Abstract
We have analyzed the placement of sites of integration of avian sarcoma-leukosis virus (ASLV) and human immunodeficiency virus (HIV) DNA in the draft chicken genome sequence, with the goals of assessing species-specific effects on integration and allowing comparison to the distribution of chicken endogenous retroviruses (ERVs). We infected chicken embryo fibroblasts (CEF) with ASLV or HIV and sequenced 863 junctions between host and viral DNA. The relationship with cellular gene activity was analyzed by transcriptional profiling of uninfected or ASLV-infected CEF cells. ASLV weakly favored integration in active transcription units (TUs), and HIV strongly favored active TUs, trends seen previously for integration in human cells. The ERVs, in contrast, accumulated mostly outside TUs, including ERVs related to ASLV. The minority of ERVs present within TUs were mainly in the antisense orientation; consequently, the viral splicing and polyadenylation signals would not disrupt cellular mRNA synthesis. In contrast, de novo ASLV integration sites within TUs showed no orientation bias. Comparing the distribution of de novo ASLV integration sites to ERVs indicated that purifying selection against gene disruption, and not initial integration targeting, probably determined the ERV distribution. Further analysis indicated that ERVs in humans, mice, and rats showed similar distributions, suggesting purifying selection dictated their distributions as well.
Repeated sequences—mostly remnants of genomic parasites—comprise a much larger fraction of vertebrate genomes than do the protein coding exons. In humans, for example, repeated sequences comprise at least 45% of the genome, with exons contributing only about 1.5% (16, 29). Most of the vertebrate repeated sequences are inactive fossils that replicated via reverse transcription (7, 9, 27). We have investigated the forces dictating the genomic placement of one such group, the endogenous retroviruses (ERVs), using chickens as a model.
The distribution of the human ERVs (HERVs) has been studied previously in some detail (1-3, 9, 13, 18). A study of HERV distribution by Arian Smit, carried out when the human genome sequence was about 10% completed, indicated that HERVs accumulated mostly outside of genes (27). For the few HERV sequences within genes, most were in the antisense orientation, which would be expected to minimize disruption of cellular mRNA synthesis because the viral poly(A) addition and splicing signals are in antisense orientation. These findings were consistent with the idea that purifying selection eliminated HERVs that disrupted the function of cellular genes (27). However, another explanation for the distribution, noted by Smit, was that the initial targeting of HERV DNA integration might have accounted for the observed distribution. Initial integration of HERV DNA has not been studied; indeed, no replication-competent HERVs have been identified, making characterization of de novo integration targeting impossible. In chickens, however, there is a large class of endogenous retroviruses, members of the ERVK group, that are related to the replication-competent avian sarcoma-leukosis viruses (ASLV). For this virus group, it is possible to compare the placement of de novo sites of integration generated experimentally to the preexisting distribution of ERVK sequences, allowing the forces dictating the accumulation of ERVs to be specified more precisely.
The availability of draft vertebrate genome sequences has allowed retroviral integration targeting to be assessed in detail by cloning and sequencing large numbers of integration sites and then analyzing their distribution relative to other features mapped on the genome sequences. Surprisingly, studies of integration targeting in human cells have shown that different retroviruses favor integration near quite different chromosomal features. Studies of several thousand sites of human immunodeficiency virus (HIV) integration in human cells indicated that transcription units (TUs) are favored for integration, and comparison to transcriptional profiles from target cells indicates that active TUs are particularly favored (19, 26, 31). Murine leukemia virus (MLV), in contrast, showed an integration bias in favor of CpG islands and transcription start sites (31) and a weaker preference for transcription units. ASLV showed a nearly random distribution of integration sites in the human genome, with TUs favored only weakly (19, 22). A variety of factors have been proposed to influence integration targeting in chromosomes (7-10, 17, 23). The finding that integration targeting by the three retroviruses differed suggests that each type of integration complex may bind specific chromosomal proteins, a model consistent with studies of related retroelements in the yeast Saccharomyces cerevisiae (4, 8, 25, 32, 33).
Here we have analyzed de novo integration targeting for ASLV and HIV in the chicken genome. Six hundred fifty-eight sites of ASLV integration were sequenced and compared to 205 sites of HIV integration. Activity of the chicken TUs was monitored by transcriptional profiling. One reason for analyzing integration in chickens was to investigate possible species-specific factors influencing integration targeting. In particular, the previous finding that integration by ASLV favored TUs only weakly in the human genome could potentially have been explained by a lack of a chicken-specific integration targeting factor. Similarly for HIV, analyzing integration in a distantly related genome allowed us to ask whether species-specific factors were important for targeting to active TUs. We found, however, that the integration target preferences in the chicken genome were generally similar to the patterns in humans for both viruses, indicating that any cellular factors important for targeting are apparently conserved between chickens and humans.
To investigate factors that determine the distribution of ERVs in the chicken genome, we compared the distributions of ERVs to the distribution of de novo ASLV integration sites. Chicken ERVs are enriched in gene-sparse chromosomes and in intergenic regions. For the minority of ERVs present within genes, most were in the antisense orientation; consequently, ERV sequences for RNA splicing or polyadenylation within introns do not interfere with host gene function. In contrast, for sites made by de novo infection with ASLV, integration was favored in TUs, and sites within genes showed no orientation bias. This indicates that the present-day distribution of chicken ERVs is likely dominated by purifying selection after integration. Further analysis of the distribution of ERVs in mice and rats and an updated analysis for humans revealed that ERVs in these organisms showed the same biases as in chickens, suggesting that purifying selection dominates the placement of ERVs in many vertebrates.
MATERIALS AND METHODS
Cell lines.
DF-1 (ATCC CRL-12203) and 293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA), supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. Chicken embryo fibroblasts (CEF) were obtained from a line 0 chicken (an inbred line from the USDA Regional Poultry Laboratory, East Lansing, Michigan). The CEF cell culture was carried out in M199 medium with Earle's salts (1×), 1% chicken serum (heat inactivated), 5% fetal bovine serum (heat inactivated), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). CEF cells used in these studies were thawed once and split 1:4 prior to infections.
Preparation of the HIV-1 and ASLV vector particles.
HIV type 1 (HIV-1)/vesicular stomatitis virus G (VSV-G) vector particles (pseudotyped with VSV-G) were generated in 293T cells by the cotransfection of three plasmids: p156RRLsinPPTCMVGFPWPRE (11) (carrying the HIV vector segment), pCMVdeltaR9 (21) (packaging construct), and pMD.G (21) (carrying the VSV-G gene). Forty-eight hours after transfection, supernatants containing the viral particles were harvested and centrifuged for 5 min at 350 × g at 4°C. Supernatants were then filtered through a 0.45-μm filter and concentrated by centrifugation at 23,000 × g for 2 h at 4°C. The viral pellets were resuspended in ∼1/100 volume of fresh medium. The concentration of HIV vector particles in stocks was determined by p24 enzyme-linked immunosorbent assay.
ASLV particles were generated by transfecting DF-1 cells with the plasmid pRCASBP(A)GFP (from Steve Hughes, National Cancer Institute, Frederick, Maryland; see http://home.ncifcrf.gov/hivdrp/RCAS/plasmid.html). Forty-eight hours after transfection, supernatants containing the viral particles were harvested, centrifuged, filtered, and concentrated as for the HIV-1 viral particles.
Virus infections.
Prior to infection, HIV-1/VSV-G preparations were digested with DNase I (0.2 U/μl) for 1 h at 37°C. CEF cells (3 × 106) at ∼40% confluence were infected with HIV-1/VSV-G (500 ng p24) or an aliquot of ASLV-containing supernatant for 48 h at 37°C with 5% CO2. Flow cytometry analysis showed that the percentages of cells expressing green fluorescent protein were 27% and 69% for HIV-1/VSV-G and ASLV, respectively.
Cloning integration sites from chicken cells.
Integration sites were cloned by ligation-mediated PCR essentially as described previously (26). Briefly, DNA was extracted using the DNeasy tissue kit (QIAGEN, Valencia, CA) 48 h postinfection and digested with DpnI to eliminate possible plasmid vector carryover, followed by digestions with AvrII, NheI, and SpeI. Linker DNA was ligated to the digested ends, and products were cleaved with SacI to prevent amplification of an internal fragment. This was followed by DNA amplification with two rounds of PCR, cloning, and sequencing. For a summary of restriction enzymes used in analyzing integration sites, see Table S4 in the supplemental material; for sequences of oligonucleotides used in this work, see Table S5 in the supplemental material.
Microarray analysis.
RNA was harvested from CEF cells in log-phase growth. ASLV vector infections were carried out as described above except that the RNA was harvested 24 h after infection. Labeling of RNA was performed as described by Affymetrix (Santa Clara, CA). To study the effect of ASLV infection on gene activity, three chips with independent RNA samples were analyzed for uninfected cells and another three chips were analyzed for independently infected cells. Significance Analysis of Microarray software was used to analyze changes in gene activity after ASLV infection (28) via the permuted t test method. For the ASLV integration sites, analysis using mRNA, Unigene, and Ensembl calls returned 249 probe sets. For HIV, a similar analysis returned 117.
Bioinformatic analysis.
Integration site locations were identified using the BLAT feature in the Chicken Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway) against the February 2004 freeze of the chicken genome sequence. Integration site sequences were judged to be of acceptable quality if (i) the match to the genome began within 3 bp of the 5′-CA-3′ terminus of the viral DNA, (ii) the match proximal to the long terminal repeat (LTR) end showed an identity of at least 98%, and (iii) this match yielded a unique best hit using default parameters in the client-server BLAT (15) ranking (galGal2 draft of chicken genome; hg17 draft of human genome). An integration site was scored as present in a TU if it lay in DNA between the base pairs encoding the 5′ and 3′ ends of the mRNA as defined by Ensembl or mRNA annotation (http://genome.ucsc.edu/cgi-bin/hgGateway). Analysis of previously published retroviral integration sites into the human genome was updated using the hg17 draft (May 2004) of the human genome. New matched random controls were also generated.
For the expression analysis in Fig. 3, integration site sequences were accepted as within a TU if the sequence was present in the Ensembl, mRNA, or Unigene catalog. For cases where multiple probe sets queried a single TU, all probe sets were accepted in the analysis. Matched random control sites were handled similarly.
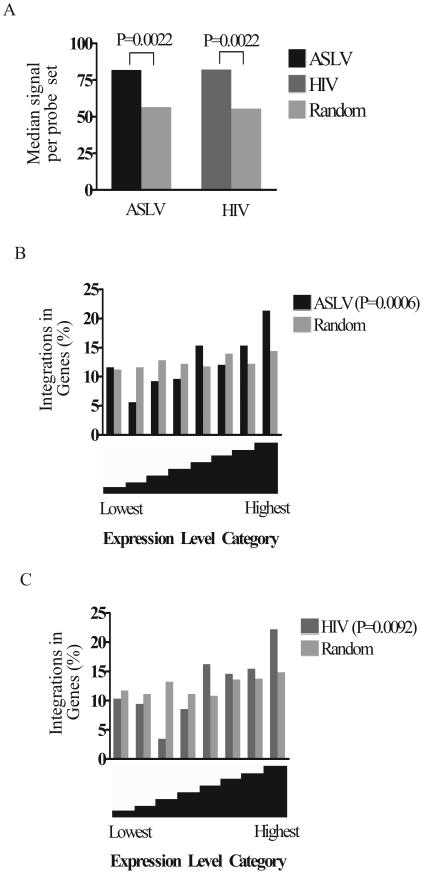
FIG. 3.
Relationship between gene activity and integration frequency. (A) ASLV and HIV show a bias towards integration into active TUs. The relative expression levels of genes in CEF cells were assayed on six Affymetrix chicken genome arrays, and the relative expression levels were averaged over the six arrays. The median expression signals for ASLV and HIV were plotted and compared to all the genes queried on the chip (P value on figure). In addition, comparison of genes targeted for integration to those in the matched random control also showed significance (P = 0.021 for ASLV and P = 0.0066 for HIV; Mann-Whitney test). In another analytical approach, the signals for genes hosting integration events were compared to the signals for genes not hosting integration events, and this similarly showed a significant difference (data not shown). (B and C) Analysis of integration frequency as a function of gene expression intensity for ASLV (B) and HIV (C). All genes assayed on the Affymetrix microarrays were divided into eight “bins” according to their relative levels of expression (x axis). The leftmost bin contains genes with the lowest expression levels, and the rightmost bin contains the highest. Genes hosting integration events were distributed into the corresponding expression bins and summed, and then the data wereplotted as the percentages of all integrations in that bin (y axis). P values were determined using the chi-square test for trend by comparison to the null hypothesis of no bias due to expression level.
For the analysis of ERV distributions relative to ASLV integration sites, the Pearson correlation was tested for a series of window sizes. The most significant P values (cited in the text) were for window sizes of 3 Mb (ERVK) and 0.1 Mb (ERVL).
An artifact was encountered in analyzing ASLV integration sites in the chicken genome. A portion of infecting retroviral DNA becomes circularized by autointegration or other reactions, and such forms could be cloned by the ligation-mediated PCR method used. These scored as integration sites clustering at chromosome 1 position 29522444 to 29532218, which is an ASLV-related ERVK. These events could be shown unambiguously to be artifacts, since apparent integration target sites matched exactly to the RCAS vector used and differed detectably from the genomic ERVK copy. These sites were removed from the analysis.
Nucleotide sequence accession numbers.
Integration site sequences in the chicken genome have been deposited in GenBank under accession numbers CZ905239 to CZ905918 (ASLV integration sites) and CZ905919 to CZ906101 (HIV integration sites).
RESULTS
Cloning of ASLV and HIV integration sites in the chicken genome.
We investigated initial retroviral integration targeting in the chicken genome by sequencing 863 sites of integration generated by acute infection of CEF with ASLV- or HIV-based vectors. CEF were chosen because they are a primary cell type and so are not subject to the chromosomal abnormalities typical of transformed lines. To permit HIV infection, the HIV-based vector particles were pseudotyped with the pantropic VSV-G envelope protein (6). Cells were grown after being infected for 48 h to allow reverse transcription and integration to take place, and then genomic DNA was isolated.
Cloning of integration sites was carried out using ligation-mediated PCR (26, 31). Genomic DNA was digested with restriction enzymes and ligated to DNA linkers. DNA containing junctions between retroviral DNA and target DNA was amplified by PCR using one primer complementary to the linker and a second complementary to the viral DNA end. A second round of PCR was performed with nested primers. PCR products were then cloned and sequenced, yielding 658 unique ASLV and 205 unique HIV integration sites. Integration sites were mapped on the February 2004 draft chicken genome sequence (Fig. 1), and local features at each integration site were quantified. Integration sites in the chicken genome were compared to integration site data sets for HIV, ASLV, and MLV in the human genome (Table 1). All analyses of the human genome were updated to the hg17 draft (May, 2004).
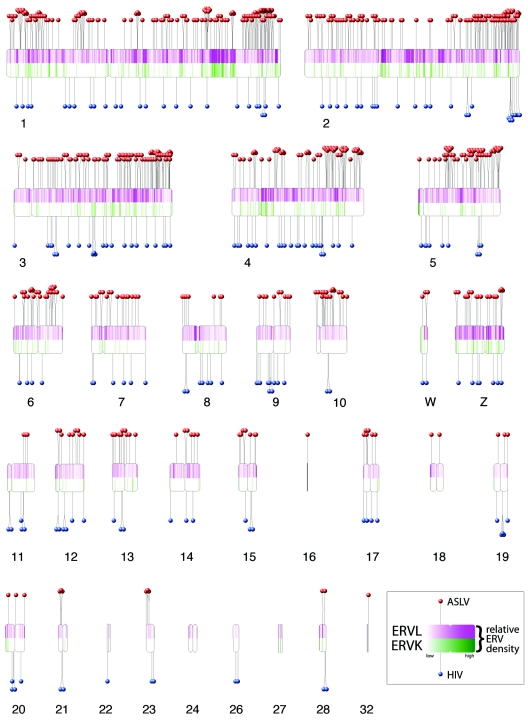
FIG. 1.
De novo integration targeting in the chicken genome and its relationship to ERVs. The chicken chromosomes are shown numbered (macrochromosomes 1 to 5, intermediate chromosomes 6 to 10, microchromosomes 11 to 32, and sex chromosomes W and Z). Note that, due to the incomplete status of the draft chicken genome sequence, some of the microchromosomes are not represented. Each de novo integration site is shown as a “lollipop” (ASLV, red; HIV, blue). Endogenous ERVL sequences are shown by the purple shading in the upper half of each chromosome, and ERVK is shown below by green. The most intense purple shading represents 85 ERVL integrations per 250 kbp; the most intense green shading represents 12 ERVK integrations per 250 kbp. Centromere locations are denoted by chromosomal indentations. The centromere positions for chromosomes 6, 9, 13 to 16, 18 to 22, 24, 27, and 32 are currently unavailable and have been arbitrarily placed at the chromosomal midpoints. The software used to draw the ideogram was obtained and adapted from http://www.uni-essen.de/∼bt0756/cc/.
TABLE 1.
Comparison of the distribution of integration sites in TUs
| Virus or vector | Cell type | No. of integration sites | Ratio in TUs (experimental/matched random control)a
|
Source or reference | |
|---|---|---|---|---|---|
| mRNA | Ensembl | ||||
| ASLV vector | Chicken CEF | 658 | 1.24** | 1.06 | This report |
| HIV vector | Chicken CEF | 205 | 1.72*** | 1.56*** | This report |
| HIV vector | Human PBMCb | 569 | 1.79*** | 2.07*** | 19 |
| HIV vector | Human IMR-90 | 504 | 1.56*** | 1.71*** | 19 |
| HIV vector | Human SupT1 | 583 | 1.66*** | 1.96*** | 26 |
| HIV | Human SupT1 | 50 | 2.00*** | 2.18*** | 26 |
| HIV | Human H9 | 174 | 1.45*** | 1.67*** | 31 |
| HIV vector | Human HeLa | 321 | 1.76*** | 2.10*** | 31 |
| ASLV vector | Human 293-Tva | 712 | 1.18*** | 1.27*** | 14 |
| ASLV vector | Human HeLa | 106 | 1.28* | 1.31* | 22 |
| MLV vector | Human HeLa | 959 | 1.22*** | 1.23*** | 31 |
P values are for comparison of each integration site population to a matched random control. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001 (chi-square test).
PBMC, peripheral blood mononuclear cells.
Statistical analysis using matched random control sites.
For statistical analysis, the set of de novo integration sites was compared to a set of about 26,000 randomly generated sites in the chicken genome. The statistical analysis then proceeded by assessing the significance of differences between the random sites and the experimentally determined integration sites.
An issue in this analysis was the possible bias introduced into experimentally determined integration sites by the reliance on restriction enzyme cleavage during cloning. Analysis indicates that integration sites closer to appropriate restriction sites in genomic DNA are more readily cloned by the ligation-mediated PCR protocol used but that the fraction of sites sampled is reasonably representative of the genome as a whole (C. Berry, J. R. Ecker, and F. D. Bushman, unpublished). We nevertheless modified our statistical procedure to account for restriction site bias by generating a matched set of random control sites. For each experimental integration site, 40 random sites were generated in silico that were constrained to be the same distance away from a restriction site in the genome that could have been used for cloning. For example, if an experimental site was 300 bp from the restriction site used for linker ligation, then 40 sites were generated that each were exactly 300 bp from such a restriction site but randomly scattered around the genome. A few random sites landed in sequence gaps and were removed from subsequent analysis. Statistical analysis was then carried out by comparing the pooled experimental sites to the pooled matched random controls.
The importance of this procedure was tested by comparing the matched random control set to an unmatched collection of random chromosomal sites. This revealed that the matching procedure resulted in slight but significant differences in the detection of several features (see Table S1 in the supplemental material).
Relationship of de novo integration sites to TUs in the chicken genome.
ASLV and HIV integration sites were mapped onto the chicken genome and the proximity to TUs quantified (Table 1). A complication arises due to the relatively early stage and incomplete analysis available for TUs in the chicken genome. In an effort to obtain a robust overview, we repeated the analysis using both the Ensembl and “mRNA” catalogs, which are the two most populated collections (Table 1). ASLV showed slightly increased integration in each collection of TUs: 1.24- and 1.06-fold, respectively, relative to the matched random control. The results achieved statistical significance for the mRNA data but not for Ensembl (Table 1). The analysis was then extended by pooling data for mRNA and Ensembl calls. This revealed that 62.2% of ASLV sites were within TUs compared to only 55.2% of matched random sites, a highly significant difference (P = 0.0055). For HIV, the bias was more pronounced (1.72- and 1.56-fold) and the difference highly significant (P < 0.0001 for both mRNA and Ensembl calls analyzed individually). Analysis of the pooled data showed that 88.3% of HIV integration sites were called as in TUs. We conclude that ASLV shows a weak bias and HIV shows a strong bias in favor of integration in TUs in chicken cells.
The relationship of integration sites to TU boundaries was then assessed (Fig. 2). Integration in transcription start regions is of particular interest because MLV integration in human cells strongly favored transcription start regions (31). One motive for studying ASLV integration in chickens was to determine whether ASLV adopted such a target preference in its natural host. However, no significant bias was seen for ASLV integration in extragenic regions extending 5 kb from TU 5′ and 3′ boundaries, though a slight surplus of integration events in these sequences was seen (Fig. 2A). Similarly for HIV, no significant biases were seen in favor of integration in 5′ and 3′ flanking regions (Fig. 2B).
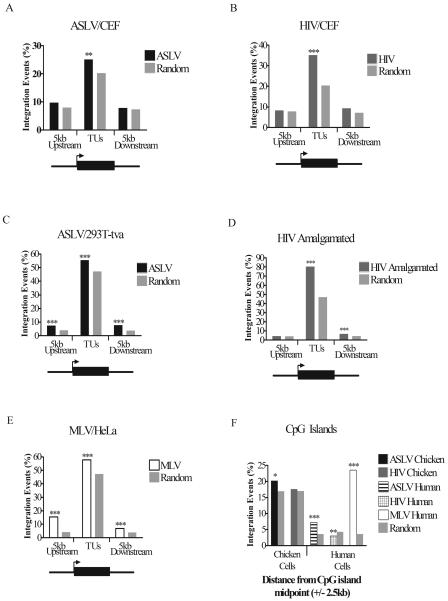
FIG. 2.
Frequency of retroviral integration in and around TUs and CpG islands: influence of cell type and retrovirus studied. The percentages of total integrations into TUs (using the “mRNA” gene catalog) and regions of DNA 5 kb upstream of the transcription start site and 5 kb downstream of the transcription end sites were plotted separately for each virus. The viruses and cell type studied are as marked above each graph. (A) ASLV in CEF cells; (B) HIV in CEF cells; (C) ASLV in human 293T-Tva cells; (D) HIV data from several human cell types (see Table 1); (E) MLV in human HeLa cells. The diagram below each graph shows the regions in and around TUs that were scored for integration events. The arrow represents the transcription start site, and the black box represents the TU. (F) The percentage of total integrations for each virus within 2.5 kb upstream and 2.5 kb downstream of CpG island midpoints compared to matched random controls. Comparison of the data on matched random control sites for human and chicken shows that a much larger fraction of the chicken genome is annotated as CpG island, perhaps an artifact of the higher G/C content of the chicken genome. If CpG islands are in fact “overcalled” in the chicken genome, then this will reduce our ability to detect biases in integration in these sites due to increased noise. P values were determined using the chi-square test and comparison to matched random controls. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
Data for ASLV, HIV, and MLV integration in human cells were then compared. The analysis was updated to the hg17 human genome draft, including generation of matched random control sites on that draft. ASLV and HIV favored integration in TUs (Fig. 2C and D), though the extent of the effect was much greater for HIV. This analysis revealed a slight but significant favoring of 5′ and 3′ flanking regions for ASLV in human cells (Fig. 2C). A similar trend could be seen in chicken cells, but there the trend was weaker and did not achieve significance. For HIV integration data, a modest but significant increase in integration in regions 5 kb downstream of TU 3′ ends could be seen (Fig. 2D). Possibly continued transcription beyond the poly(A) addition signal leads to transcription of these regions, causing favored integration.
Integration by MLV (Fig. 2E) showed a much stronger bias for upstream regions than either ASLV or HIV, with about fivefold more integration sites present than expected by chance, paralleling the original report (31). MLV also favored integration in TUs and downstream regions, though the extent of the bias was more modest than for the 5′ bias (Fig. 2E).
The frequency of integration near CpG islands was also assessed. CpG islands are often associated with promoter regions and are strongly favored for MLV integration in human cells (31) but disfavored for HIV integration (19) (Fig. 2F). In chicken cells, ASLV showed a slight but significant bias in favor of integration near these features, as seen previously in human cells (19). HIV did not show any significant bias for or against integration in CpG islands in chicken cells (19).
In summary, ASLV integration and HIV integration in chicken and human cells were generally similar with respect to TUs, flanking sites, and CpG islands. ASLV integration in flanking sites was slightly favored in both chickens and humans, but the data only achieved significance in humans. At present we attribute the difference in significance to the early stage of gene annotation of the chicken genome and not any major biological difference. ASLV did not adopt an MLV-like integration pattern in its natural host.
Effect of gene activity on de novo integration in the chicken genome.
We next analyzed the relationship between gene activity and retroviral integration in CEF cells. Steady-state levels of mRNA were quantified using Affymetrix chicken genome microarrays, which query the activity of about 33,000 transcripts in the chicken genome. In the following, we used the steady-state mRNA levels as an approximation of gene activity, though we note that differential mRNA processing, turnover, etc., could also affect these measurements. Uninfected cells were compared to cells 24 h after infection with ASLV.
Three arrays were analyzed with labeled RNA from uninfected cells and three with RNA from infected cells. Changes in gene activity due to infection were analyzed using the Significance Analysis of Microarray package (28). Infection was found to have very little influence on cellular gene activity: only 20 genes were called as significantly affected even when a relatively high false-discovery rate was accepted (25%). The most significantly affected gene was GFP, which had a 15.7-fold higher expression value after infection. The GFP gene was introduced into cells by infection with the ASLV vector used and so provides a positive control. See Table S2 in the supplemental material for a summary of further genes affected by infection. We conclude that there was little or no effect of infection with the ASLV vector on CEF cell transcription under the conditions studied. Results for all six arrays were thus pooled to improve the statistical resolution in subsequent analysis.
The expression levels of TUs hosting integration events were then compared to that of the remainder of genes queried on the array (Fig. 3A). TUs hosting integration events for both viruses were more active than the average of TUs on the chip (P = 0.0022 for each; Mann-Whitney test). To examine the relationship between transcriptional activity and integration frequency in more detail, the chicken TUs analyzed by the Affymetrix microarray were divided into classes by expression level and the frequency of genes hosting integration events in each class was determined (Fig. 3B and C). One reason for carrying out such an analysis was to examine whether highly transcribed genes were disfavored for integration, as has been suggested in previous studies of ASLV integration in two model genes in quail cells (17, 30). For both ASLV and HIV, comparison to the matched random control indicated that TUs in the lower expression categories were less favored for integration and TUs in the higher expression classes were more favored. There was no reduction in integration frequency in the highest expression class of genes for either virus; in fact, integration was favored. Evidently, under the conditions of this study, even very-high-level transcription favored integration.
Biased integration at the chromosomal level.
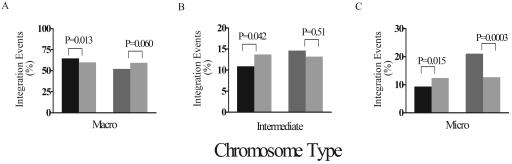
An unexpected trend was observed in the distribution of integration sites at the chromosomal level. The chicken genome is composed of macrochromosomes, intermediate chromosomes, and microchromsomes (14) (Fig. 1). The larger macrochromosomes are gene sparse, A/T rich, repeat dense, and CpG island sparse and have genes with longer introns. The smaller microchromosomes are opposite in these tendencies, and the intermediate chromosomes are intermediate. Thus the tendency for ASLV and HIV to favor integration in TUs predicts that the microchromosomes would be favored targets. This was seen for HIV, as expected, but not for ASLV (Fig. 4). Paradoxically, ASLV favored integration in the macrochromosomes and disfavored integration in the microchromosomes. Inspection of Fig. 1 indicates that some of the excess integration sites in macrochromosomes appear to have accumulated in the telomere-proximal regions of the longer chromosome arms. The reason for this bias is unknown.
FIG. 4.
Relative integration frequency in macrochromosomes, intermediate chromosomes, and microchromosomes. Chicken chromosomes were grouped into gene-sparse macrochromosomes (chromosomes 1 to 5; (A), intermediate chromosomes (chromosomes 6 to 10; (B), and gene-dense microchromosomes (chromosomes 11 to 32; (C). Integrations into each chromosomal group were summed and expressed as the percentage of total integrations for each virus. The distribution of ASLV (black bars) integrations shows an association with macrochromosomes, whereas HIV (dark grey bars) integration shows an association with microchromosomes. P values were determined by chi-square test (comparison to the matched random control [light grey bars]).
Relationship of de novo integration sites to chicken ERVs.
The chicken genome contains ERVs that are members of the ASLV group (5, 9, 14), allowing the relationship of de novo ASLV integration sites to be compared to the endogenous ASLVs. We note that, here and below, we make no effort to distinguish between integration by retroviral infection and integration via retrotransposition; indeed, it may be that some elements replicate by both means (7, 9). The two are considered together in the analysis below.
Chicken ERVs consist of two main groups, the ERVKs and ERVLs, and a smaller miscellaneous group designated “ERV.” An analysis of the positions of each ERV group relative to TUs showed that each has accumulated primarily outside TUs (Table 2). Only 11.2% of ERVKs and 19% of ERVLs were present in TUs, while 37.1% of de novo ASLV integration events were within TUs (P < 0.0001 versus ERVK or ERVL; chi-square test). The distributions of de novo integration events and ERVs are compared in Fig. 1. The genomic distributions of de novo ASLV integration sites could also be compared directly to the distributions of ERVKs and ERVLs. This showed a detectable anticorrelation for ERVKs and a strong anticorrelation for ERVLs (P = 0.0251 and P = 0.0004, respectively; Pearson correlation).
TABLE 2.
Percentages of endogenous retroviruses integrated within TUs
| Cell source | % in TUs of ERV groupa:
|
Ensembl TUs in genome (%) | |||
|---|---|---|---|---|---|
| ERVK | ERVL | ERV | ERV1 | ||
| Chicken | 11.2*** | 19.0*** | 8.9*** | NAb | 32.0 |
| Human | 24.5*** | 19.2*** | 25.9*** | 21.6*** | 35.0 |
| Mouse | 20.4*** | 25.3*** | 20.0 | 20.0*** | 27.7 |
| Rat | 14.4** | 17.0*** | 12.1 | 12.4*** | 20.4 |
**, 0.001 < P < 0.01; ***, P < 0.001 (chi-square test).
NA, not applicable.
We next analyzed the distribution of the ERVs most closely related to ASLV, since these ERVs were most likely to have been initially integrated in an ASLV-like fashion. ASLV-related proviruses in the chicken genome were identified using data summarized in references 5 and 9 (for a quantitative analysis of these relationships see reference 5) and by manual sequence alignment (see Table S3 in the supplemental material). Of these, only 10.9% were in TUs, thus displaying a significant bias against TUs and an opposite pattern of accumulation compared to de novo ASLV integration (P < 0.0001; chi-square test).
Of the integration events in TUs, most of which are within introns, ERVKs and ERVLs showed a significant bias in the orientation of the proviral genome relative to the host TUs. ERVK and ERVL sequences were much more commonly in the antisense orientation (73% and 56.9%, respectively; Table 3; P < 0.0001, binomial test). In the antisense orientation the ERV sequences are expected to have minimal effects on the host cell TUs, because the proviral signals for RNA splicing and poly(A) addition are in reverse orientation and so nonfunctional. For de novo integration, the orientation of ASLV and HIV sequences within TUs did not show a significant bias (50.0% and 48.8%, respectively, P > 0.05). For the subset of ERVs most closely related to ASLVs, the antisense bias was highly significant (78.3% in the antisense orientation; P < 0.0001, binomial test). These findings support the idea that purifying selection against gene disruption dominates the distribution of the chicken ERVs. Further analysis indicates that ERVs and related LTR retrotransposons in many vertebrates show similar biases in their distributions (discussed below) (2, 3, 18, 27).
TABLE 3.
Orientation bias of endogenous retroviruses within TUs
| Cell source | % Antisense directiona for ERV group:
|
|||
|---|---|---|---|---|
| ERVK | ERVL | ERV | ERV1 | |
| Chicken | 73.0*** | 56.9*** | 75.7*** | NAb |
| Human | 65.8*** | 69.9*** | 60.3** | 71.8*** |
| Mouse | 66.1*** | 65.8*** | 48.5 | 68.8*** |
| Rat | 64.6*** | 66.8*** | 56.3 | 68.7*** |
**, 0.001 < P < 0.01 (binomial test, two tail); ***, P < 0.0001 (binomial test, two tail).
NA, not applicable.
DISCUSSION
We have investigated the distribution of retroviral sequences newly introduced into the chicken genome by infection and asked how their distribution compares to that of chicken ERVs. Eight hundred sixty-three sites of de novo infection with HIV or ASLV integration were determined and mapped on the draft chicken genome sequence. Transcriptional activity was monitored in the CEF target cells using transcriptional profiling. Analysis of these data is presented below. Future analysis of these data sets should also make possible additional studies of the relationship between integration and newly annotated features as they are posted on the chicken genome.
Parallels in integration targeting among data sets.
A key question in interpreting the data presented here is that of how widely conclusions can be generalized. Do different cell types show major differences in integration targeting when infected by the same retrovirus? How different is integration targeting among related retroviruses? The available data are sparse, but a few generalizations can be tentatively proposed. For HIV integration, comparison among human cell types shows only slight differences (19, 26, 31). Tissue-specific transcription influences targeting detectably, but only modestly (19). For ASLV, similar patterns of integration targeting have been reported for two human cell types (19, 22). Given the similarity among the targeting patterns in two human cell types and CEF cells, we infer that the integration pattern in chicken germ cells, the cell type relevant to ERV formation, would probably be similar. Another question is that of whether related but nonidentical ASLVs would be expected to have similar patterns of integration. Again the data are sparse, but for lentiviruses it has been shown that simian immunodeficiency virus in rhesus monkey cells has a similar targeting preference to that of HIV in human cells (12), suggesting that integration targeting may be similar across the lentiviral group. We thus propose that ASLV-related ERVs likely had the same integration targeting preferences as the ASLV vector studied here, though more data would be helpful to strengthen this point.
Integration targeting in cells of different species.
Do species-specific factors influence integration targeting? In human cells, ASLV showed only a weak preference for integration in TUs: statistically significant biases could be detected, but they were quantitatively modest (Fig. 2C). One possible reason is that human cells lacked a species-specific factor for targeting ASLV integration. According to this idea, a chicken-specific factor might have tethered ASLV integration complexes to chromosomal DNA, resulting in highly selective integration, perhaps resembling MLV in human cells. However, no such bias was detected in de novo integration by ASLV in CEF cells: ASLV integration in chickens resembled ASLV integration in humans (compare Fig. 2A and C). While it is not ruled out that ASLV tethering factors might exist in chicken cell types other than the CEFs studied, our findings do not provide evidence for species-specific factors that more precisely target ASLV integration.
Similarly for HIV, we sought to test whether human-specific factors influenced the distribution of HIV integration sites. However, we found that HIV strongly favored integration in active TUs in chicken cells as well as human cells. These findings indicated that potential cellular factors important for HIV integration targeting are conserved between chickens and humans. This may be technically helpful in studies of HIV integration; for any candidate targeting factors identified in human cells, it may be useful to confirm that the chicken counterpart is also functional.
Integration and transcriptional activity in chicken cells.
The relationship between integration and transcriptional activity was assessed using newly available Affymetrix microarrays, which contain 38,392 probe sets querying the activity of about 33,000 chicken TUs. For both ASLV and HIV, the TUs hosting integration events showed a significantly higher median expression level than those targeted in the matched random control, indicating favored integration in active genes (19, 26, 31). The trend in chicken cells may differ slightly from that in human cells (19), since the association with gene activity in chicken cells was somewhat stronger for ASLV and weaker for HIV compared to the same trend in human cells. However, given the much earlier state of assembly and analysis of the chicken genome sequence it is uncertain whether these slight differences are meaningful.
Published studies analyzing ASLV integration frequency in two model genes in quail cells have suggested that induction of transcription reduced integration in these model genes (17, 30). In our study, ASLV integration frequency was significantly increased in the most highly expressed class of TUs (P = 0.0022 for the comparison of ASLV integration sites in Fig. 3B, rightmost bin, to the matched random control; chi-square test). It should be noted, however, that the available data are somewhat sparse: our collection of highly expressed TUs targeted for integration consisted of only 53 probe sets (Fig. 3B, rightmost bin, ASLV data), and the data set described in reference 17 contained 55 experimental and control integration sites. Perhaps the divergent results are explained by sparse sampling or by the use of different cell types or analytical methods in the two studies.
Integration site distribution in the chicken chromosomes.
The distribution of ASLV integration sites in the chicken chromosomes was unexpected. It would be expected for both ASLV and HIV that integration would be more frequent in the gene-dense microchromosomes and disfavored in the gene-sparse macrochromosomes, since both viruses favor integration in TUs. The expected favoring of microchromosomes was seen for HIV. However, for unclear reasons, ASLV significantly favored integration in the gene-sparse macrochromosomes. The finding that macrochromosomes are enriched in all types of chicken repeated sequences, including new ASLV integrations, points to a possible factor causing new integration events to favor these larger chromosomes. Perhaps HIV showed the opposite bias because it responded more strongly to active transcription.
Comparing de novo integration and ERVs in the chicken genome.
The distribution of de novo ASLV integration sites was quite different from that of related ERVs, suggesting models for the forces determining which ERVs persist in the genome. Both total chicken ERVs and the ASLV-related ERV subset have accumulated outside of TUs, and the minority of ERVs within TUs were typically in antisense orientation relative to host cell transcription. For ERVs in the antisense orientation, the viral splicing and polyadenylation signals do not affect mRNA synthesis by the host gene, thereby minimizing the genetic damage of integration. Comparison of the de novo distribution of ASLV integration sites to related ERVs indicates that the present distribution of chicken ERVs relative to TUs was likely not determined by the initial integration targeting, but rather by selective pressures against gene disruption. This trend has been noted previously for human ERVs (27), but the analysis of ASLV presented here provides the first case where the de novo pattern of integration was experimentally determined, allowing the observed biases in ERV distribution to be attributed to forces acting after integration.
Forces dictating the placement of ERV sequences in vertebrate genomes.
Tables 2 and 3 present a comparison of ERVs in chickens to those in rats (24), mice (20), and humans (16, 29). Earlier analyses of ERVs in the human genome (18, 27) are updated here to the most recent genome draft (hg17). For some of the annotated ERVs, it is not clear whether they have replicated as retroviruses or LTR retrotransposons (7, 9); our analysis did not attempt to distinguish between these possibilities. For all the vertebrates examined, most ERV families show a highly significant bias in favor of accumulation outside of TUs (Table 2). For the minority of ERVs within TUs, most are present in the antisense orientation relative to host gene transcription (Table 3).
In mice and rats, a small group of ERV sequences, designated LTR55 and MER95, did not show an orientation bias when present within TUs (Table 3). An analysis of these ERV sequences showed them to be quite short, apparently representing single LTRs. Such solo LTRs are known to be formed from integrated retroviruses by intrachromosomal recombination between LTRs (9). An analysis of the LTR55 and MER95 sequences present in the sense orientation in TUs showed that none contained the 5′-AATAAA-3′ sequence that directs poly(A) addition. Retroviral LTRs typically also lack splicing signals. Thus the LTR55 and MER95 sequences in sense orientation would not be expected to affect cellular mRNA synthesis, likely resulting in a lack of purifying selection and thereby explaining the lack of orientation bias.
Thus the analysis of the LTR55 and MER95 sequences reinforces the idea that selection acts to remove genes that are impaired for mRNA synthesis due to retroviral DNA integration. Overall, our findings indicate that selection against gene disruption has been the major force dictating the distribution of ERV sequences in chickens and that similar biases in ERV distribution were found in three other vertebrates.
Supplementary Material
Acknowledgments
We thank Paul Bates, Charles Berry, Robert Doms, and members of the Bushman laboratory for helpful discussions.
This work was supported by NIH grants AI52845 and AI34786, the James B. Pendleton Charitable Trust, and Robin and Frederic Withington (to F.D.B.) and the Fritz B. Burns Foundation (to J.R.E.). S.B. was supported by a fellowship from the Alberta Heritage Foundation for Medical Research and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 26:861-868. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, R., A. Katzourakis, J. Paces, A. Burt, and M. Tristem. 2005. High copy number in human endogeneous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22:814-817. [DOI] [PubMed] [Google Scholar]
- 3.Belshaw, R., V. Perieira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. [DOI] [PubMed] [Google Scholar]
- 5.Borisenko, L., and A. Rynditch. 2004. Complete nucleotide sequences of ALV-related endogenous retroviruses available from the draft chicken genome sequence. Folia Biologica (Prague) 50:136-141. [PubMed] [Google Scholar]
- 6.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman, F. D. 2001. Lateral DNA transfer: mechanisms and consequences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135-138. [DOI] [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 10.Engelman, A. 2005. The ups and downs of gene expression and retroviral DNA integration. Proc. Natl. Acad. Sci. USA 102:1275-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Follenzi, A., L. E. Ailes, S. Bakovic, M. Gueuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 12.Hematti, P., B.-K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, and B. Calmels. 23. November 2004, posting date. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2:e423. [Online.] http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101:1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Chicken Genome Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695-716. [DOI] [PubMed] [Google Scholar]
- 15.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lander, E., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 17.Maxfield, L. F., C. D. Fraize, and J. M. Coffin. 2005. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc. Natl. Acad. Sci. USA 102:1436-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medstrand, P., L. N. van de Lagematt, and D. L. Mager. 2002. Retroelement distributions in the human genome: variations associate with age and proximity to genes. Genome Res. 12:1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell, R., B. Beitzel, A. Schroder, P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. D. Bushman. 17. August 2004, posting date. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [Online.] http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 21.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 22.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panet, A., and H. Cedar. 1977. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell 11:933-940. [DOI] [PubMed] [Google Scholar]
- 24.Rat Genome Sequencing Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493-521. [DOI] [PubMed] [Google Scholar]
- 25.Sandmeyer, S. 2003. Integration by design. Proc. Natl. Acad. Sci. USA 100:5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroder, A., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. D. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 27.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 28.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venter, J. C. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 30.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, Y., J. Dai, P. G. Fuerst, and D. F. Voytas. 2003. Controlling integration specificity of yeast retrotransposon. Proc. Natl. Acad. Sci. USA 100:5891-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, Y., S. Zou, D. A. Wright, and D. F. Voytas. 1999. Tagging chromatin with retrotransposons: target specificity of the Sacchromyces Ty5 retrotransposon with the chromosomal localization of Sir3p and Sir4p. Genes Dev. 13:2738-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.