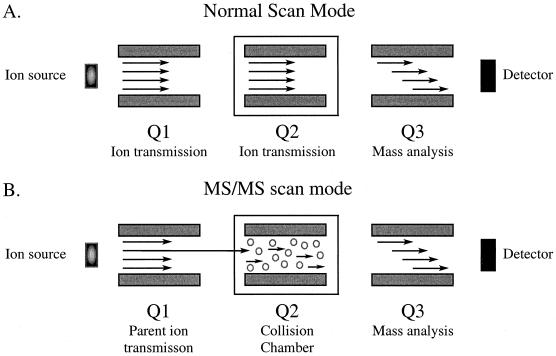
Triple-quadrupole mass spectrometers are most commonly used to obtain amino acid sequences. In the first stage of analysis, the machine is operated in MS scan mode and all ions above a certain m/z ratio are transmitted to the third quadrupole for mass analysis (Fig. 6) (82, 173). In the second stage, the mass spectrometer is operated in MS/MS mode and a particular peptide ion is selectively passed into the collision chamber. Inside the collision chamber, peptide ions are fragmented by interactions with an inert gas by a process known as collision-induced dissociation or collisionally activated dissociation. The peptide ion fragments are then resolved on the basis of their m/z ratio by the third quadrupole (Fig. 6). Since two different mass spectra are obtained in this analysis, it is referred to as tandem mass spectrometry (MS/MS). MS/MS is used to obtain the amino acid sequence of peptides by generating a series of peptides that differ in mass by a single amino acid (71, 73).
FIG. 6.
MS/MS. Conventional and MS/MS modes of analysis in a triple-quadrupole mass spectrometer are shown. (A) In the normal scanning mode, all ions of a certain m/z range are transmitted through the first two quadrupoles for mass analysis in the third quadrupole. From this MS spectrum, a parent ion is selected for fragmentation in the collision cell. (B) In MS/MS mode, the parent ion is selectively transmitted into the collision chamber and fragmented, and the resulting daughter ions are resolved in the third quadrupole.
(b) Quadrupole-TOF.
In recent years, several “hybrid” mass spectrometers have emerged from the combination of different ionization sources with mass analyzers. One example is the quadrupole-TOF mass spectrometer (111, 112, 162). In this machine, the first quadrupole (Q1) and the quadrupole collision cell (q) of a triple-quadrupole machine have been combined with a time-of-flight analyzer (TOF) (145). The main applications of a QqTOF mass spectrometer are protein identification by amino acid sequencing and characterization of protein modifications. However, because it is coupled to electrospray, it is not typically utilized for large-scale proteomics.
(c) MALDI-TOF.
The principal application of a MALDI-TOF mass spectrometer is peptide mass fingerprinting because it can be completely automated, making it the method of choice for large-scale proteomics work (48). Because of its speed, MALDI-TOF is frequently used as a first-pass instrument for protein identification. If proteins cannot be identified by fingerprinting, they can then be analyzed by electrospray and MS/MS. A MALDI-TOF machine can also be used to obtain the amino acid sequence of peptides by a method known as post-source decay (152). However, peptide sequencing by post-source decay is not as reliable as sequencing with competing electrospray methods because the peptide fragmentation patterns are much less predictable (85, 111).
(d) MALDI-QqTOF.
The MALDI-QqTOF mass spectrometer was developed to permit both peptide mass fingerprinting and amino acid sequencing (97, 147). It was formed by the combination of a MALDI ion source with a QqTOF mass analyzer (63, 91, 97, 147, 162). Thus, if a sample is not identified by peptide mass fingerprinting in the first step, the amino acid sequence can then be obtained without having to use a different mass spectrometer. However, in our experience, the amino acid sequence information obtained using this instrument was more difficult to interpret than that obtained from a nanospray-QqTOF mass spectrometer.
(e) FT-ICR.
A Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer is an ion-trapping instrument that can achieve higher mass resolution and mass accuracy than any other type of mass spectrometer (10). Recently, FT-ICR has been employed in the analysis of biomolecules ionized by both ESI and MALDI. The unique abilities of FT-ICR provide certain advantages compared to other mass spectrometers. For example, because of its high resolution, FT-ICR can be used for the analysis of complex mixtures. FT-ICR, coupled to ESI, is also being employed in the study of protein interactions and protein conformations. A high-throughput, large-scale proteomics approach involving FT-ICR has recently been developed by Smith et al. (150). For a review of the operating principles of FT-ICR and its applications, the reader is directed to reference 104.
(v) Peptide fragmentation.
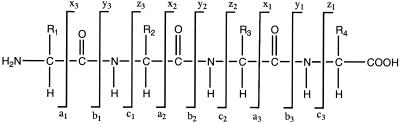
As peptide ions are introduced into the collision chamber, they interact with the collision gas (usually nitrogen or argon) and undergo fragmentation primarily along the peptide backbone (71, 73, 172). Since peptides can undergo multiple types of fragmentation, nomenclature has been created to indicate what type of ions have been generated (Fig. 7). If, after peptide bond cleavage, the charge is maintained on the N-terminus of the ion, it is designated a b-ion, whereas if the charge is maintained on the C terminus, it is a y-ion (Fig. 7) (18, 135, 173). The difference in mass between adjacent y- or b-ions corresponds to that of an amino acid. This can be used to identify the amino acid and hence the peptide sequence, with the exception of isoleucine and leucine, which are identical in mass and therefore indistinguishable (103). Both y- and b-type ions can also eliminate NH3 (−17 Da), H2O (−18 Da) and CO (−28 Da), resulting in pairs of signals observed in the mass spectrum (Fig. 7). In addition to fragmentation along the peptide backbone, cleavage can occur along amino acid side chains, and this information can be used to distinguish isoleucine and leucine (172).
FIG. 7.
Peptide ion fragmentation nomenclature. Low-energy collisions promote fragmentation of a peptide primarily along the peptide backbone (73). Peptide fragmentation which maintains the charge on the C terminus is designated a y-ion, whereas fragmentation which maintains the charge on the N terminus is designated a b-ion. Additional types of fragmentation are also indicated.
(vi) Our approach to mass spectrometry.
The sensitivity of a mass spectrometer is probably the single most important feature of the instrument. What is the sensitivity of a modern mass spectrometer? How much protein is needed to make an unambiguous identification? Many factors can affect sensitivity, such as sample preparation, sample ionization, the type of mass spectrometer used, the sample itself, and the type of database search employed. In our laboratory, we rely on 1- or 2-DE electrophoresis for the isolation and visualization of protein targets. We typically stain our gels with either Coomassie blue or silver stain. For most proteins, staining with Coomassie blue will give a dark band for ∼1μg of protein and a discernible one for ∼200 ng. With silver staining, we can detect a dark band at ∼50 ng and faint yet discernible bands at ∼5 to 10 ng. However, a significant number of proteins do not stain well by these methods and larger proteins tend to bind more stain (mole/mole) than small proteins. In addition, MS is not a quantitative technique because peptide ionization is not quantitative. Therefore, some proteins that are barely visible on gels can give stronger signals by MS than do some darkly staining proteins. For example, one of the most frequently sequenced proteins in MS is human keratin, a component of dust. It is a contaminant that will often appear on polyacrylamide gels as faint silver-stained bands with a variety of molecular weights. It can be introduced simply from the glass plates or gel combs used for protein gels; therefore, it is a good idea to wash these items in concentrated acid before use.
We have found in our laboratory that most proteins applied to the gel at 5 to 10 ng (100 to 200 fmol for a 50-kDa protein) can be identified by MS. However, the ability to identify a protein depends on the protein itself and its presence in the database. Below 5 to 10 ng, the success rate decreases because fewer peptides are obtained for sequencing. Several prominent MS laboratories routinely report record-breaking sequencing sensitivity to the attomolar level. However, this sensitivity is usually toward a purified peptide sample that is directly introduced into the mass spectrometer. Since most proteins are isolated from gels for identification, this is not an accurate measure of sensitivity. In another case, it was reported that an amino acid sequence was obtained after the in-gel digestion of 25 fmol (1.7 ng) of pure bovine serum albumin (90). Again, since the protein was known before the analysis began, this is not a fair assessment of sensitivity. For unknown proteins, more protein is required because several peptides have to be sequenced before a confident assignment can be made.
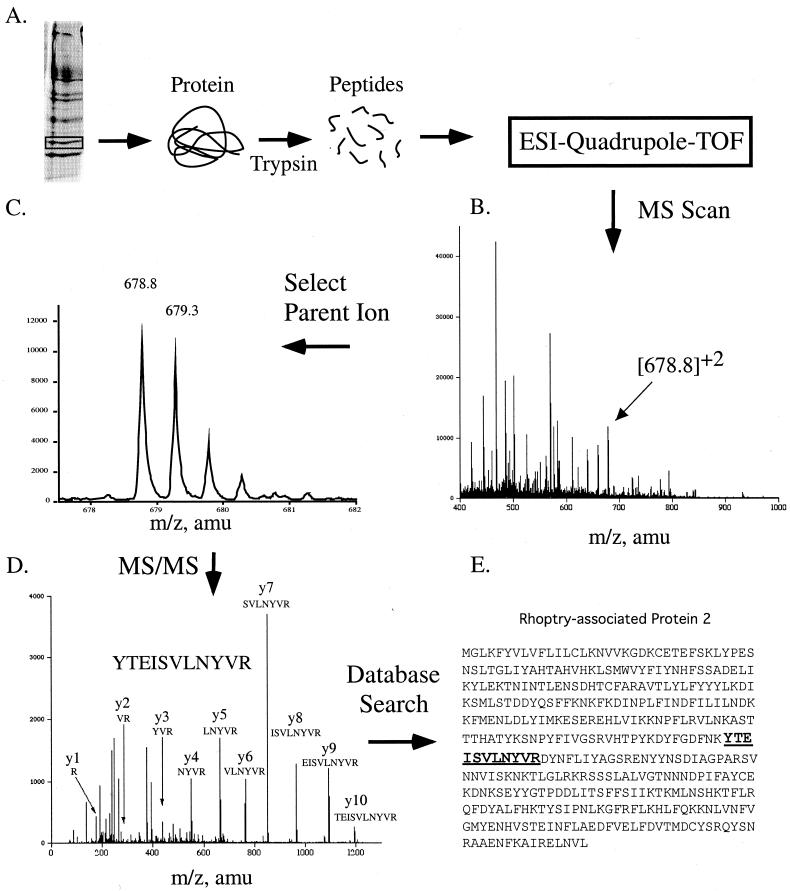
A typical approach to protein identification in our laboratory is outlined in Fig. 8. Protein from a polyacrylamide gel is excised and then in-gel digested with trypsin by the method of Wilm et al. (170). Following peptide extraction from the gel, we purify the peptides on Poros R2 (149, 169) in microcapillary tubes by using the method described on the website http://www.protana.com/products/applicationnotes/purification/default.asp. We use the API QSTAR Pulsar mass spectrometer (AB/MDS-SCIEX) with nanospray ionization to obtain an MS scan of the peptide mixture. From the MS scan, a peptide ion is selected for MS/MS based on its signal strength and charge state, which allow it to be distinguished from the background ions. In nanospray ionization, most peptide ions are either doubly or triply charged whereas the background ions are singly charged. This peptide ion is also known as the parent ion. MS/MS of a parent ion is performed, and amino acid sequence information for the peptide is obtained. As shown in Fig. 8, a single peptide was sequenced and found to match rhoptry-associated protein 2 (RAP-2) from Plasmodium falciparum. Since matching multiple peptides to a protein increases the confidence of identification (106), we typically sequence several peptides for each sample. For RAP-2, a total of four peptides were found to match the protein. Because the staining intensity on gels is not always a good indicator of the signal obtained by MS and because gel bands often contain protein mixtures, additional criteria can aid in protein identification. For example, if the major protein excised from the gel was 50 kDa, does the protein identified match in molecular mass? Is the protein from the expected species? If a protein is isolated from a 2-D gel, does it match the expected isoelectric point as exhibited on the gel?
FIG. 8.
Protein identification by MS/MS. (A) Protein from P. falciparum was resolved on a one-dimensional polyacrylamide gel, excised, and in-gel digested with trypsin. The resulting peptides were ionized by electrospray and analyzed by a Quadrupole-TOF mass spectrometer. (B) The MS spectrum produced was scanned, and a parent ion of 678.8 was selected for fragmentation. (C) Enlargement of the parent ion peak at 678 shown in panel B. The multiplet of peaks is due to the contribution in mass from the naturally occurring isotope 13C. A mass difference between the peaks of 0.5 Da indicates that the peptide is doubly charged. (D) MS/MS scan of the 678 parent ion and analysis of the daughter ions produced. All y-ions (except for y-11) produced from fragmentation of the peptide are shown. (E) Identification of rhoptry-associated protein-2 using BioAnalyst software (Applied Biosystems, Foster City, Calif.).
Database Utilization
Databases allow protein structural information harvested from Edman sequencing or MS to be used for protein identification. The goal of database searching is to be able to quickly and accurately identify large numbers of proteins (132). The success of database searching depends on the quality of the data obtained in the mass spectrometer, the quality of the database searched, and the method used to search the database. What is the best way to identify an unknown protein? What type of database search engine should be used?
Peptide mass fingerprinting database searching.
One method of protein identification is peptide mass fingerprinting (77, 79, 102, 125, 175). In this method, the masses of peptides obtained from the proteolytic digestion of an unknown protein are compared to the predicted masses of peptides from the theoretical digestion of proteins in a database (Fig. 9). If enough peptides from the real mass spectrum and the theoretical one overlap, a protein identification can be made. The principal advantage of peptide mass fingerprinting is speed. The analysis and database search can be fully automated.
FIG. 9.
Strategy of protein identification by peptide mass fingerprinting. (A) The unknown protein is excised from a gel and converted to peptides by the action of a specific protease. The mass of the peptides produced is then measured in a mass spectrometer. (B) The mass spectrum of the unknown protein is searched against theoretical mass spectra produced by computer-generated cleavage of proteins in the database.
The single biggest disadvantage of peptide mass fingerprinting is ambiguity in protein identification. This is because of peptide mass redundancy. For example, a peptide of 5 amino acids can have the same mass by simple rearrangement of its constitutive amino acids; e.g., peptide VAGSE has the same mass as AVGSE or AEVGS and so on. For this technique to be successful, the masses of a large number of peptides must be obtained to provide enough specificity in the search, and this is not always possible. Mass redundancy occurs with greater frequency in large genomes. Moreover, peptide mass fingerprinting is effective only in the analysis of proteins from organisms whose genome is small, completely sequenced, and well annotated (131). It has limited use against unannotated or untranslated DNA databases such as the human genome. Because mass fingerprinting is not error tolerant, several factors in addition to mass redundancy contribute to its limited use, including sequencing errors, conservative substitutions, polymorphisms, and six possible translations at the DNA level.
Another factor affecting the success of peptide mass fingerprinting is mass accuracy (32, 62). Because it is critical to obtain an accurate measurement of the masses of multiple peptides, factors that alter the masses of those peptides can reduce the success of the method. One such example is the posttranslational modification of proteins. If the unknown protein is extensively modified, the peptides produced from that protein will not match the unmodified protein in the database. Recent improvements in the mass accuracy of mass spectrometers has increased the success rate of protein identification by this method (32, 54).
Finally, peptide mass fingerprinting does not work well with protein mixtures. As a protein mixture is converted to a mixture of peptides, it increases the complexity of the peptide mass fingerprint. The process of protein identification can be hindered if even two or three proteins are present in the sample (107). Several search methods have emerged to accommodate peptide mixtures in the mass spectrum. One example is a program called ProFound, which enables protein identification in simple protein mixtures (176). However, the lack of ability to analyze protein mixtures remains a major limitation of this method. A variety of tools for database searching now exist on the World Wide Web (Table 1). The ExPASy server provides a variety of tools for proteomics and programs for protein identification (reviewed in reference 165). Search programs used for peptide mass fingerprinting include PepSea (102), PeptIdent/MultiIdent (165), MS-Fit (32), MOWSE (125), and ProFound (176).
Amino acid sequence database searching.
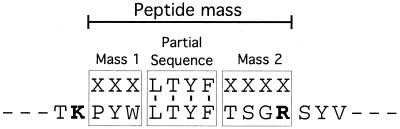
The most specific type of database searching for protein identification uses peptide amino acid sequence. If the amino acid sequence of a peptide can be identified, it can be used to search databases to find the protein from which it was derived. One method which utilizes this information is peptide mass tag searching. In this method, a partial amino acid sequence is obtained by interpretation of the MS/MS spectrum (the sequence tag) and this information is combined with the mass of the peptide and the masses of the peptide on either side of the sequence tag where the sequence is not known (Fig. 10). Also included in the search is the type of protease used to produce the peptides. Peptide mass tag searching is a more specific tool for protein identification than peptide mass fingerprinting (49, 103, 115, 170). In addition, one of the biggest advantages of utilizing MS/MS to obtain peptide amino acid sequence is that, unlike peptide mass fingerprinting, it is compatible with protein mixtures. The ability to identify proteins in mixtures is one of the great advantages of using MS as a protein identification tool. For example, in our laboratory we frequently identify multiple proteins from what appears to be a single band on an SDS-gel. In fact, in the majority of proteomics experiments, proteins are present in mixtures at the time of analysis.
FIG. 10.
Peptide mass tag searching. Shown is a schematic of how information from an unknown peptide (top) is matched to a peptide sequence in a database (bottom) for protein identification. The partial amino acid sequence or “tag” obtained by MS/MS is combined with the peptide mass (parent mass), the mass of the peptide at the start of the sequence (mass tag 1), and the mass of the peptide at the end of the sequence (mass tag 2). The specificity of the protease used (trypsin is shown) can also be included in the search (103).
The major disadvantage of performing MS/MS is that the process is not easily automated. As a result, considerable time is expended in performing the analysis and interpreting the mass spectrum. Although computer programs can assist in the interpretation of the spectrum, they currently are not able to make accurate assignments without some guidance. In addition, when searching a database with peptide mass tags, there is a lack of flexibility in the search programs. If a single mistake is made in the assignment of a y- or b-ion (which can happen quite frequently), the amino acid sequence will be incorrect and the database search will bring up irrelevant proteins. Often it is necessary to confirm that the peptide sequence obtained from the database matches the sequence obtained in the mass spectrometer. This can be done by performing a theoretical fragmentation of the peptide from the database and comparing the two mass spectra. Additional clues can also be used, such as verifying if the peptide obtained from the database ends in amino acids consistent with the type of protease used.
De novo peptide sequence information.
Another approach to protein identification is to obtain de novo sequence data from peptides by MS/MS and then use all the peptide sequences to search appropriate databases. Multiple peptide sequences can be used for protein identification by searching databases with the FASTS program (Mackey et al., submitted) (Fig. 5). The single biggest advantage of this method is the capability of searching peptide sequence information across both DNA and protein databases. This is because the search engine utilized exhibits a certain amount of flexibility in the assignment of protein scores. This search method is useful for organisms that do not have well-annotated databases such as Xenopus laevis or human. However, because this method requires several peptide amino acid sequences of 3 or 4 amino acids, it is not the first choice for peptide identification. Rather, the much faster methods of peptide mass fingerprinting or peptide mass tag searching can be used first. If these search methods fail, de novo sequence information can be obtained and used to identify the protein.
Uninterpreted MS/MS data searching.
A large number of programs are now available for the identification of proteins by using uninterpreted MS/MS data. Examples include programs such as Mascot (129), SONAR (53), and SEQUEST (49) (Table 1). However, searches against unannotated or untranslated DNA databases with uninterpreted MS/MS data are likely to suffer from the same pitfalls associated with mass fingerprinting. In particular, polymorphisms, sequencing errors, and conservative substitutions will probably contribute to failure to accurately identify a protein. The development of uninterpreted MS/MS search algorithms that are error tolerant may overcome some of these shortcomings, provided that they assign some form of statistical scoring to the identified proteins.
PROTEOMICS APPLICATIONS
The single most common application of proteomics is protein identification. Most investigators use proteomics approaches to isolate and display proteins based on their own specific criteria and then identify the proteins. Protein identification provides immediate information that will direct subsequent experimentation. For example, the identity of a protein can reveal an expected result, validate a proteomics approach, provide completely unexpected information, or reveal that your biochemical method is not working at all. We feel that the most critical stage of any proteomics approach is the strategic design for the isolation of protein targets. In recent years, as the technology of MS has improved, there has been a de-emphasis on the “front-end” of proteomics experiments compared to data analysis. This can result in the isolation of hundreds of irrelevant proteins for identification, consuming both time and effort. Our general strategy is to devise techniques that enrich for low-abundance proteins and then analyze only the proteins that appear on differential display or are isolated by affinity chromatography. To accomplish this, we use affinity columns and other strategies to select for protein targets. In each case, protein samples are subjected to a series of precolumns and high-stringency washes to remove nonspecific proteins. This reduces the number of irrelevant proteins for analysis.
Characterization of Protein Complexes
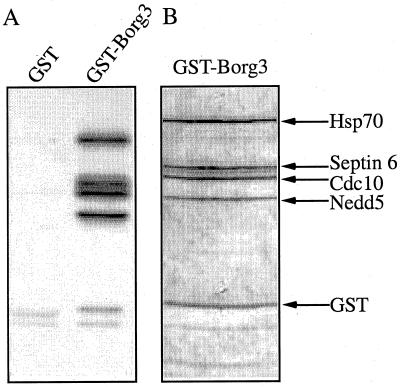
Many laboratories are now engaged in an effort to characterize protein complexes by MS. Examples include Link et al. utilizing multidimensional LC and MS/MS to identify proteins (95) or Mann and colleagues identifying proteins present after immunoprecipitation of protein complexes (124). Recently, Macara, Haystead, and coworkers used MS to identify interacting proteins with the Cdc42 effector, Borg3 (80). In this case, the “bait” protein, Borg3, was produced as a glutathione S-transferase (GST) fusion in E. coli and then mixed with NIH 3T3 cell lysate. Four interacting proteins were identified by mixed-peptide sequencing: heat shock protein Hsp70 and three septins including Septin6, Cdc10, and Nedd5 (Fig. 11). None of these proteins were present in the GST-only control sample. Although the interaction with Hsp70 was not pursued, it was shown from coimmunoprecipitation studies that endogenous Borg3 interacts with endogenous Cdc10 and Nedd5 (80). Additional proof from expression and structure-function studies confirmed a role for the Borg proteins as regulators of septin organization. It should be noted that although several proteins were quickly identified as Borg3 interactors by the pull-down experiment, it took several more months of work to confirm this interaction.
FIG. 11.
Identification of novel protein interactions by protein coprecipitation. (A) Pull-down experiment with a control (GST) or target (GST-Borg3) protein using 35S-labeled NIH 3T3 cell lysate. (B) Large-scale affinity purification of GST-Borg3 from the NIH 3T3 lysate. Individual proteins were microsequenced by mixed-peptide sequencing and identified by database searching with the FASTF algorithm (101).
Protein Expression Profiling
The largest application of proteomics continues to be protein expression profiling. Through the use of two-dimensional gels or novel techniques such as ICAT, the expression levels of proteins or changes in their level of modification between two different samples can be compared and the proteins can be identified. This approach can facilitate the dissection of signaling mechanisms or identify disease-specific proteins.
Expression profiling by two-dimensional electrophoresis.
Currently, the majority of protein expression profiling studies are performed by 2-DE. Several diseases have been studied, including heart disease (44) and cancer (30). Cancer cells are good candidates for proteomics studies because they can be compared to their nontransformed counterparts. Analysis of differentially expressed proteins in normal versus cancer cells can (i) identify novel tumor cell biomarkers that can be used for diagnosis, (ii) provide clues to mechanisms of cancer development, and (iii) identify novel targets for therapeutic intervention. Protein expression profiling has been used in the study of breast (121), esophageal (121), bladder (30) and prostate (114) cancer. From these studies, tumor-specific proteins were identified and 2-D protein expression databases were generated. Many of these 2-D protein databases are now available on the World Wide Web (15).
Isotope-coded affinity tags.
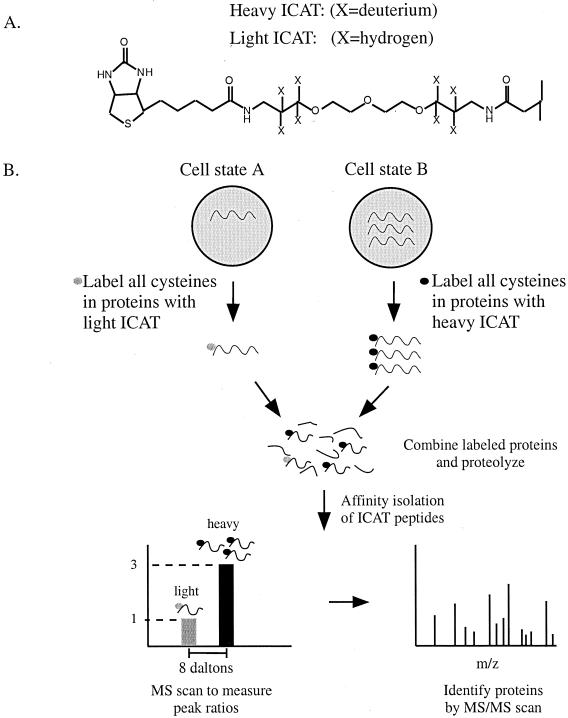
Recently, a novel method for protein expression profiling was introduced that does not depend on the separation of proteins by 2-DE. This method is known as isotope-coded affinity tags (ICAT) and relies on the labeling of protein samples from two different sources with two chemically identical reagents that differ only in mass as a result of isotope composition (66). Differential labeling of samples by mass allows the relative amount of protein between two samples to be quantitated in the mass spectrometer. An example of the methodology of ICAT is shown in Fig. 12. Cell extract from two different samples is reacted with one of two forms of the ICAT reagent, an isotopically light form in which the linker contains eight hydrogens or a heavy form in which the linker contains eight deuterium atoms. The ICAT reagent reacts with cysteine residues in proteins via a thiol-reactive group and contains a biotin moiety to facilitate purification (Fig. 12). Peptides are recovered on the basis of the biotin tag by avidin affinity chromatography and are then analyzed by MS. The difference in peak heights between heavy and light peptide ions directly correlates with the difference in protein abundance in the cells. Thus, if a protein is present at a threefold higher level in one sample, this will be reflected in a threefold difference in peak heights. Following quantitation of the peptides, they can be fragmented by MS/MS and the amino acid sequence can be obtained. Thus, using this approach, proteins can be identified and their expression levels can be compared in the same analysis.
FIG. 12.
The ICAT method for measuring differential protein expression. (A) Structure of the ICAT reagent. ICAT consists of a biotin affinity group, a linker region that can incorporate heavy (deuterium) or light (hydrogen) atoms, and a thiol-reactive end group for linkage to cysteines. (B) ICAT strategy. Proteins are harvested from two different cell states and labeled on cysteine residues with either the light or heavy form of the ICAT reagent. Following labeling, the two protein samples are mixed and digested with a protease such as trypsin. Peptides labeled with the ICAT reagent can be purified by virtue of the biotin tag by using avidin chromatography. Following purification, ICAT-labeled peptides can be analyzed by MS to quantitate the peak ratios and proteins can be identified by sequencing the peptides with MS/MS.
The single biggest advantage of this method is the elimination of the 2-D gel for protein quantitation. As a result, an increased amount of sample can be used to enrich for low-abundance proteins. Alternatively, the cell lysate can be fractionated prior to reaction with the ICAT reagent. This can allow the enrichment of low-abundance proteins before the analysis begins. The main disadvantages are that currently this method works only for proteins containing cysteine, even though this includes the majority of proteins (68). In addition, peptides must contain appropriately spaced protease cleavage sites flanking the cysteine residues. Finally, the ICAT label is large (∼500 kDa) and remains with each peptide throughout the analysis. This can make database searching more difficult, especially for small peptides with limited sequence (4, 65). Sensitivity may also be of concern since tagged peptides derived from low-copy proteins are likely to be poorly recovered during the affinity step as a result of nonspecific interactions with avidin-Sepharose. Studies have been performed to optimize the labeling of proteins with the ICAT reagent (151).
Protein arrays.
Protein arrays are undergoing rapid development for the detection of protein-protein interactions and protein expression profiling (17, 98, 180, 181). Recently, protein microarrays were created using ordinary laboratory equipment (98). Proteins were immobilized by being covalently attached to glass microscope slides, and the protein microarrays were shown to be capable of interacting with other proteins, small molecules, and enzyme substrates (98). In another report, 5,800 yeast proteins were expressed and printed onto microscope slides. These protein microarrays were used to identify novel calmodulin- and phospholipid-interacting proteins (180). These reports indicate that protein arrays hold great promise for the global analysis of protein-protein and protein-ligand interactions. Undoubtedly, these arrays will improve as the technology for their creation is developed and refined.
Proteomics Approach to Protein Phosphorylation
Posttranslational modification of proteins is a fundamental regulatory mechanism, and characterization of protein modifications is paramount for understanding protein function. MS is one of the most powerful tools for the analysis of protein modifications because virtually any type of protein modification can be identified. Although we focus here on protein phosphorylation, the analysis of other types of protein modification by MS has been described (25). Protein phosphorylation is one of the most common of all protein modifications and has been found in nearly all cellular processes (74, 88, 153). MS can be used to identify novel phosphoproteins, measure changes in the phosphorylation state of proteins in response to an effector, and determine phosphorylation sites in proteins. Identification of phosphorylation sites can provide information about the mechanism of enzyme regulation and the protein kinases and phosphatases involved. A proteomics approach to protein phosphorylation has the advantage that instead of studying changes in the phosphorylation of a single protein in response to some perturbation, one can study all the phosphoproteins in a cell (the phosphoproteome) at the same time. A common approach to studying protein phosphorylation events is the use of in vivo labeling of phosphoproteins with inorganic 32P. The phosphoproteomes of cells that differ in some way (e.g., normal versus diseased) can be analyzed by growing cells in inorganic 32P and creating cell lysates. Changes in the phosphorylation state of proteins can then be examined by 2-DE and autoradiography. Proteins of interest are excised from the gel and microsequenced by MS. A major limitation of this approach is that while many phosphorylated proteins can be visualized by autoradiography, they cannot be identified because of their low abundance. One solution to this problem is enrichment of the phosphoproteome.
Phosphoprotein enrichment.
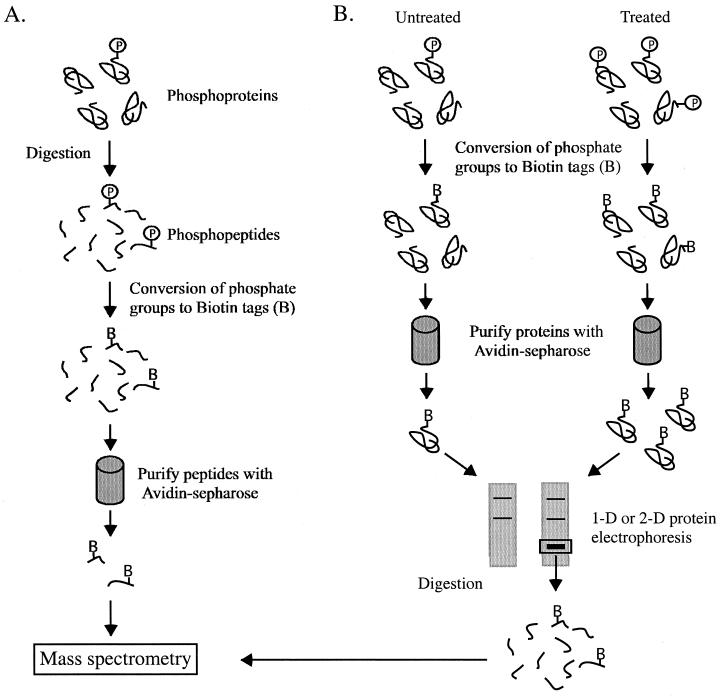
Enrichment of the phosphoproteome of a cell can allow the identification of low-copy phosphoproteins that would otherwise go undetected. In one approach, phosphoproteins were enriched by conversion of phosphoserine residues to biotinylated residues (118). This method is an extension of techniques originally developed by Hielmeyer and colleagues (108) and more recently by our laboratory (51) for the identification of phosphorylation sites using Edman sequencing. Following derivatization, proteins that were formerly phosphorylated can be isolated by avidin affinity chromatography (118). Proteins immobilized on avidin beads can then be eluted with biotin, theoretically resulting in the isolation of the entire phosphoserine proteome (Fig. 13). By increasing the amount of cell lysate used for avidin affinity chromatography, low-abundance phosphoproteins can be enriched. However, this technique does not work for phosphotyrosine and the reactivity of phosphothreonine by this method is very poor (118). Tyrosine-phosphorylated proteins can be isolated by the use of antiphosphotyrosine antibodies (124). As an alternative, another method for phosphopeptide enrichment was devised to allow the recovery of proteins phosphorylated on serine, threonine, and tyrosine (179). In this method, a protein or mixture of proteins is digested to peptides with a protease and then subjected to a multistep procedure for the conversion of phosphoamino acids into free sulfhydryl groups. To capture the derivatized peptides, the free sulfhydryl groups in the peptides are then reacted with iodoacetyl groups immobilized on glass beads. Using this method, several phosphopeptides were recovered from β-casein and from a yeast cell extract, although it was unclear whether all the proteins isolated from the yeast extract were phosphoproteins (179).
FIG. 13.
Phosphopeptide and phosphoprotein enrichment. (A) Enrichment of phosphopeptides. Phosphoproteins are digested with a protease, and the phosphate groups are converted to biotin tags (119). Once biotinylated, the peptides can be selectively recovered with avidin-Sepharose and analyzed by MS. (B) Differential display of phosphoproteins. The phosphate groups present in proteins derived from two different samples are converted to biotin tags, and the phosphoproteins are purified on avidin-Sepharose in an identical manner. The phosphoproteins are then compared by 1- or 2-DE, and the target proteins are digested and analyzed by MS.
Enrichment of the phosphoproteome can also be combined with protein profiling by 1- or 2-DE. In this way, changes in protein amount observed on electrophoresis will reflect the level of protein phosphorylation (Fig. 13). Recently, the principle of protein quantitation by ICAT has been combined with phosphoprotein enrichment (60). This was accomplished by the introduction of isotopic label into ethanedithiol, the reagent used to convert the alkene created by β-elimination of phosphoserine into a free sulfhydryl group. In this way, the differences in the amount of phosphoproteins in extracts can be analyzed quantitatively in the mass spectrometer (60). It should be noted that because of the chemistry used in both of these methods, these techniques are relatively insensitive and require tens of picomoles of phosphoprotein. As a result, we have found that these methods as currently designed are impractical for the isolation and enrichment of low-abundance phosphoproteins.
Phosphorylation site determination by Edman degradation.
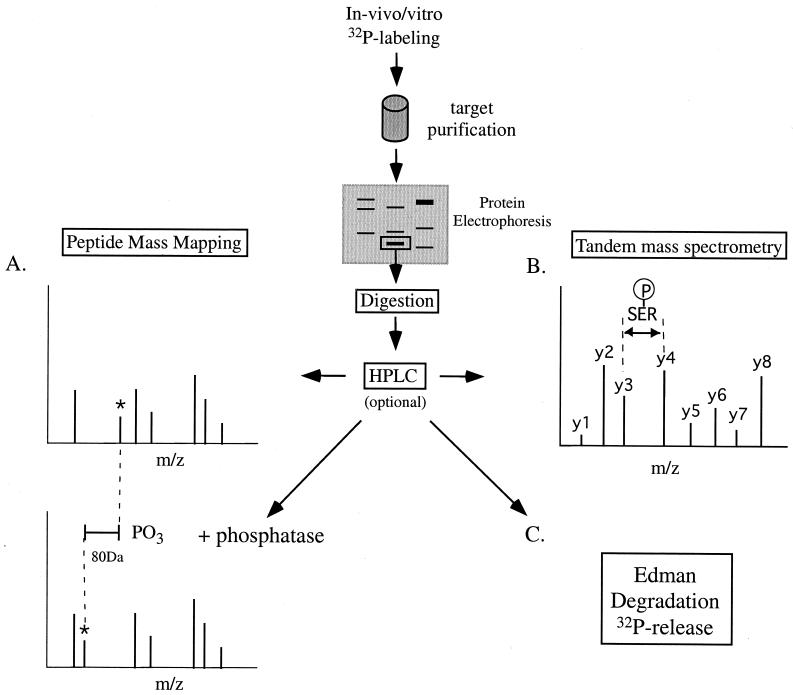
Edman sequencing is still a widely used method for determining phosphorylation sites in proteins labeled with 32P, either in vitro or in vivo (5, 22, 164). This is because sites can be determined at the sub-femtomolar level if enough radioactivity can be incorporated into the phosphoprotein of interest. In our hands, this can be as little as 1,000 cpm (not ideal). Briefly, a 32P-labeled protein is digested with a protease and the resulting phosphopeptides are separated and purified by reverse-phase HPLC or thin-layer chromatography (TLC) (Fig. 14). The isolated peptides are then cross-linked via their C termini to an inert membrane (e.g. Immobilon P; PerSeptive Biosystems). The radioactive membrane is subjected to several rounds of Edman cycles, and radioactivity is collected after the cleavage step. The released 32P is counted in a scintillation counter. This method positionally places the phosphoamino acid within the sequenced phosphopeptide. Of course, this is meaningful only if the sequence of the phosphopeptide is already known. In addition, the analysis ceases to become quantitative beyond 30 Edman cycles (even with efficient, modern Edman machines) due to well-understood issues with repetitive yield associated with Edman chemistry.
FIG. 14.
Strategies for determination of phosphorylation sites in proteins. Proteins phosphorylated in vitro or in vivo can be isolated by protein electrophoresis and analyzed by MS. (A) Identification of phosphopeptides by peptide mass fingerprinting. In this method, phosphopeptides are identified by comparing the mass spectrum of an untreated sample to that of a sample treated with phosphatase. In the phosphatase-treated sample, potential phosphopeptides are identified by a decrease in mass due to loss of a phosphate group (80 Da). (B) Phosphorylation sites can be identified by peptide sequencing using MS/MS. (C) Edman degradation can be used to monitor the release of inorganic 32P to provide information about phosphorylation sites in peptides.
Recently, our laboratory has extended the usefulness of phosphorylation site characterization by Edman chemistry through the development of the cleaved radioactive peptide (CRP) program (J. A. MacDonald, A. J. Mackay, W. R. Pearson, and T. A. J. Haystead, submitted for publication). In CRP analysis, one requires only that the sequence of the protein be known. Purification and sequencing of individual peptides is not required. Radiolabeled proteins (isolated following immunoprecipitation from 32P-labeled cells, for example) are cleaved at predetermined residues by the action of a protease. The phosphopeptides are then separated by HPLC or TLC (if only one site is present, no peptide separation is required), cross-linked to the inert membrane, and carried through 25 to 30 Edman cycles. The sequence of the target protein is entered into the CRP program. This program predicts how many Edman cycles are required to cover 100% of all the serines, threonines, and tyrosines from the site of cleavage. Generally, one round of CRP analysis narrows the number of possible sites to 5 to 10 for most proteins. Phosphoamino acid analysis can be used to reduce the number of possibilities still further. The CRP analysis is then repeated following cleavage with a second protease (usually one cutting at R, but M and F are alternatives). The second round of CRP usually unambiguously localizes the phosphoamino acid to one possible site. The technique does not work if sites are more than 30 amino acids away from all possible cleavage sites. The finding that CRP analysis is not applicable may in itself confine a phosphorylation site to a segment of the protein that is likely to produce very large proteolytic fragments. The Cleavage of Radioactive Proteins (CRP) program is accessible at http://fasta.bioch.virginia.edu/crp/ and was written in collaboration with Aaron Mackey and Bill Pearson of the University of Virginia (MacDonald et al., submitted).
Phosphorylation site determination by mass spectrometry.
Because of its sensitivity, MS can allow the direct sequencing of phosphopeptides, resulting in unambiguous phosphorylation site identification. Below, a brief overview of some common methods for phosphorylation site determination by MS are given. A more complete discussion of this topic is provided by Mitchelhill and Kemp (110). Identification of phosphorylation sites in proteins provides several unique challenges for the mass spectrometrist. For example, unlike in protein identification, where analysis of any peptide within the protein can be informative, phosphorylation site analysis requires that the phosphorylated peptide be analyzed. This means that considerably more protein is required for analysis. In addition, phosphorylation can alter the cleavage pattern of a protein and the resulting phosphopeptides may require different purification methods. To isolate and purify the phosphopeptides of interest, it may be necessary to alter the way in which the phosphoprotein is digested and to alter the pH or the chromatographic material used for peptide purification (27, 110, 116).
(i) Phosphopeptide sequencing by MS/MS.
In our laboratory, we have found that a combination of HPLC, Edman degradation, and phosphopeptide sequencing by MS/MS provides the best results for phosphorylation site determination (Fig. 14). Following excision and digestion of a 32P-labeled protein, the peptides are resolved by HPLC. By monitoring HPLC fractions for radioactivity, the phosphopeptides can be selected for analysis. This reduces the complexity of the peptide mixture before MS is performed and facilitates phosphopeptide identification (Fig. 14).
Phosphopeptides can be identified from a mixture of peptides by a method known as precursor ion scanning (116). In this method, the second mass analyzer in the mass spectrometer is set at the mass of the reporter ion for the phospho group (PO3−) of m/z = 79. Peptides are sprayed under neutral or basic conditions, and phosphopeptides are identified in the precursor ion scan only if their fragmentation yields an ion of m/z = 79. Once a phosphopeptide is identified, the peptide mixture is sprayed under acidic conditions and the phosphopeptide is sequenced by conventional tandem MS/MS. On fragmentation of the phosphopeptide, phosphoserine can be identified by the formation of dehydroalanine (69 Da), the β-elimination product of phosphoserine. Similarly, phosphothreonine can be identified by the formation of its β-elimination product, dehydroamino-2-butyric acid at 83 Da (116).
(ii) Analysis of phosphopeptides by MALDI-TOF.
MALDI-TOF mass spectrometry can also be used to identify phosphopeptides (81, 130, 177, 178). When phosphorylated peptides are subjected to ionization by MALDI, phosphate groups are frequently liberated from the peptides. This is the case for phosphoserine- and phosphothreonine-containing peptides, which can liberate HPO3 or H3PO4, resulting in a neutral loss of 80 and 98 Da, respectively. Careful examination of the TOF spectrum for differences in peptide masses of 80 Da that are not found in the unphosphorylated peptide control can identify phosphopeptides. Phosphopeptides can also be identified by treating one of two identical samples with protein phosphatase to liberate phosphate groups (Fig. 14). Once a phosphopeptide is identified, it can be sequenced by MS/MS for identification of the phosphorylation site (178).
Yeast Genomics and Proteomics
One of the most exciting applications of proteomics involves combining this technology with the power of yeast genetics to delineate signaling events in vivo. Our laboratory has published two papers using this strategy to identify in vivo targets for protein phosphatases (9, 40). In one study (9), we identified physiological substrates for the Glc7p-Reg1p complex by examining the effects of deletion of the REG1 gene on the yeast phosphoproteome. In S. cerevisiae, PP-1 (Glc7p) and its binding protein, Reg1p, are essential for the regulation of glucose repression pathways. The target for this phosphatase complex was not known. Analysis by 2-D phosphoprotein mapping identified two distinct proteins that were greatly increased in phosphate content in reg1Δ mutants. Mixed-peptide sequencing identified these proteins as hexokinase II (Hxk2p) and the E1α subunit of pyruvate dehydrogenase. We then went on to validate these findings in a comprehensive biochemical study. Consistent with increased phosphorylation of Hxk2p in response to REG1 deletion, fractionation of yeast extracts by anion-exchange chromatography identified a Hxk2p phosphatase activity in wild-type strains that was selectively lost in the reg1Δ mutant. Having carried out these studies, we attempted to rescue the reg1Δ phosphoprotein phenotype by overexpressing both wild-type and mutant Reg1p in the deletion strains. Here, both the phosphorylation state of Hxk2p and Hxk2p phosphatase activity were restored to wild-type levels in the reg1Δ mutant by expression of a LexA-Reg1p fusion protein. In contrast, expression of a LexA-Reg1p protein containing mutations at phenylalanine in a putative PP-1C (the catalytic subunit) binding site motif (K/R)(X)(I/V)XF was unable to rescue Hxk2p dephosphorylation in intact yeast or restore Hxk2p phosphatase activity. These results demonstrate that Reg1p targets PP-1C to dephosphorylate Hxk2p in vivo and that the peptide motif (K/R)(X)(I/V)XF is necessary for its PP-1 targeting function. These studies therefore demonstrate how a proteomics approach can be used to first identify enzyme targets in cells and then direct all further analysis to verify the findings. It should be pointed out that often 6 to 12 months of work ensues following the initial sequencing of the targeted proteins. Nevertheless, clearly a combined proteomics and genetics approach greatly enhances one's ability to directly answer key biological questions. We believe that a similar strategy could be adopted with transgenic or knockout mouse work, particularly in cases where there is no obvious phenotype.
Proteome Mining
Proteome mining is a functional proteomics approach used to extract protein information from the analysis of specific subproteomes. The strategy of proteome mining is shown in Fig. 15. The principles of proteome mining are based on the assumption that all drug-like molecules selectively compete with a natural cellular ligand for a binding site on a protein target. In a proteome mine, natural ligands are immobilized on beads at high density and in an orientation that sterically favors interaction with their protein targets. The immobilized ligand is then exposed to whole-animal or tissue extract, and bound proteins are evaluated for specificity by protein sequencing. In the prototypic example from our laboratory, ATP is immobilized in the “protein kinase orientation” (via its gamma phosphate). Microsequencing of the proteins that were eluted with free ATP demonstrated that the nucleotide selectively recovered purine binding proteins including protein kinases, dehydrogenases, various purine-dependent metabolic enzymes, DNA ligases, heat shock proteins, and a variety of miscellaneous ATP-utilizing enzymes (P. R. Graves, J. Kwiek, P. Fadden, R. Ray, K. Hardeman, and T. A. J. Haystead, submitted for publication). This immobilized proteome represents ∼4% of the expressed eukaryotic genome.
FIG. 15.
Proteome-mining strategy. Proteins are isolated on affinity column arrays from a cell line, organ, or animal source and purified to remove nonspecific adherents. Then, compound libraries are passed over the array and the proteins eluted are analyzed by protein electrophoresis. Protein information obtained by MS or Edman degradation is then used to search DNA and protein databases. If a relevant target is identified, a sublibrary of compounds can be evaluated to refine the lead. From this analysis, both a protein target and a drug lead can be simultaneously identified.
We have utilized this captured proteome (the purine binding cassette proteome) to test the selectivity of purine analogs that inhibit protein kinases and stress-induced ATPases in vitro. Using a proteome-mining ATP affinity array apparatus constructed in our laboratory, sufficient biomass was applied to ensure the recovery, per column, of 1 fmol of any protein expressed at 100 copies/cell (107 cells). After washing, each column in the array is eluted in parallel with molecules from a purine-based iterative library and fractions are collected. Eluates are screened for protein, and positive fractions generally contain a single protein, a small number of structurally related proteins, or a complex mixture. Only the first two categories are sequenced, since the third resulted from elution with a nonselective inhibitor. Once one has identified an eluted protein, one has all the necessary information on how to proceed. The first decision is biological relevance. Does the eluted protein(s) in any given fraction have relevance to any human disease? If the protein has no obvious use as a drug target, it is ignored. If the protein is deemed relevant, one immediately has a lead molecule and a defined target. In cases where a single protein is eluted, the lead is likely to be selective because it had an equal opportunity to interact with the rest of the captured proteome (∼4% of the genome). Selectivity can be tested by increasing the concentration of the lead compound during elution from nanomolar to micromolar. Information concerning potential toxicity can be gained by sequencing other proteins that are simultaneously eluted or eluted at higher concentrations. If some of these are undesirable targets, iterative substitutions can be made around the lead scaffold to improve selectivity. Proof of principle of this technology was obtained by using an iterative library derived from the heat shock protein 90 inhibitor geldanamycin, and a new physiological target, ADE2, was identified (P. Fadden, V. J. Davisson, L. Neckers, and T. A. J. Haystead, unpublished data). Screening Combichem libraries through a proteome-mining approach exploits the serendipitous nature of drug discovery to its maximum, merely because it accelerates the hit rate over a conventional screen by a factorial of the proteome that is bound. In the case of purine binding proteins, this may be several hundredfold. Protein microsequencing, the data contained within the various genome projects, and the ability to instantly search the literature for relevance enable one to interpret the outcomes in a rationale way.
We are currently using proteome mining to discover new antimalarial drugs that target purine binding proteins in the blood stage of infection. Because of the essential roles of purine-utilizing enzymes in cellular function, it is our hypothesis that these proteins are attractive candidates for a new generation of antimalarial drugs. In our malaria project, the P. falciparum (blood stage) and human red blood cell purine binding proteome are captured on ATP affinity arrays and simultaneously screened against purine-based combinatorial libraries. Combining both proteomes enables the selectivity and potential toxicity of a lead molecule to be measured early in the discovery process. Microsequencing enables human proteins to be readily discriminated from malarial ones. An additional benefit of mining the entire malarial purine binding cassette proteome is that multiple leads and their targets will be identified. Combined therapies that target multiple genes simultaneously are likely to exert such tremendous selective pressure on the targeted pathogen that it cannot develop resistance. We are currently expanding our immobilized natural-ligand library in order to apply proteome mining to other areas of biology.
Challenges for Proteomics
The study of proteins, in contrast to that of DNA, presents a number of unique challenges. For example, there is no equivalent of PCR for proteins, so the analysis of low-abundance proteins remains a major challenge. In addition, in protein interaction studies, native conformations of proteins must be maintained to obtain meaningful results. Can proteins be studied on a large scale with speed, sensitivity, and reliability? In the last several years, recognition of the limitations of proteomics are beginning to point the field in new directions.
Although the technology for the analysis of proteins is rapidly progressing, it is still not feasible to study proteins on a scale equivalent to that of the nucleic acids. Most of proteomics relies on methods, such as protein purification or PAGE, that are not high-throughput methods. Even performing MS can require considerable time in either data acquisition or analysis. Although hundreds of proteins can be analyzed quickly and in an automated fashion by a MALDI-TOF mass spectrometer, the quality of data is sacrificed and many proteins cannot be identified. Much higher quality data can be obtained for protein identification by MS/MS, but this method requires considerable time in data interpretation. In our opinion, new computer algorithms are needed to allow more accurate interpretation of mass spectra without operator intervention. In addition, to access unannotated DNA databases across species, these algorithms should be error tolerant to allow for sequencing errors, polymorphisms, and conservative substitutions. New technologies will have to emerge before protein analysis on a large-scale (such as mapping the human proteome) becomes a reality.
Another major challenge for proteomics is the study of low-abundance proteins. In some eukaryotic cells, the amounts of the most abundant proteins can be 106-fold greater than those of the low-abundance proteins. Many important classes of proteins (that may be important drug targets) such as transcription factors, protein kinases, and regulatory proteins are low-copy proteins. These low-copy proteins will not be observed in the analysis of crude cell lysates without some purification. Therefore, new methods must be devised for subproteome isolation. Despite these limitations, proteomics, when combined with other complementary technologies such as molecular biology, has enormous potential to provide new insight into biology. The ability to study complex biological systems in their entirety will ultimately provide answers that cannot be obtained from the study of individual proteins or groups of proteins.
Acknowledgments
We are very grateful to Elizabeth Herrick for figure making and design, and we thank Patrick Fadden, Justin MacDonald, and Timothy Wadkins for critical review of the manuscript.
REFERENCES
- 1.Abbott, A. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 2.Abbott, A. 1999. A post-genomic challenge: learning to read patterns of protein synthesis. Nature 402:715-720. [DOI] [PubMed] [Google Scholar]
- 3.Adams, M. D., S. E. Celniker, R. A. Holt, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold, R., B. Rist, and S. P. Gygi. 2000. Quantitative proteome analysis: methods and applications. Ann. N. Y. Acad. Sci. 919:33-47. [DOI] [PubMed] [Google Scholar]
- 5.Aebersold, R., J. D. Watts, H. D. Morrison, and E. J. Bures. 1991. Determination of the site of tyrosine phosphorylation at the low picomole level by automated solid-phase sequence analysis. Anal. Biochem. 199:51-60. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold, R. H., J. Leavitt, R. A. Saavedra, L. E. Hood, and S. B. Kent. 1987. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc. Natl. Acad. Sci. USA 84:6970-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aebersold, R. H., G. Pipes, L. E. Hood, and S. B. Kent. 1988. N-terminal and internal sequence determination of microgram amounts of proteins separated by isoelectric focusing in immobilized pH gradients. Electrophoresis 9:520-530. [DOI] [PubMed] [Google Scholar]
- 8.Aebersold, R. H., D. B. Teplow, L. E. Hood, and S. B. Kent. 1986. Electroblotting onto activated glass. High efficiency preparation of proteins from analytical sodium dodecyl sulfate-polyacrylamide gels for direct sequence analysis. J. Biol. Chem. 261:4229-4238. [PubMed] [Google Scholar]
- 9.Alms, G. R., P. Sanz, M. Carlson, and T. A. Haystead. 1999. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 18:4157-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amster, J. 1996. Fourier transform mass spectrometry. J. Mass Spectrom. 31:1325-1337. [Google Scholar]
- 11.Andersen, J. S., and M. Mann. 2000. Functional genomics by mass spectrometry. FEBS Lett. 480:25-31. [DOI] [PubMed] [Google Scholar]
- 12.Anderson, L., and J. Seilhamer. 1997. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18:533-537. [DOI] [PubMed] [Google Scholar]
- 13.Anderson, N. G., and N. L. Anderson. 1996. Twenty years of two-dimensional electrophoresis: past, present and future. Electrophoresis 17:443-453. [DOI] [PubMed] [Google Scholar]
- 14.Anderson, N. L., J. J. Edwards, C. S. Giometti, K. E. Willard, S. L. Tollaksen, S. L. Nance, B. J. Hickman, K. E. Taylor, B. Coulter, A. Scandora, and N. G. Anderson. 1980. Electrophoresis '79. Walter de Gruyter, New York, N.Y.
- 15.Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1999. 2-D databases on the World Wide Web. Methods Mol. Biol. 112:383-391. [DOI] [PubMed] [Google Scholar]
- 16.Appel, R. D., and D. F. Hochstrasser. 1999. Computer analysis of 2-D images. Methods Mol. Biol. 112:363-381. [DOI] [PubMed] [Google Scholar]
- 17.Arenkov, P., A. Kukhtin, A. Gemmell, S. Voloshchuk, V. Chupeeva, and A. Mirzabekov. 2000. Protein microchips: use for immunoassay and enzymatic reactions. Anal. Biochem. 278:123-131. [DOI] [PubMed] [Google Scholar]
- 18.Biemann, K. 1988. Contributions of mass spectrometry to peptide and protein structure. Biomed. Environ. Mass Spectrom. 16:99-100. [DOI] [PubMed] [Google Scholar]
- 19.Binz, P. A., M. Muller, D. Walther, W. V. Bienvenut, R. Gras, C. Hoogland, G. Bouchet, E. Gasteiger, R. Fabbretti, S. Gay, P. Palagi, M. R. Wilkins, V. Rouge, L. Tonella, S. Paesano, G. Rossellat, A. Karmime, A. Bairoch, J. C. Sanchez, R. D. Appel, and D. F. Hochstrasser. 1999. A molecular scanner to automate proteomic research and to display proteome images. Anal. Chem. 71:4981-4988. [DOI] [PubMed] [Google Scholar]
- 20.Bjellqvist, B., C. Pasquali, F. Ravier, J. C. Sanchez, and D. Hochstrasser. 1993. A nonlinear wide-range immobilized pH gradient for two-dimensional electrophoresis and its definition in a relevant pH scale. Electrophoresis 14:1357-1365. [DOI] [PubMed] [Google Scholar]
- 21.Blackstock, W. P., and M. P. Weir. 1999. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 17:121-127. [DOI] [PubMed] [Google Scholar]
- 22.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two- dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 23.Broder, S., and J. C. Venter. 2000. Sequencing the entire genomes of free-living organisms: the foundation of pharmacology in the new millennium. Annu. Rev. Pharmacol. Toxicol. 40:97-132. [DOI] [PubMed] [Google Scholar]
- 24.Burley, S. K., S. C. Almo, J. B. Bonanno, M. Capel, M. R. Chance, T. Gaasterland, D. Lin, A. Sali, F. W. Studier, and S. Swaminathan. 1999. Structural genomics: beyond the human genome project. Nat. Genet. 23:151-157. [DOI] [PubMed] [Google Scholar]
- 25.Burlingame, A. L., R. K. Boyd, and S. J. Gaskell. 1998. Mass spectrometry. Anal. Chem. 70:647R-716R. [DOI] [PubMed]
- 26.Campos, M., P. Fadden, G. Alms, Z. Qian, and T. A. Haystead. 1996. Identification of protein phosphatase-1-binding proteins by microcystin-biotin affinity chromatography. J. Biol. Chem. 271:28478-28484. [DOI] [PubMed] [Google Scholar]
- 27.Carr, S. A., M. J. Huddleston, and R. S. Annan. 1996. Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal. Biochem. 239:180-192. [DOI] [PubMed] [Google Scholar]
- 28.Cash, P. 2000. Proteomics in medical microbiology. Electrophoresis 21:1187-1201. [DOI] [PubMed] [Google Scholar]
- 29.Celis, J. E., and P. Gromov. 1999. 2D protein electrophoresis: can it be perfected? Curr. Opin. Biotechnol. 10:16-21. [DOI] [PubMed] [Google Scholar]
- 30.Celis, J. E., M. Ostergaard, H. H. Rasmussen, P. Gromov, I. Gromova, H. Varmark, H. Palsdottir, N. Magnusson, I. Andersen, B. Basse, J. B. Lauridsen, G. Ratz, H. Wolf, T. F. Orntoft, P. Celis, and A. Celis. 1999. A comprehensive protein resource for the study of bladder cancer: http://biobase.dk/cgi-bin/celis. Electrophoresis 20:300-309. [DOI] [PubMed] [Google Scholar]
- 31.Celis, J. E., G. P. Ratz, and A. Celis. 1987. Secreted proteins from normal and SV40 transformed human MRC-5 fibroblasts: toward establishing a database of human secreted proteins. Leukemia 1:707-717. [PubMed] [Google Scholar]
- 32.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 33.Colledge, M., and J. D. Scott. 1999. AKAPs: from structure to function. Trends Cell Biol. 9:216-221. [DOI] [PubMed] [Google Scholar]
- 34.Cooks, R. G., G. L. Glish, S. A. McLuckey, and R. E. Kaiser. 1991. Ion trap mass spectrometry. Chem. Eng. Newsl. 25:26-41. [Google Scholar]
- 35.Cordwell, S. J., A. S. Nouwens, N. M. Verrills, D. J. Basseal, and B. J. Walsh. 2000. Subproteomics based upon protein cellular location and relative solubilities in conjunction with composite two-dimensional electrophoresis gels. Electrophoresis 21:1094-1103. [DOI] [PubMed] [Google Scholar]
- 36.Corthals, G. L., V. C. Wasinger, D. F. Hochstrasser, and J. C. Sanchez. 2000. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis 21:1104-1115. [DOI] [PubMed] [Google Scholar]
- 37.Courchesne, P. L., R. Luethy, and S. D. Patterson. 1997. Comparison of in-gel and on-membrane digestion methods at low to sub- pmol level for subsequent peptide and fragment-ion mass analysis using matrix-assisted laser-desorption/ionization mass spectrometry. Electrophoresis 18:369-381. [DOI] [PubMed] [Google Scholar]
- 38.Damer, C. K., J. Partridge, W. R. Pearson, and T. A. Haystead. 1998. Rapid identification of protein phosphatase 1-binding proteins by mixed peptide sequencing and data base searching. Characterization of a novel holoenzymic form of protein phosphatase 1. J. Biol. Chem. 273:24396-24405. [DOI] [PubMed] [Google Scholar]
- 39.Davis, M. T., and T. D. Lee. 1998. Rapid protein identification using a microscale electrospray LC/MS system on an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 9:194-201. [DOI] [PubMed] [Google Scholar]
- 40.de Nadal, E., R. P. Fadden, A. Ruiz, T. Haystead, and J. Arino. 2001. A role for the Ppz Ser/Thr protein phosphatases in the regulation of translation elongation factor 1Bα. J. Biol. Chem. 276:14829-14834. [DOI] [PubMed] [Google Scholar]
- 41.Deterding, L. J., M. A. Moseley, K. B. Tomer, and J. W. Jorgenson. 1991. Nanoscale separations combined with tandem mass spectrometry. J. Chromatogr. 554:73-82. [DOI] [PubMed] [Google Scholar]
- 42.Drumm, M. L., and F. S. Collins. 1993. Molecular biology of cystic fibrosis. Mol. Genet. Med. 3:33-68. [DOI] [PubMed] [Google Scholar]
- 43.Dunham, I., N. Shimizu, B. A. Roe, et al. 1999. The DNA sequence of human chromosome 22. Nature 402:489-495. [DOI] [PubMed] [Google Scholar]
- 44.Dunn, M. J. 2000. Studying heart disease using the proteomic approach. Drug Discov. Today 5:76-84. [DOI] [PubMed] [Google Scholar]
- 45.Edman, P. 1949. A method for the determination of the amino acid sequence of peptides. Arch. Biochem. Biophys. 22:475-483. [PubMed] [Google Scholar]
- 46.Eisenberg, D., E. M. Marcotte, I. Xenarios, and T. O. Yeates. 2000. Protein function in the post-genomic era. Nature 405:823-826. [DOI] [PubMed] [Google Scholar]
- 47.Eisenstein, E., G. L. Gilliland, O. Herzberg, J. Moult, J. Orban, R. J. Poljak, L. Banerjei, D. Richardson, and A. J. Howard. 2000. Biological function made crystal clear—annotation of hypothetical proteins via structural genomics. Curr. Opin. Biotechnol. 11:25-30. [DOI] [PubMed] [Google Scholar]
- 48.Ekstrom, S., P. Onnerfjord, J. Nilsson, M. Bengtsson, T. Laurell, and G. Marko-Varga. 2000. Integrated microanalytical technology enabling rapid and automated protein identification. Anal. Chem. 72:286-293. [DOI] [PubMed] [Google Scholar]
- 49.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Fadden, P., and T. A. Haystead. 1995. Quantitative and selective fluorophore labeling of phosphoserine on peptides and proteins: characterization at the attomole level by capillary electrophoresis and laser-induced fluorescence. Anal. Biochem. 225:81-88. [DOI] [PubMed] [Google Scholar]
- 52.Fenn, J. B., M. Mann, C. K. Meng, S. F. Wong, and C. M. Whitehouse. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 246:64-71. [DOI] [PubMed] [Google Scholar]
- 53.Fenyo, D. 2000. Identifying the proteome: software tools. Curr. Opin. Biotechnol 11:391-395. [DOI] [PubMed] [Google Scholar]
- 54.Fenyo, D., J. Qin, and B. T. Chait. 1998. Protein identification using mass spectrometric information. Electrophoresis 19:998-1005. [DOI] [PubMed] [Google Scholar]
- 55.Figeys, D., G. L. Corthals, B. Gallis, D. R. Goodlett, A. Ducret, M. A. Corson, and R. Aebersold. 1999. Data-dependent modulation of solid-phase extraction capillary electrophoresis for the analysis of complex peptide and phosphopeptide mixtures by tandem mass spectrometry: application to endothelial nitric oxide synthase. Anal. Chem. 71:2279-2287. [DOI] [PubMed] [Google Scholar]
- 56.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 57.Garrels, J. I., B. Futcher, R. Kobayashi, G. I. Latter, B. Schwender, T. Volpe, J. R. Warner, and C. S. McLaughlin. 1994. Protein identifications for a Saccharomyces cerevisiae protein database. Electrophoresis 15:1466-1486. [DOI] [PubMed] [Google Scholar]
- 58.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed]
- 59.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 60.Goshe, M. B., T. P. Conrads, E. A. Panisko, N. H. Angell, T. D. Veenstra, and R. D. Smith. 2001. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal. Chem. 73:2578-2586. [DOI] [PubMed] [Google Scholar]
- 61.Reference deleted.
- 62.Green, M. K., M. V. Johnston, and B. S. Larsen. 1999. Mass accuracy and sequence requirements for protein database searching. Anal. Biochem. 275:39-46. [DOI] [PubMed] [Google Scholar]
- 63.Griffin, T. J., S. P. Gygi, B. Rist, R. Aebersold, A. Loboda, A. Jilkine, W. Ens, and K. G. Standing. 2001. Quantitative proteomic analysis using a MALDI quadrupole time-of-flight mass spectrometer. Anal. Chem. 73:978-986. [DOI] [PubMed] [Google Scholar]
- 64.Gygi, S. P., G. L. Corthals, Y. Zhang, Y. Rochon, and R. Aebersold. 2000. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA 97:9390-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gygi, S. P., B. Rist, and R. Aebersold. 2000. Measuring gene expression by quantitative proteome analysis. Curr. Opin. Biotechnol. 11:396-401. [DOI] [PubMed] [Google Scholar]
- 66.Gygi, S. P., B. Rist, S. A. Gerber, F. Turecek, M. H. Gelb, and R. Aebersold. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17:994-999. [DOI] [PubMed] [Google Scholar]
- 67.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haynes, P. A., and J. R. Yates III. 2000. Proteome profiling—pitfalls and progress. Yeast 17:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinke, M. Y., C. H. Wheeler, J. X. Yan, V. Amin, D. Chang, R. Einstein, M. J. Dunn, and C. G. dos Remedios. 1999. Changes in myocardial protein expression in pacing-induced canine heart failure. Electrophoresis 20:2086-2093. [DOI] [PubMed] [Google Scholar]
- 70.Henzel, W. J., T. M. Billeci, J. T. Stults, and S. C. Wong. 1993. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl. Acad. Sci. USA 90:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunt, D. F., A. M. Buko, J. M. Ballard, J. Shabanowitz, and A. B. Giordani. 1981. Sequence analysis of polypeptides by collision activated dissociation on a triple quadrupole mass spectrometer. Biomed. Mass Spectrom. 8:397-408. [DOI] [PubMed] [Google Scholar]
- 72.Hunt, D. F., R. A. Henderson, J. Shabanowitz, K. Sakaguchi, H. Michel, N. Sevilir, A. L. Cox, E. Appella, and V. H. Engelhard. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255:1261-1263. [DOI] [PubMed] [Google Scholar]
- 73.Hunt, D. F., J. R. Yates, 3rd, J. Shabanowitz, S. Winston, and C. R. Hauer. 1986. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 83:6233-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hunter, T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225-236. [DOI] [PubMed] [Google Scholar]
- 75.Ideker, T., V. Thorsson, J. A. Ranish, R. Christmas, J. Buhler, J. K. Eng, R. Bumgarner, D. R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929-934. [DOI] [PubMed] [Google Scholar]
- 76.James, P. 1997. Protein identification in the post-genome era: the rapid rise of proteomics. Q. Rev. Biophys. 30:279-331. [DOI] [PubMed] [Google Scholar]
- 77.James, P., M. Quadroni, E. Carafoli, and G. Gonnet. 1993. Protein identification by mass profile fingerprinting. Biochem. Biophys. Res. Commun. 195:58-64. [DOI] [PubMed] [Google Scholar]
- 78.Jansen, M., C. H. de Moor, J. S. Sussenbach, and J. L. van den Brande. 1995. Translational control of gene expression. Pediatr. Res. 37:681-686. [DOI] [PubMed] [Google Scholar]
- 79.Jensen, O. N., A. V. Podtelejnikov, and M. Mann. 1997. Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal. Chem. 69:4741-4750. [DOI] [PubMed] [Google Scholar]
- 80.Joberty, G., R. R. Perlungher, P. J. Sheffield, M. Kinoshita, M. Noda, T. Haystead, and I. G. Macara. 2001. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat. Cell Biol. 3:861-866. [DOI] [PubMed] [Google Scholar]
- 81.Jonscher, K. R., and J. R. Yates III. 1997. Matrix-assisted laser desorption ionization/quadrupole ion trap mass spectrometry of peptides. Application to the localization of phosphorylation sites on the P protein from Sendai virus. J. Biol. Chem. 272:1735-1741. [DOI] [PubMed] [Google Scholar]
- 82.Jonscher, K. R., and J. R. Yates III. 1997. The quadrupole ion trap mass spectrometer—a small solution to a big challenge. Anal. Biochem. 244:1-15. [DOI] [PubMed] [Google Scholar]
- 83.Jung, E., M. Heller, J. C. Sanchez, and D. F. Hochstrasser. 2000. Proteomics meets cell biology: the establishment of subcellular proteomes. Electrophoresis 21:3369-3377. [DOI] [PubMed] [Google Scholar]
- 84.Karas, M., and F. Hillenkamp. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60:2299-2301. [DOI] [PubMed] [Google Scholar]
- 85.Kaufmann, R., P. Chaurand, D. Kirsch, and B. Spengler. 1996. Post-source decay and delayed extraction in matrix-assisted laser desorption/ionization-reflectron time-of-flight mass spectrometry. Are there trade-offs? Rapid Commun. Mass Spectrom. 10:1199-1208. [DOI] [PubMed] [Google Scholar]
- 86.Kirschner, M. 1999. Intracellular proteolysis. Trends Cell Biol. 9:M42-M45. [PubMed] [Google Scholar]
- 87.Klose, J. 1975. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26:231-243. [DOI] [PubMed] [Google Scholar]
- 88.Krebs, E. G. 1994. The growth of research on protein phosphorylation. Trends Biochem. Sci. 19:439.. [DOI] [PubMed] [Google Scholar]
- 89.Krishna, R. G., and F. Wold. 1993. Post-translational modification of proteins. Adv. Enzymol. Relat. Areas Mol. Biol. 67:265-298. [DOI] [PubMed] [Google Scholar]
- 90.Kristensen, D. B., K. Imamura, Y. Miyamoto, and K. Yoshizato. 2000. Mass spectrometric approaches for the characterization of proteins on a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer. Electrophoresis 21:430-439. [DOI] [PubMed] [Google Scholar]
- 91.Krutchinsky, A. N., W. Zhang, and B. T. Chait. 2000. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J. Am. Soc. Mass Spectrom. 11:493-504. [DOI] [PubMed] [Google Scholar]
- 92.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 93.Lander, E. S., L. M. Linton, B. Birren, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 94.Lewis, T. S., J. B. Hunt, L. D. Aveline, K. R. Jonscher, D. F. Louie, J. M. Yeh, T. S. Nahreini, K. A. Resing, and N. G. Ahn. 2000. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6:1343-1354. [DOI] [PubMed] [Google Scholar]
- 95.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 96.Link, A. J., L. G. Hays, E. B. Carmack, and J. R. Yates III. 1997. Identifying the major proteome components of Haemophilus influenzae type-strain NCTC 8143. Electrophoresis 18:1314-1334. [DOI] [PubMed] [Google Scholar]
- 97.Loboda, A. V., A. N. Krutchinsky, M. Bromirski, W. Ens, and K. G. Standing. 2000. A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun. Mass Spectrom. 14:1047-1057. [DOI] [PubMed] [Google Scholar]
- 98.MacBeath, G., and S. L. Schreiber. 2000. Printing proteins as microarrays for high-throughput function determination. Science 289:1760-1763. [DOI] [PubMed] [Google Scholar]
- 99.MacDonald, J. A., M. A. Borman, A. Muranyi, A. V. Somlyo, D. J. Hartshorne, and T. A. Haystead. 2001. Identification of the endogenous smooth muscle myosin phosphatase-associated kinase. Proc. Natl. Acad. Sci. USA 98:2419-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reference deleted.
- 101.Mackey, A. J., T. A. Haystead, and W. R. Pearson. Getting more from less: algorithms for rapid protein identification with multiple short peptide sequences. Mol. Proteomics, in press. [DOI] [PubMed]
- 102.Mann, M., P. Hojrup, and P. Roepstorff. 1993. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom. 22:338-345. [DOI] [PubMed] [Google Scholar]
- 103.Mann, M., and M. Wilm. 1994. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66:4390-4399. [DOI] [PubMed] [Google Scholar]
- 104.Marshall, A. G., C. L. Hendrickson, and G. S. Jackson. 1998. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 17:1-35. [DOI] [PubMed] [Google Scholar]
- 105.Matsumoto, T., J. Wu, T. Baba, Y. Katayose, K. Yamamoto, K. Sakata, M. Yano, and T. Sasaki. 2001. Rice genomics: current status of genome sequencing. Novartis Found. Symp. 236:28-38. [DOI] [PubMed] [Google Scholar]
- 106.McCormack, A. L., D. M. Schieltz, B. Goode, S. Yang, G. Barnes, D. Drubin, and J. R. Yates III. 1997. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69:767-776. [DOI] [PubMed] [Google Scholar]
- 107.McDonald, W. H., and J. R. Yates III. 2000. Proteomic tools for cell biology. Traffic 1:747-754. [DOI] [PubMed] [Google Scholar]
- 108.Meyer, H. E., E. Hoffmann-Posorske, and L. M. Heilmeyer, Jr. 1991. Determination and location of phosphoserine in proteins and peptides by conversion to S-ethylcysteine. Methods Enzymol. 201:169-185. [DOI] [PubMed] [Google Scholar]
- 109.Miller, P. E., and M. B. Denton. 1986. The quadrupole mass filter: basic operating concepts. J. Chem. Ed. 63:617-622. [Google Scholar]
- 110.Mitchelhill, K. I., and B. E. Kemp. 1999. Phosphorylation site analysis by mass spectrometry, 2nd ed. Oxford University Press, New York, N.Y.
- 111.Morris, H. R., T. Paxton, A. Dell, J. Langhorne, M. Berg, R. S. Bordoli, J. Hoyes, and R. H. Bateman. 1996. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 10:889-896. [DOI] [PubMed] [Google Scholar]
- 112.Morris, H. R., T. Paxton, M. Panico, R. McDowell, and A. Dell. 1997. A novel geometry mass spectrometer, the Q-TOF, for low-femtomole/attomole-range biopolymer sequencing. J. Protein Chem. 16:469-479. [DOI] [PubMed] [Google Scholar]
- 113.Myers, E. W. G. G. Sutton, A. L. Delcher, et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196-2204. [DOI] [PubMed] [Google Scholar]
- 114.Nelson, P. S., N. Clegg, B. Eroglu, V. Hawkins, R. Bumgarner, T. Smith, and L. Hood. 2000. The prostate expression database (PEDB): status and enhancements in 2000. Nucleic Acids Res. 28:212-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Neubauer, G., A. Gottschalk, P. Fabrizio, B. Seraphin, R. Luhrmann, and M. Mann. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94:385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Neubauer, G., and M. Mann. 1999. Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal. Chem. 71:235-242. [DOI] [PubMed] [Google Scholar]
- 117.Newman, A. 1998. RNA splicing. Curr. Biol. 8:R903-R905. [DOI] [PubMed] [Google Scholar]
- 118.Oda, Y., T. Nagasu, and B. T. Chait. 2001. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 19:379-382. [DOI] [PubMed]
- 119.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 120.Opiteck, G. J., K. C. Lewis, J. W. Jorgenson, and R. J. Anderegg. 1997. Comprehensive on-line LC/LC/MS of proteins. Anal. Chem. 69:1518-1524. [DOI] [PubMed] [Google Scholar]
- 121.Page, M. J., B. Amess, R. R. Townsend, R. Parekh, A. Herath, L. Brusten, M. J. Zvelebil, R. C. Stein, M. D. Waterfield, S. C. Davies, and M. J. O'Hare. 1999. Proteomic definition of normal human luminal and myoepithelial breast cells purified from reduction mammoplasties. Proc. Natl. Acad. Sci. USA 96:12589-12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pandey, A., and F. Lewitter. 1999. Nucleotide sequence databases: a gold mine for biologists. Trends Biochem. Sci. 24:276-280. [DOI] [PubMed] [Google Scholar]
- 123.Pandey, A., and M. Mann. 2000. Proteomics to study genes and genomes. Nature 405:837-846. [DOI] [PubMed] [Google Scholar]
- 124.Pandey, A., A. V. Podtelejnikov, B. Blagoev, X. R. Bustelo, M. Mann, and H. F. Lodish. 2000. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. USA 97:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pappin, D. D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass finger printing. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 126.Patton, W. F. 2000. A thousand points of light: the application of fluorescence detection technologies to two-dimensional gel electrophoresis and proteomics. Electrophoresis 21:1123-1144. [DOI] [PubMed] [Google Scholar]
- 127.Pawson, T., and P. Nash. 2000. Protein-protein interactions define specificity in signal transduction. Genes Dev 14:1027-1047. [PubMed] [Google Scholar]
- 128.Pennisi, E. 2001. New genomes shed light on complex cells. Science 292:1280-1281. [DOI] [PubMed] [Google Scholar]
- 129.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 130.Qin, J., and B. T. Chait. 1997. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal. Chem. 69:4002-4009. [DOI] [PubMed] [Google Scholar]
- 131.Qin, J., D. Fenyo, Y. Zhao, W. W. Hall, D. M. Chao, C. J. Wilson, R. A. Young, and B. T. Chait. 1997. A strategy for rapid, high-confidence protein identification. Anal. Chem. 69:3995-4001. [DOI] [PubMed] [Google Scholar]
- 132.Quadroni, M., and P. James. 1999. Proteomics and automation. Electrophoresis 20:664-677. [DOI] [PubMed] [Google Scholar]
- 133.Rain, J. C., L. Selig, H. De Reuse, V. Battaglia, C. Reverdy, S. Simon, G. Lenzen, F. Petel, J. Wojcik, V. Schachter, Y. Chemama, A. Labigne, and P. Legrain. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211-215. [DOI] [PubMed] [Google Scholar]
- 134.Rappsilber, J., S. Siniossoglou, E. C. Hurt, and M. Mann. 2000. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 72:267-275. [DOI] [PubMed] [Google Scholar]
- 135.Roepstorff, P., and J. Fohlman. 1984. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11:601.. [DOI] [PubMed] [Google Scholar]
- 136.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 137.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rubin, G. M. M. D. Yandell, J. R. Wortman, et al. 2000. Comparative genomics of the eukaryotes. Science 287:2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sanchez, J. C., and D. F. Hochstrasser. 1999. High-resolution, IPG-based, mini two-dimensional gel electrophoresis. Methods Mol. Biol. 112:227-233. [DOI] [PubMed] [Google Scholar]
- 140.Scheele, G. A. 1975. Two-dimensional gel analysis of soluble proteins. Characterization of guinea pig exocrine pancreatic proteins. J. Biol. Chem. 250:5375-5385. [PubMed] [Google Scholar]
- 141.Scheler, C., X. P. Li, J. Salnikow, M. J. Dunn, and P. R. Jungblut. 1999. Comparison of two-dimensional electrophoresis patterns of heat shock protein Hsp27 species in normal and cardiomyopathic hearts. Electrophoresis 20:3623-3628. [DOI] [PubMed] [Google Scholar]
- 142.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 143.Shalon, D., S. J. Smith, and P. O. Brown. 1996. A DNA microarray system for analyzing complex DNA samples using two- color fluorescent probe hybridization. Genome Res. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 144.Shaw, A. C., M. Rossel Larsen, P. Roepstorff, A. Holm, G. Christiansen, and S. Birkelund. 1999. Mapping and identification of HeLa cell proteins separated by immobilized pH-gradient two-dimensional gel electrophoresis and construction of a two-dimensional polyacrylamide gel electrophoresis database. Electrophoresis 20:977-983. [DOI] [PubMed] [Google Scholar]
- 145.Shevchenko, A., I. Chernushevich, W. Ens, K. G. Standing, B. Thomson, M. Wilm, and M. Mann. 1997. Rapid ‘de novo' peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 11:1015-1024. [DOI] [PubMed] [Google Scholar]
- 146.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shevchenko, A., A. Loboda, W. Ens, and K. G. Standing. 2000. MALDI quadrupole time-of-flight mass spectrometry: a powerful tool for proteomic research. Anal. Chem. 72:2132-2141. [DOI] [PubMed] [Google Scholar]
- 148.Shevchenko, A., M. Wilm, and M. Mann. 1997. Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J. Protein Chem. 16:481-490. [DOI] [PubMed] [Google Scholar]
- 149.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 150.Smith, R. D., L. Pasa-Tolic, M. S. Lipton, P. K. Jensen, G. A. Anderson, Y. Shen, T. P. Conrads, H. R. Udseth, R. Harkewicz, M. E. Belov, C. Masselon, and T. D. Veenstra. 2001. Rapid quantitative measurements of proteomes by Fourier transform ion cyclotron resonance mass spectrometry. Electrophoresis 22:1652-1668. [DOI] [PubMed] [Google Scholar]
- 151.Smolka, M. B., H. Zhou, S. Purkayastha, and R. Aebersold. 2001. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal. Biochem. 297:25-31. [DOI] [PubMed] [Google Scholar]
- 152.Spengler, B., D. Kirsch, R. Kaufmann, and E. Jaeger. 1992. Peptide sequencing by matrix-assisted laser-desorption mass spectrometry. Rapid Commun. Mass Spectrom. 6:105-108. [DOI] [PubMed] [Google Scholar]
- 153.Sun, H., and N. K. Tonks. 1994. The coordinated action of protein tyrosine phosphatases and kinases in cell signaling. Trends Biochem. Sci. 19:480-485. [DOI] [PubMed] [Google Scholar]
- 154.Tabata, S. 2000. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408:823-826. [DOI] [PubMed] [Google Scholar]
- 155.Tong, W., A. Link, J. K. Eng, and J. R. Yates III. 1999. Identification of proteins in complexes by solid-phase microextraction/multistep elution/capillary electrophoresis/tandem mass spectrometry. Anal. Chem. 71:2270-2278. [DOI] [PubMed] [Google Scholar]
- 156.Traini, M., A. A. Gooley, K. Ou, M. R. Wilkins, L. Tonella, J. C. Sanchez, D. F. Hochstrasser, and K. L. Williams. 1998. Towards an automated approach for protein identification in proteome projects. Electrophoresis 19:1941-1949. [DOI] [PubMed] [Google Scholar]
- 157.Uetz, P. L. Giot, G. Cagney, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 158.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071-2077. [DOI] [PubMed] [Google Scholar]
- 159.Vandekerckhove, J., G. Bauw, M. Puype, J. Van Damme, and M. Van Montagu. 1985. Protein-blotting on Polybrene-coated glass-fiber sheets. A basis for acid hydrolysis and gas-phase sequencing of picomole quantities of protein previously separated on sodium dodecyl sulfate/polyacrylamide gel. Eur. J. Biochem. 152:9-19. [DOI] [PubMed] [Google Scholar]
- 160.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 161.Venter, J. C., M. D. Adams, E. W. Myers, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 162.Verentchikov, A. N., W. Ens, and K. G. Standing. 1994. Reflecting time-of-flight mass spectrometer with an electrospray ion source and orthogonal extraction. Anal. Chem. 66:126-133. [DOI] [PubMed] [Google Scholar]
- 163.Wasinger, V. C., S. J. Cordwell, A. Cerpa-Poljak, J. X. Yan, A. A. Gooley, M. R. Wilkins, M. W. Duncan, R. Harris, K. L. Williams, and I. Humphery-Smith. 1995. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 16:1090-1094. [DOI] [PubMed] [Google Scholar]
- 164.Wettenhall, R. E., R. H. Aebersold, and L. E. Hood. 1991. Solid-phase sequencing of 32P-labeled phosphopeptides at picomole and subpicomole levels. Methods Enzymol. 201:186-199. [DOI] [PubMed] [Google Scholar]
- 165.Wilkins, M. R., E. Gasteiger, A. Bairoch, J. C. Sanchez, K. L. Williams, R. D. Appel, and D. F. Hochstrasser. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112:531-552. [DOI] [PubMed] [Google Scholar]
- 166.Wilkins, M. R., E. Gasteiger, A. A. Gooley, B. R. Herbert, M. P. Molloy, P. A. Binz, K. Ou, J. C. Sanchez, A. Bairoch, K. L. Williams, and D. F. Hochstrasser. 1999. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 289:645-657. [DOI] [PubMed] [Google Scholar]
- 167.Wilkins, M. R., J. C. Sanchez, A. A. Gooley, R. D. Appel, I. Humphery-Smith, D. F. Hochstrasser, and K. L. Williams. 1995. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 13:19-50. [DOI] [PubMed] [Google Scholar]
- 168.Wilkins, M. R., J. C. Sanchez, K. L. Williams, and D. F. Hochstrasser. 1996. Current challenges and future applications for protein maps and post-translational vector maps in proteome projects. Electrophoresis 17:830-838. [DOI] [PubMed] [Google Scholar]
- 169.Wilm, M., and M. Mann. 1996. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68:1-8. [DOI] [PubMed] [Google Scholar]
- 170.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 171.Yan, J. X., J. C. Sanchez, L. Tonella, K. L. Williams, and D. F. Hochstrasser. 1999. Studies of quantitative analysis of protein expression in Saccharomyces cerevisiae. Electrophoresis 20:738-742. [DOI] [PubMed] [Google Scholar]
- 172.Yates, J. R., III. 1998. Mass spectrometry and the age of the proteome. J. Mass Spectrom. 33:1-19. [DOI] [PubMed] [Google Scholar]
- 173.Yates, J. R., III. 1996. Protein structure analysis by mass spectrometry. Methods Enzymol. 271:351-377. [DOI] [PubMed] [Google Scholar]
- 174.Yates, J. R., III, A. L. McCormack, D. Schieltz, E. Carmack, and A. Link. 1997. Direct analysis of protein mixtures by tandem mass spectrometry. J. Protein Chem. 16:495-497. [DOI] [PubMed] [Google Scholar]
- 175.Yates, J. R., III, S. Speicher, P. R. Griffin, and T. Hunkapiller. 1993. Peptide mass maps: a highly informative approach to protein identification. Anal. Biochem. 214:397-408. [DOI] [PubMed] [Google Scholar]
- 176.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
- 177.Zhang, W., A. J. Czernik, T. Yungwirth, R. Aebersold, and B. T. Chait. 1994. Matrix-assisted laser desorption mass spectrometric peptide mapping of proteins separated by two-dimensional gel electrophoresis: determination of phosphorylation in synapsin I. Protein Sci. 3:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang, X., C. J. Herring, P. R. Romano, J. Szczepanowska, H. Brzeska, A. G. Hinnebusch, and J. Qin. 1998. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal. Chem. 70:2050-2059. [DOI] [PubMed] [Google Scholar]
- 178a.Zhas, S., et al. 2001. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 11:1736-1745. [DOI] [PMC free article] [PubMed]
- 179.Zhou, H., J. D. Watts, and R. Aebersold. 2001. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 19:375-378. [DOI] [PubMed] [Google Scholar]
- 180.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]
- 181.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283-289. [DOI] [PubMed] [Google Scholar]