Abstract
Translation of mitochondrial mRNAs in Saccharomyces cerevisiae depends on mRNA-specific translational activators that recognize the 5′ untranslated leaders (5′-UTLs) of their target mRNAs. We have identified mutations in two new nuclear genes that suppress translation defects due to certain alterations in the 5′-UTLs of both the COX2 and COX3 mRNAs, indicating a general function in translational activation. One gene, MRP21, encodes a protein with a domain related to the bacterial ribosomal protein S21 and to unidentified proteins of several animals. The other gene, MRP51, encodes a novel protein whose only known homolog is encoded by an unidentified gene in S. kluyveri. Deletion of either MRP21 or MRP51 completely blocked mitochondrial gene expression. Submitochondrial fractionation showed that both Mrp21p and Mrp51p cosediment with the mitochondrial ribosomal small subunit. The suppressor mutations are missense substitutions, and those affecting Mrp21p alter the region homologous to E. coli S21, which is known to interact with mRNAs. Interactions of the suppressor mutations with leaky mitochondrial initiation codon mutations strongly suggest that the suppressors do not generally increase translational efficiency, since some alleles that strongly suppress 5′-UTL mutations fail to suppress initiation codon mutations. We propose that mitochondrial ribosomes themselves recognize a common feature of mRNA 5′-UTLs which, in conjunction with mRNA-specific translational activation, is required for organellar translation initiation.
While mitochondrial ribosomes exhibit distinct similarities to bacterial (eubacterial) ribosomes (30, 69), the yeast mitochondrial translation system has many intriguing differences from bacterial systems. Mitochondrial ribosomes have more proteins than do bacterial ribosomes (23, 43). Of the mitochondrial ribosomal proteins whose sequences are known, some are simple homologs of their bacterial counterparts, others have domains homologous to bacterial ribosomal proteins attached to domains with no recognizable homology to any known proteins, and still others are completely unrelated to bacterial ribosomal proteins (reviewed in reference 32). Saccharomyces cerevisiae mitochondrial mRNAs generally have long, A+U-rich 5′ untranslated leaders (5′-UTLs) lacking a typical Shine-Dalgarno sequence (11, 27, 31). While the mechanism of start site selection remains obscure in this system, translation initiation on most or all yeast mitochondrial mRNAs requires membrane-bound mRNA-specific activator proteins whose targets lie in the 5′-UTLs (reviewed in reference 27). These mRNA-specific activators appear to play a dual role in mitochondrial gene expression: tethering the synthesis of the very hydrophobic mitochondrial gene products to the inner membrane (27) and modulating the translation levels of individual mRNAs (63).
We have focused on translation of the COX2 and COX3 mRNAs, which encode subunits II and III of cytochrome c oxidase, respectively. Previous studies have identified their mRNA-specific translational activators and established functional interactions among activator proteins, their mRNA targets, and other components of the mitochondrial translation system. The COX2-specific translational activator protein is encoded by the nuclear gene PET111 (46, 54), while the COX3-specific activator is a complex containing three proteins encoded by the nuclear genes PET54, PET122, and PET494 (6, 9, 14, 38). Using suppressor analysis, we have shown that one subunit of the COX3-specific activator, Pet122p, interacts functionally with the small subunit of mitochondrial ribosomes (33, 35, 42) and that each translational activator interacts functionally with the 5′-UTL of its target mRNA (12, 45, 67). These findings suggested that yeast mitochondrial ribosomes were unable to recognize mRNAs unless the ribosome-mRNA interaction was mediated by translational activators that recognized sites unique to each mRNA.
Here we report that certain mutations in the COX2 and COX3 mRNA 5′-UTLs that are suppressible by alterations of mRNA-specific activators can also be suppressed by mutations in nuclear genes encoding two mitochondrial ribosomal small-subunit proteins. The suppression is allele specific, indicating that the ribosomes play an active role in the recognition of translation start signals. Surprisingly, however, suppression is not gene specific, indicating that the ribosomes are recognizing features of the 5′-UTLs that are common to at least several mRNAs. One of these yeast mitochondrial ribosomal proteins is homologous to bacterial S21 and to the products of unidentified genes from several animals.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The S. cerevisiae strains used in this study are listed in Table 1. All the strains were isogenic or congenic to the wild-type strain D273-10B (ATCC 25657). The media and genetic methods used were as described previously (60). Respiratory growth was assessed on YPEG medium (3% ethanol, 3% glycerol, 1% yeast extract, 2% Bacto Peptone, 2% agar). Second-site suppressors of 5′-UTL mutations were selected in the strains JJM120, JJM156, MCC199, and MCC200 (Table 1).
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| DAU1 | MATα ade2 ura3 [rho+] | 41 |
| DL1 | MATα lys2 [rho+] | 45 |
| JJM113 | MATa lys2 ura3 [rho+ cox2-10] | 47 |
| JJM120 | MATα lys2 [rho+ cox2-11] | 45 |
| JJM156 | MATα lys2 ura3 [rho+ cox2-11] | 45 |
| JJM158 | MATα lys2 ura3 [rho+ cox2-12] | 45 |
| JJM173 | MATα MRP51-5 lys2 ura3 [rho+ cox2-11] | 44 |
| LSF75 | MATα lys2 ura3 [rho+ cox3-1] | 26 |
| MCC199 | MATα lys2 [rho+ cox3-15] | This study |
| MCC200 | MATα ade2 [rho+ cox3-15] | 12 |
| MCC211 | MATα MRP21-1 ade2 ura3 [rho+ cox3-15] | This study |
| MCC267 | MATα MRP21-1 MRP51-3 [rho+ cox2-11] | This study |
| MCC291 | MATα MRP21-HA-TRP1 ura3-52 trp1-Δ1 [rho+] | This study |
| NSG50 | MATα MRP51-3 lys2 [rho+ cox2-11] | This study |
| NSG59 | MATa MRP51-3 ade2 ura3 [rho+ cox2-10] | This study |
| NSG63 | MATa mrp51Δ::URA3 lys2 ura3Δ [rho0] | This study |
| NSG78 | MATα MRP51-3 lys2 [rho+] | This study |
| NSG83 | MATα MRP51-3 lys2 [rho+ cox3-1] | This study |
| PTY11 | MATα ura3-52 trp1-Δ1 [rho+] | P. E. Thorsness |
| PTY22rho0 | MATa ura3-52 ade2 leu2-3,112 [rho0] | P. E. Thorsness |
| TF210 | MATa ura3-52 leu2-3,112 [rho+ cox3-15] | 12 |
Mitochondrial genotypes are in brackets; genes not in brackets are nuclear.
Plasmid manipulations, nucleotide sequencing, and computer analysis.
Plasmids were constructed and transformed into Escherichia coli DH5αF′IQ by standard techniques (58). Nucleotide sequencing was performed either with the Sequenase version 2.0 DNA-sequencing kit (U.S. Biochemicals) or by DNA Services, Cornell University, with an ABI 371 DNA sequencer. Nucleotide sequence data were analyzed with Lasergene biocomputing software (DNAStar, Inc.). The Basic Local Alignment Search Tool (BLAST) (2) program was accessed through the National Center for Biotechnology Information or the Saccharomyces Genome Database to search for nucleotide and protein sequence similarities.
Cloning of the MRP21 and MRP51 genes.
Nuclear DNA from a strain carrying both the MRP21-1 and MRP51-3 suppressor genes (MCC267 [Table 1]) was prepared as described previously (28) and partially digested with Sau3AI; the partial digestion products were separated by size in a 10 to 40% sucrose gradient as described by Rose and Broach (56), and 6- to 10-kb fragments were pooled. The genomic DNA fragments were ligated to BamHI-cleaved YEp24 (5) to create a library of approximately 20,000 independent E. coli transformants, which was amplified by standard methods (56).
To clone MRP21-1, the cox3-15 mutant strain TF210 (Table 1) was transformed with the MCC267 library. Transformants were selected on minimal medium and then printed to YPEG medium and incubated at 13.5°C. The transformants that grew on YPEG medium at 13.5°C were analyzed to verify that cold resistance segregated with the plasmid. Six plasmids carrying a region of the genome near the ROX3 gene and 13 plasmids carrying PET494 were isolated (see Results). To determine whether MRP21 was on the plasmids carrying the ROX3 region, a 4.4-kb BamHI-EcoRI fragment from this region was subcloned from plasmid pBSROX3BR (57) into the integrating vector YIp5 (64), which carries the URA3 gene, creating the plasmid pMC327. pMC327 was cut at a single XbaI site in the insert and integrated into the genome of strain TF210 by transformation and homologous recombination; the integrant strain was crossed to the MRP21-1 cox3-15 strain MCC211, and respiratory growth of the meiotic progeny was analyzed.
To determine the nucleotide sequences of the MRP21 suppressor alleles, the MRP21 coding sequence was PCR amplified from genomic DNA of strains carrying each of the suppressor alleles. The nucleotide sequence of the entire PCR product from each strain was determined, and in each case a single nucleotide difference from the wild-type sequence (GenBank accession no. Z35851) was observed. MRP21-1 and MRP21-2 alleles had the same mutation, a G-to-A change at nucleotide 343 of the MRP21 coding region. In the MRP21-3 allele position 363 of the MRP21 coding sequence was changed from C to G.
To clone MRP51-3, the cox2-12 mutant strain JJM158 was transformed with the MCC267 library. The transformants were selected on minimal medium and then printed to YPEG medium. Plasmids were isolated from respiratory-competent transformants and transformed back into cox2-12 and cox2-11 strains to verify that suppression was plasmid linked. Fourteen overlapping plasmids were isolated. To determine whether MRP51 was on the suppressing plasmids, a 1,975-bp SalI fragment, including 276 bp of the vector, YEp24, was subcloned from the suppressing library plasmid pB-14 into the BamHI site of the integrating vector pRS306 (62), creating plasmid pNSG17. pNSG17 was cut at either a unique AflII site or a unique MunI-MfeI site in the insert and used for integrative transformation of an MRP51-5/MRP51 cox2-11 diploid strain (JJM173 × PTY22rho0). The integrant strain was sporulated, and the meiotic progeny were analyzed.
The sequence of MRP51-3 was determined by direct sequencing of the suppressing library plasmid. It corresponded to the wild-type sequence (YPL118W; coordinates 16771 to 17805 of GenBank no. U43503) except for the single nucleotide change from C to G at position 782 of the coding sequence. The other MRP51 suppressor alleles (except MRP51-8) were cloned by gap repair (52), and the ability of gap-repaired plasmids to suppress cox2-11 was confirmed. For MRP51-2 and MRP51-4, the sequence of the entire open reading frame was determined, partly from the gap-repaired plasmids and partly from PCR products amplified from the genomic DNAs of the suppressor strains; for MRP51-1 and MRP51-5, the sequence of the entire gene except the 3′ 114 bp was determined in the same manner. For MRP51-8, the sequence was determined from PCR products amplified from genomic DNA. In MRP51-1, position 704 was changed from T to C; in MRP51-2, position 721 was changed from A to C; MRP51-4 had the same change as the independently isolated MRP51-3 allele (see above); in MRP51-5, position 779 was changed from C to T; and in MRP51-8, positions 835 and 836 were changed from GA to AG.
In vivo labeling of mitochondrial translation products.
In vivo labeling was performed as described previously (28). Cells were grown in galactose-containing minimal medium containing the 35S-labeled E. coli hydrolysate labeling reagent Tran 35S-label (ICN Radiochemicals) in the presence of cycloheximide. Crude mitochondria were subjected to electrophoresis on 16% polyacrylamide gels (prepared from a stock solution containing 29.2% acrylamide and 0.8% bisacrylamide) containing 10% glycerol in the presence of 0.1% sodium dodecyl sulfate. The gels were dried and autoradiographed.
Generation of null alleles, and epitope tagging of Mrp21p.
An mrp21 null allele was generated by removing a 459-bp ClaI-BglII fragment internal to the structural gene and inserting a hisG::URA3::hisG cassette (1). An mrp51 null allele was generated by removing an internal 550-bp MunI-BglII fragment and inserting the same cassette. To tag the MRP21 gene with three copies of the sequence encoding the influenza virus hemagglutinin (HA) epitope (25, 65) at its 3′ end, we used the plasmid pCS124 (59), an integrative plasmid carrying three copies of the HA sequence and the TRP1 gene. The 3′ 294 bp of MRP21 was amplified by PCR and inserted into pCS124 in frame with the HA sequence. The resulting plasmid, pMC343, was cut at a unique EcoRI site within the MRP21 coding sequence, and the linearized DNA was used to transform strain PTY11 to Trp+. In the resulting integrative transformant, MCC291, the only complete copy of MRP21 was the tagged allele, MRP21-HA.
Mitochondrial isolation and fractionation.
Mitochondria were prepared from cells grown to late exponential phase in complete medium (yeast-peptone [YP] medium) containing 2% galactose as described previously (29), except that spheroplasts were disrupted with a Parr-Bomb (Parr Instrument Co., Moline, Ill.) as described previously (17). Mitochondria were purified by equilibrium density gradient centrifugation in Nycodenz [5-(N-2,3-dihydroxypropylacetamido)-2,4,6-triiodo-N,N′-bis(2,3-dihydroxypropyl)-isophthalimide; Sigma, St. Louis, Mo.] step gradients as described previously (29). Mitochondrial ribosomes were analyzed by sucrose gradient centrifugation (15 to 30% sucrose in 0.5 M NH4Cl, 10 mM Tris [pH 7.4], 10 mM magnesium acetate, 7 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, and 2 μg each of the protease inhibitors antipain, aprotinin, bestatin, chymostatin, E-64, leupeptin, pepstatin A, and phosphorhamidon per ml) directly from disrupted mitochondria as described previously (53). The clarified lysate obtained from 2 mg of whole mitochondria was applied to a 36-ml gradient; 1-ml fractions were collected, and 0.2 ml of each was precipitated with trichloroacetic acid and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% polyacrylamide gels.
Antisera and immunological methods.
Mouse monoclonal antibodies against Mrp7p (24) and Mrp13p (53) were obtained from T. L. Mason. Mouse monoclonal antibody 12CA5 against the HA epitope was purchased from BAbCo (Berkeley, Calif.).
Anti-Mrp51p polyclonal rabbit antiserum was prepared as described previously (36) with histidine-tagged Mrp51p as an antigen. An MRP51 gene with six His codons at the 3′ end of the coding sequence was generated by PCR and inserted into pQE-30 (Qiagen), with an additional six His codons added to the 5′ end of the coding sequence. The resulting plasmid, pNSG29, was transformed into E. coli and induced with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the fusion protein was affinity purified with Ni-nitrilotriacetic acid resin as directed by the manufacturer (Qiagen) (37).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed by standard techniques (36). Antigen-antibody complexes were visualized by using horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G or goat anti-rabbit immunoglobulin G (Gibco BRL, Bethesda, Md.) secondary antibody and the enhanced chemiluminescence system (Amersham Life Science Inc., Arlington Heights, Ill.).
RESULTS
Selection of second-site suppressors of cox2 and cox3 5′-UTL mutations.
The cox2-11 mutation is a single-base deletion (deletion of the G residue at −24) in the COX2 5′-UTL that does not affect the stability of the COX2 mRNA but greatly decreases its translation, causing weak respiratory growth (45). In a previous study, six independent dominant nuclear suppressors of the cox2-11 mutation were isolated (45). One of these affected PET111, the known specific translational activator for the COX2 mRNA, while the other five did not (45). To characterize further the unknown mutations, a strain carrying one of the suppressors was crossed to each of the other four suppressor strains. Analysis of the respiratory growth of the meiotic progeny showed that the first suppressor was tightly linked to each of the other four. Thus, all five cox2-11 suppressors were in a single gene, now called MRP51 (MRP51-1, MRP51-2, MRP51-3, MRP51-4, and MRP51-5). Respiratory growth of the cox2-11 strains containing these suppressors remained dependent upon the function of the PET111 gene.
The cox3-15 mutation, which consists of two deletions in the COX3 5′-UTL, causes cold-sensitive respiratory growth (12). The respiratory defect is due to cold-sensitive translation, since the cox3-15 mRNA is present at wild-type levels in cells grown in the cold but Cox3p is not synthesized (12). Translation of the cox3-15 mRNA at the permissive temperature is still dependent on the COX3-specific translational activator complex (12).
Six spontaneous cold-resistant revertants of a cox3-15 mutant strain were isolated previously (12), and for this study an additional 20 revertants were isolated. The revertant strains were characterized genetically as previously described (12) to determine whether the dominant suppressor mutations were nuclear or mitochondrial and whether they were linked to any known genes whose products are involved in translational activation. Of the 26 revertants analyzed, 17 had nuclear suppressor mutations. Nine of these suppressors mapped to the PET122 gene, which encodes a subunit of the COX3-specific translational activator (10, 12, 38); three mapped to a new gene we have called MRP21 (MRP21-1, MRP21-2, and MRP21-3); and one mapped to the MRP51 gene (MRP51-8), also identified above as a suppressor of the cox2-11 mutation (the four remaining nuclear suppressors were relatively weak and were not studied further). The wild-type function of the COX3-specific translational activators PET54, PET122, and PET494 was required for respiratory growth of the cox3-15 strains carrying MRP21 or MRP51 suppressor alleles. Thus, the selection of suppressors of 5′-UTL mutations that specifically blocked the translation of particular mitochondrial mRNAs yielded not only mutations in the corresponding specific translational activators (12, 45) but also mutations in two previously unidentified genes whose products interact functionally with the same regions of these 5′-UTLs.
Specificity of the MRP21 and MRP51 suppressors.
To characterize the specificity of the two novel suppressors, they were combined with a variety of nuclear and mitochondrial mutations affecting translation and the double-mutant phenotypes were analyzed. Respiratory growth, a phenotype we have repeatedly found to be a sensitive indicator of Cox2p and Cox3p synthesis in mutant strains (6, 12, 13, 22, 26, 45), was assessed for all combinations of alleles (Tables 2 and 3; Fig. 1). To confirm that the observed respiratory growth phenotypes reflected the synthesis rates of mitochondrial proteins, in vivo labeling of mitochondrial translation products in the presence of cycloheximide was performed for selected strains (Fig. 2).
TABLE 2.
Allele specificity of the MRP21 suppressor mutations
| Relevant genotype | Growth with MRP21 allelea:
|
||
|---|---|---|---|
| MRP21 | MRP21-1 | MRP21-3 | |
| COX3, COX2 | ++++ | ++++ | ++++ |
| cox3-15 | ± | +++ | +++ |
| pet54-A244, cox3-15 | − | + | ND |
| cox3-438 | − | − | − |
| cox3-1 | + | ± | ++ |
| cox2-11 | ± | ++ | ++ |
| cox2-105 | ± | + | ++ |
| cox2-12 | − | − | − |
| cox2-10 | ± | ± | + |
The number of + signs indicates relative level of respiratory growth, determined by measuring the colony size on YPEG medium at 30°C, except for cox3-15 mutants, which were incubated at 13.5°C. −, no growth; ND, not determined. Suppression was tested in haploids, except with cox2-12, cox2-105, and cox3-438, for which dominant suppression in heterozygous diploids is reported. References for the alleles listed in the table are as follows: cox3-15 (12); pet54-A244 (6); cox3-438 (67); cox3-1 (26); cox2-11 (45); cox2-105 (22); cox2-12 (45); cox2-10 (47). The following alleles showed no genetic interaction (neither suppression nor synthetic defects) with MRP21-1: cox3-13 and -14 (67); cox3-209 (13); cox2-13 (45); cox2-103, cox2-106, cox2-107, and cox2-108 (22); MRP51-1, MRP51-2, MRP51-3, MRP51-4, and MRP51-5 (this study); pet54-A244 (6); PET122-L195 (12); PET122-I175 and PET122-V211 (6); oli1-h45 (51).
TABLE 3.
Allele specificity of the MRP51 suppressor mutations
| Relevant genotype | Growth with MRP51 allelea:
|
|||||
|---|---|---|---|---|---|---|
| MRP51 | MRP51-1 | MRP51-2 | MRP51-3 | MRP51-5 | MRP51-8 | |
| COX2 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| cox2-11 | ± | ++ | ++ | +++ | ++ | − |
| cox2-12 | − | − | − | ++ | ± | − |
| cox2-13 | − | − | − | − | − | − |
| cox2-10 | + | ++ | +++ | + | +++ | ± |
| cox2-105 | ± | + | + | +++ | ++ | ± |
| cox3-1 | + | + | + | + | + | + |
| cox3-15 | ± | ± | + | + | ++ | ++ |
| cox3-438 | − | − | − | − | − | − |
The number of + signs indicates the relative level of respiratory growth, determined by measuring the colony size on YPEG medium at 30°C, except for cox3-15 mutants, which were incubated at 13.5°C. −, no growth. Suppression was tested in haploids except with cox2-105 and cox3-438, for which dominant suppression in heterozygous diploids is reported. References for the alleles listed in the table are indicated in the legend to Table 2. MRP51 suppressors did not exhibit dominant suppression of the cox2-103, cox2-106, cox2-107, and cox2-108 alleles (22) and the oli1-h45 alleles (51).
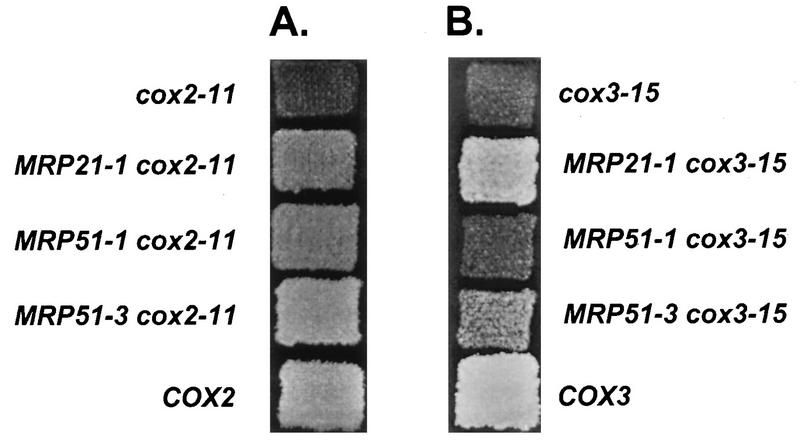
FIG. 1.
Suppression of selected cox2 and cox3 5′-UTL mutations by MRP21 and MRP51 mutations. Cells were grown on glucose-containing medium (YPD), printed to nonfermentable medium (YPEG medium supplemented with 0.02 mg of adenine per ml), and incubated at 30°C for 2 days (A) or at 13.5°C for 12 days (B). Relevant genotypes are shown. Where it is not indicated otherwise, MRP21 and MRP51 are wild type.
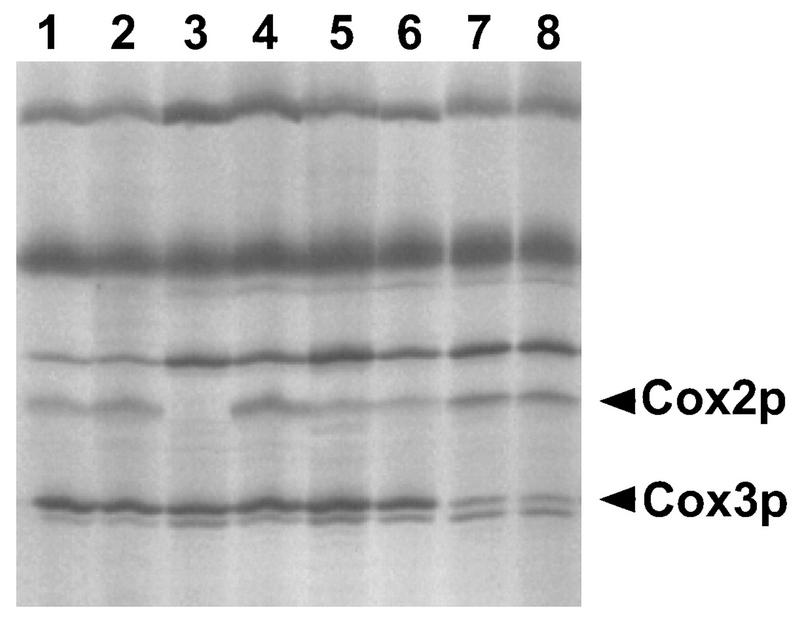
FIG. 2.
Effects of the MRP51-3 suppressor on Cox2p and Cox3p synthesis. Mitochondrial translation products were radioactively labeled in the presence of cycloheximide and subjected to electrophoresis as described in Materials and Methods. The positions of Cox2p and Cox3p are indicated. Strain names and relevant genotypes are as follows: lane 1, DL1 (wild-type); lane 2, NSG78 (MRP51-3); lane 3, JJM120 (5′-UTL mutation cox2-11); lane 4, NSG50 (MRP51-3 cox2-11); lane 5, JJM113 (initiation codon mutation cox2-10); lane 6, NSG59 (MRP51-3 cox2-10); lane 7, LSF75 (initiation codon mutation cox3-1); lane 8, NSG83 (MRP51-3 cox3-1).
None of the suppressor alleles had a detectable phenotype when combined with the wild-type mitochondrial genome. However, some suppressor alleles at both MRP21 and MRP51 suppressed some 5′-UTL mutations in both COX2 and COX3 (Tables 2 and 3; Fig. 1 and 2), suggesting that Mrp21p and Mrp51p are involved in the translation of at least these two mitochondrial mRNAs.
One possible mechanism for suppression of 5′-UTL mutations in both the COX2 and COX3 mRNAs is that the alterations in Mrp21p and Mrp51p cause a general increase in the level of mitochondrial translation. To test this possibility, we asked whether the MRP21 and MRP51 suppressor alleles would improve the respiratory growth of strains with leaky initiation codon mutations (AUG to AUA) in these mitochondrial mRNAs (cox2-10 and cox3-1). Since the respiratory growth of these mutants is responsive to levels of translational activity (26, 47), we would expect that all of the MRP21 and MRP51 suppressor alleles would suppress the initiation codon mutations if the mechanism of suppression were an overall increase in translation. We might also expect that the strength of suppression of 5′-UTL mutations would correlate with the strength of suppression of initiation codon mutations for each particular suppressor allele: a strong suppressor of 5′-UTL mutations would strongly suppress initiation codon mutations, and the converse. However, this was not the case. Some alleles that suppressed 5′-UTL mutations failed to affect the initiation codon mutants, others suppressed them, and still others reduced their respiratory growth, causing a synthetic defect (Tables 2 and 3).
In the experiment in Fig. 2, this phenomenon is illustrated at the level of mitochondrial protein synthesis for the MRP51-3 allele. MRP51-3 had a dramatic effect on the cox2-11 5′-UTL mutation: in the unsuppressed strain, the level of Cox2p was greatly reduced (Fig. 2, lane 3), while in an MRP51-3 cox2-11 strain, Cox2p was synthesized at wild-type levels (lane 4). In a leaky cox2 initiation codon mutant strain, Cox2p levels were reduced from wild-type levels, whether or not the strain carried MRP51-3 (lanes 5 and 6). Similarly, MRP51-3 had no effect on Cox3p synthesis from the leaky cox3 initiation codon mutant allele (lanes 7 and 8). Levels of Cox2p and Cox3p synthesis in these strains correlated with their respiratory growth phenotypes, confirming that MRP51-3 strongly suppressed the 5′-UTL mutation while having no effect on the initiation codon mutations.
In vivo labeling of mitochondrial translation products in MRP21-1 strains with cox2 and cox3 initiation codon mutations (not shown) yielded similar results. The MRP21-1 allele, a strong suppressor of the 5′-UTL mutation cox3-15, had no effect on Cox2p synthesis in the cox2 initiation codon mutant strain. In combination with the cox3 initiation codon mutation, MRP21-1 caused reduced Cox3p synthesis, consistent with the synthetic respiratory defect observed in the double-mutant strain (Table 2).
Molecular cloning and nucleotide sequence analysis of the MRP21 and MRP51 genes.
The dominant suppression phenotypes of the MRP21 and MRP51 suppressors were used to clone both genes. A genomic library was constructed in a multicopy plasmid from a strain (MCC267; Table 1) carrying both the MRP21-1 and MRP51-3 alleles, to isolate both genes from a single library (Materials and Methods). To clone MRP21, the cox3-15 mutant strain TF210 (Table 1) was transformed with the library and transformants with cold-resistant respiratory growth were selected. To clone MRP51, the cox2-12 mutant strain JJM158 (Table 1) was transformed with the library and respiring transformants were selected.
Plasmids that conferred cold-resistant respiratory growth on the cox3-15 mutant strain TF210 fell into two distinct sets based on restriction mapping and hybridization analysis. The nucleotide sequences of small fragments from each class were determined to localize the plasmid inserts in the genome. One class of plasmids was found to carry PET494, which encodes a subunit of the COX3-specific translational activator (9, 10) and is known to suppress cox3-15 when overexpressed (12). The other class of plasmids conferring cold-resistant respiratory growth carried a region of DNA near the ROX3 gene (57) from chromosome II. To test whether the MRP21 gene was located in this region of the genome, the URA3 gene was integrated into the ROX3 region of a strain carrying the cox3-15 allele and the integrant strain was crossed to an MRP21-1 strain also carrying cox3-15 (see Materials and Methods). Among the meiotic progeny of this cross, all the spores that were able to respire at 13.5°C were Ura− whereas all Ura+ spores had cold-sensitive respiratory growth: thus, the MRP21 gene is tightly linked to the ROX3 region.
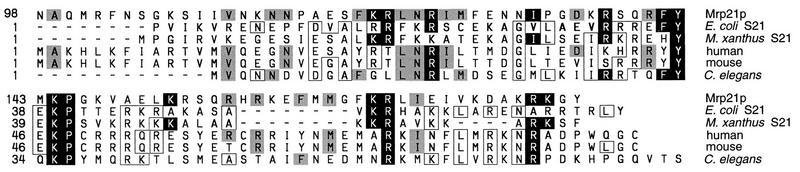
A 2.9-kb BamHI fragment which included 1.8 kb from the region common to all six plasmids obtained from the genomic library carried the complete MRP21-1 gene, as judged by its ability when subcloned to confer cold-resistant respiration on the cox3-15 strain TF210 as well as did the original library plasmids. This fragment carried two open reading frames, but only one was located entirely within the region of overlap of the library plasmids, identifying it as MRP21. The MRP21 open reading frame (YBL090W; GenBank accession no. Z35851) encodes a strongly basic (net charge of +18) 177-amino-acid protein with a predicted molecular mass of 20.4 kDa. Homology searches with the Basic Local Alignment Search Tool (BLAST) program (2) revealed weak similarity between the C-terminal region of Mrp21p and predicted proteins of unknown function from humans, mice, and Caenorhabditis elegans, all of which are highly homologous. The metazoan proteins exhibit a clear similarity to the small-subunit ribosomal S21 protein from bacteria. Alignment of the Mrp21p, higher eukaryotic, and bacterial sequences (Fig. 3) reveals clear similarities between Mrp21p and S21. Interestingly, the sequences of the suppressor alleles (see Materials and Methods) revealed that they caused missense substitutions in a small region of the S21-homologous C-terminal domain of Mrp21p. The independently isolated MRP21-1 and MRP21-2 alleles were identical (see Materials and Methods), both causing a Glu-to-Lys change at amino acid 118. The MRP21-3 allele changed Asn to Lys at amino acid 124, increasing the similarity to bacterial S21 proteins.
FIG. 3.
Alignment of the C-terminal region of Mrp21p with bacterial ribosomal S21 proteins and with sequences from higher eukaryotes. Black boxes, identities between Mrp21p and either or both of the bacterial S21 proteins; gray boxes, identities between Mrp21p and the metazoan proteins not shared by the bacterial S21 proteins; white boxes, identities between the metazoan proteins and the bacterial S21 proteins not found in Mrp21p. The percentages of identical plus similar amino acids for selected pairwise comparisons are as follows: Mrp21p-E. coli S21, 23.9%; Mrp21p-Myxococcus xanthus S21, 28.1%; Mrp21p-human, 36.1%; Mrp21p-mouse, 32.5%; Mrp21p-C. elegans, 25.3%; human-E. coli S21, 34.3%; human-M. xanthus S21, 39.1%. The accession numbers of the sequences are as follows: Mrp21p, Z35851; E. coli rpsU gene, V00346 (40); M. xanthus rpsU gene, U20669; human, coordinates 447 to 710, U79258; mouse, coordinates 73 to 336, AA050698; C. elegans gene F29B9.10, U70849.
To clone MRP51, the cox2-12 mutant strain JJM158 was transformed with the MCC267 library and respiring transformants were selected. Fourteen overlapping plasmids which conferred respiratory growth were isolated. To test whether MRP51 was linked to this region, a fragment from this plasmid was cloned into an integrating vector carrying the URA3 gene and transformed into a MRP51-5/MRP51 diploid (see Materials and Methods). When independent diploid transformants were sporulated and tetrads were dissected, the ability to respire segregated either with the Ura+ marker (integration into the suppressor chromosome) or opposite to the Ura+ marker (integration into the wild-type chromosome). Thus, the plasmid-borne sequences were tightly linked to MRP51.
Sequence analysis of the smallest library plasmid revealed two complete open reading frames: the IDI1 gene encoding isopentenyl diphosphate-dimethylallyl diphosphate isomerase, an enzyme of the isoprenoid biosynthetic pathway (3), and an unidentified gene. To determine whether the unidentified open reading frame was MRP51, it was subcloned from the suppressing plasmid after PCR amplification (Materials and Methods). The resulting plasmid, pNSG19, had suppressor activity in cox2-11 and cox2-12 mutant strains, identifying this gene as MRP51. The MRP51 open reading frame (YPL118W; coordinates 16771 to 17805 of GenBank sequence no. U43503) encodes a 344-amino-acid protein with a predicted molecular mass of 39.5 kDa. Mrp51p is predicted to be a strongly basic protein with a net charge of +22. The DNA sequences of the suppressor alleles (see Materials and Methods) revealed that they were associated with amino acid substitutions in a limited region of Mrp51p: MRP51-1, Val to Ala at position 235; MRP51-2, Asp to His at position 241; MRP51-3 and MRP51-4, Pro to Arg at position 261; MRP51-5, Pro to Leu at position 260; MRP51-8, Glu to Arg at position 279. No proteins of known function are homologous to Mrp51p. However, Mrp51p is 46% identical to an unidentified open reading frame of S. kluyveri (66) (coordinates 290 to 378 of GenBank sequence no. U83662 and coordinates 2569 to 1543 of EMBL sequence no. Z14125).
Construction and characterization of mrp21 and mrp51 null mutations.
To inactivate MRP21 and MRP51, internal fragments of both genes were removed and replaced with a hisG::URA3::hisG cassette (1) (see Materials and Methods). DNA fragments carrying each deleted and disrupted gene were used, separately, to transform a diploid strain homozygous for a ura3 mutation. In each case, the Ura+ diploid transformants respired well, but when the diploids were sporulated and tetrads were dissected, each tetrad had two respiratory-competent Ura− spores and two respiratory-deficient Ura+ spores. The Ura+ spores were unable to produce respiring diploids when mated to a nuclearly wild-type, rho0 tester strain (lacking mitochondrial DNA), indicating that deletion of either MRP21 or MRP51 caused the cells to lose their mitochondrial DNA. The destabilization of mitochondrial DNA is a hallmark of mutations that block all mitochondrial translation (24, 34, 49, 50). This suggested that both Mrp21p and Mrp51p might be required generally for mitochondrial translation.
Subcellular and submitochondrial localization of Mrp21p and Mrp51p.
To detect Mrp21p, it was tagged at the carboxy terminus with three copies of the influenza virus HA epitope (25, 65) (see Materials and Methods), which is recognized by the 12CA5 mouse monoclonal antibody. HA-tagged Mrp21p was only partially functional: strains in which the only copy of MRP21 carried the HA tag showed a mild respiratory defect. To detect Mrp51p, we raised a rabbit polyclonal antiserum to a version of Mrp51p carrying amino- and carboxy-terminal six-histidine tags (37), purified after expression in E. coli (see Materials and Methods).
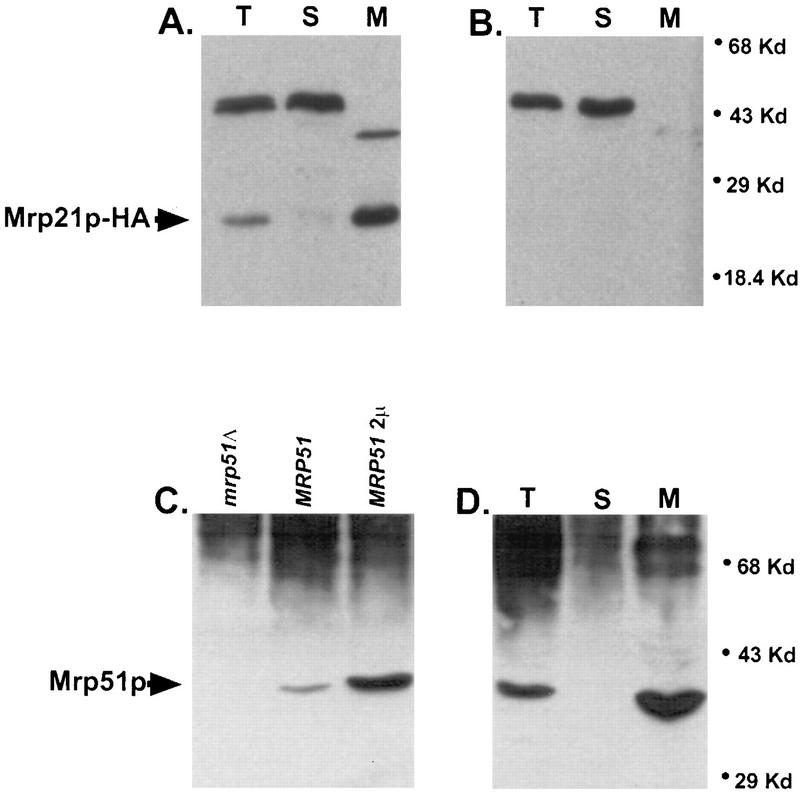
Wild-type yeast (PTY11) and a strain carrying a chromosomally integrated gene encoding HA-tagged Mrp21p (MCC291) were grown and fractionated into mitochondrial pellets and postmitochondrial supernatant fractions, after which the mitochondria were purified by buoyant density gradient centrifugation (see Materials and Methods). The fractions were analyzed by gel electrophoresis and Western blotting, probing either with the anti-HA monoclonal antibody or with the polyclonal anti-Mrp51p antiserum (Fig. 4). As expected, both Mrp21p and Mrp51p were associated specifically with mitochondria.
FIG. 4.
Mrp21p and Mrp51p are located in mitochondria. (A and B) Subcellular location of Mrp21p-HA (arrow). A 50-μg portion of protein was applied to each lane of the gel. Western blots in both panels were probed with monoclonal anti-HA antibody. (A) Subcellular fractions of an MRP21-HA strain (MCC291). Mrp21p-HA is present in whole-cell extract (lane T) and gradient-purified mitochondria (lane M) but not in the cytosol (lane S). (B) Corresponding fractions from a wild-type strain (PTY11). (C) The polyclonal anti-Mrp51p antibody detects an approximately 39-kDa protein in whole-cell extract from the wild type (MRP51; strain DAU1) that is absent in a null mutant (mrp51Δ; NSG63) and overproduced in a strain carrying MRP51 on a high-copy-number plasmid (MRP51 2μm; plasmid pNSG22 in strain DAU1), identifying this band as Mrp51p. Approximately 10 μg of total-cell protein was applied per lane. (D) Subcellular location of Mrp51p (arrow). The anti-Mrp51p antibody detects Mrp51p in whole-cell extract (lane T) and gradient-purified mitochondria (lane M) but not in the cytosol (lane S) of a wild-type strain (PTY11). The amounts of protein applied to the gel were 50 μg (lane T), 20 μg (lane S), and 20 μg (lane M).
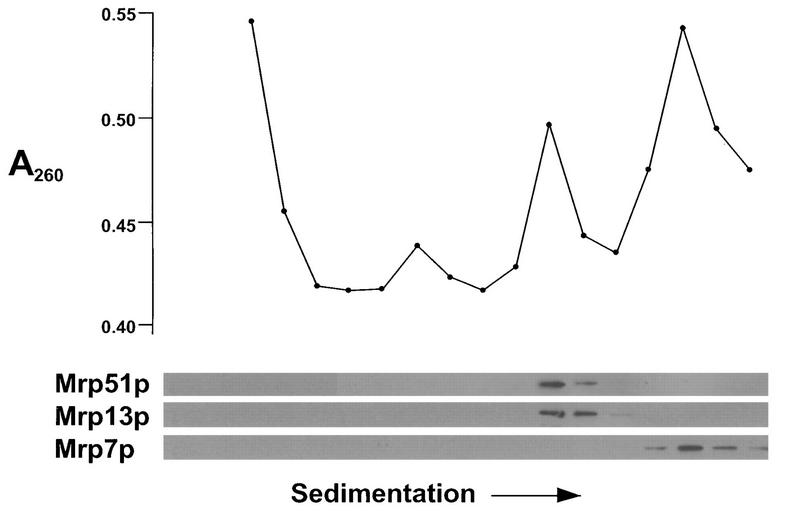
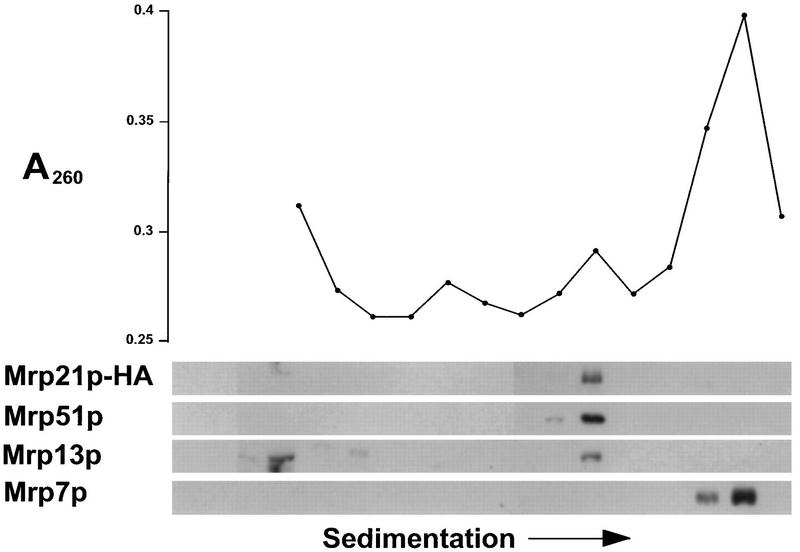
The phenotypes of the mrp21 and mrp51 null mutations suggested that they were required for all mitochondrial translation. This, as well as the similarity between Mrp21p and bacterial S21 proteins, raised the possibility that they were components of the mitochondrial ribosome. To test this possibility, gradient-purified mitochondria were solubilized with detergent and the contents were sedimented into a sucrose gradient in the presence of high salt concentrations (0.5 M NH4Cl; see Materials and Methods) to separate the subunits of mitochondrial ribosomes. The gradient fractions were analyzed for absorbance at 260 nm, to locate the rRNAs, and by gel electrophoresis and Western blotting (Fig. 5 and 6). In the experiment in Fig. 5, mitochondrial ribosomes from wild-type (PTY11) cells were subjected to this analysis. The position of Mrp51p coincided with that of the ribosomal small subunit, as identified by the smaller of two peaks of absorbance at 260 nm and by the presence of Mrp13p, a known small-subunit constituent (53). In the experiment in Fig. 6, a similar analysis was performed on mitochondrial ribosomes isolated from a strain (MCC291) in which the only functional MRP21 gene carried the HA tag at its 3′ end. As noted above, the HA-tagged Mrp21p did not function as well as the wild type, and the experiment in Fig. 6 reveals the probable reason for this. The small subunit of mitochondrial ribosomes was partially destabilized in this strain, as shown by the dramatic decrease in the peak of absorbance for the small rRNA relative to that for the large rRNA. Nevertheless, the peak of Mrp21p-HA coincided with the small ribosomal subunit. Indeed, the fact that an alteration which decreased the function of Mrp21p specifically affected the small subunit of mitochondrial ribosomes strongly supports the idea that Mrp21p is a small-subunit component.
FIG. 5.
Mrp51p cosediments with the small subunit of mitochondrial ribosomes. Nycodenz gradient-purified mitochondria of strain PTY11 were disrupted with deoxycholate, and the soluble contents were centrifuged into a sucrose gradient in the presence of 0.5 M salt (see Materials and Methods). (Top) Absorbance at 260 nm (A260) of alternate fractions. (Bottom) Western blots of alternate fractions probed with antisera against Mrp51p, the known small-subunit protein Mrp13p (53), and the known large-subunit protein Mrp7p (24).
FIG. 6.
Partially functional Mrp21p-HA cosediments with the small subunit of mitochondrial ribosomes but destabilizes it. Nycodenz gradient-purified mitochondria of strain MCC291 were analyzed as described in the legend to Fig. 5, except that the fractions were also probed with the anti-HA monoclonal antibody.
DISCUSSION
We have identified two nuclear yeast genes encoding previously unidentified mitochondrial ribosomal small-subunit proteins, Mrp21p and Mrp51p. These genes can mutate to suppress defects in the 5′-UTLs of two different mitochondrial mRNAs, COX2 and COX3, but do not bypass the mRNA-specific translational activation system. The 5′-UTL mutations are known from previous genetic analysis to alter the targets of the COX2 and COX3 mRNA-specific translational activators (12, 46). However, the functions of the MRP21 and MRP51 products are not mRNA specific. Suppressor alleles at each of these two nuclear genes were able to improve the respiratory growth of certain 5′-UTL mutations, but not others, affecting both the COX2 and COX3 mRNAs. Furthermore, deletion of either MRP21 or MRP51 prevented mitochondrial translation globally.
Suppression of 5′-UTL mutations might occur by any alteration that caused a general increase in mitochondrial translational activity. However, this does not appear to be the mechanism by which the MRP21 and MRP51 suppressors work, since many of the suppressors failed to increase the respiratory growth of strains bearing leaky COX2 and COX3 initiation codon mutations (cox2-10 and cox3-1) and also failed to increase Cox2p or Cox3p synthesis in these strains. The initiation codon mutations reduce the translation of mRNAs bearing otherwise wild-type 5′-UTLs roughly five- to sevenfold without altering the sites of initiation (26, 47). Furthermore, the growth phenotypes they cause are influenced by the levels of their respective mRNA-specific translational activators, indicating that they are sensitive to translational activity (26, 47). While some of the other suppressor alleles did improve the growth of initiation codon mutants, MRP21-1 and MRP51-8 actually reduced the respiratory growth of cox3-1 and cox2-10 mutants, respectively. Thus, we conclude that the MRP21 and MRP51 mutations do not generally increase the activity of mitochondrial ribosomes. Instead, the patterns of suppression by the MRP21 and MRP51 mutations, which are allele specific but, surprisingly, gene nonspecific, suggest that yeast mitochondrial ribosomes may recognize a common feature in mRNA 5′-UTLs. According to this hypothesis, the structure of the common element was altered by mutations in the COX2 and COX3 5′-UTLs and the suppressors altered the ribosomal small subunit to compensate for the defects.
Mrp21p resembles several other yeast mitochondrial small-subunit ribosomal proteins (4, 18, 19, 39) in that it has a domain lacking homology to any known protein and a domain identifiably homologous to a bacterial ribosomal protein. The amino-terminal 99-amino-acid sequence of Mrp21p is not similar to currently known sequences, but the carboxy-terminal 78-residue sequence exhibits clear similarity to a metazoan sequence, which in turn is clearly similar to those of bacterial ribosomal S21 proteins. This family of proteins is absent in eukaryotic cytoplasmic ribosomes (68) and those of known members of the Archaea (7). The limited homology is convincing when taken together with the facts that both Mrp21p and S21 are small-subunit ribosomal proteins and that our suppressors are missense substitutions in a small region of the S21-homologous C-terminal domain of Mrp21p. The functions of the metazoan Mrp21p homologs are unknown, but it is likely that they are also mitochondrial ribosomal proteins involved in translation initiation.
The available evidence is consistent with the idea that Mrp21p and bacterial S21 may have similar functions in promoting mRNA-ribosome interactions. Our genetic data suggest that Mrp21p may interact directly with the 5′-UTLs. Ribosomal protein-mapping studies indicate that E. coli S21 protein is in the platform region of the small subunit, the site of Shine-Dalgarno and codon-anticodon interactions (8). S21 is in very close proximity to both the 16S rRNA and the initiation region of mRNAs, as shown by cross-linking and resonance energy transfer experiments (15, 21, 48). However, the in vivo function of S21 has not been studied genetically. The only reported alleles of the E. coli gene encoding S21, rpsU, have no effect on translation (16).
Like several other mitochondrial ribosomal small-subunit proteins (33, 42, 49, 53), Mrp51p exhibits no clear homology to any known ribosomal proteins. The only known homolog is the product of an unidentified open reading frame in the yeast S. kluyveri, which probably also encodes a mitochondrial ribosomal protein. Interestingly, the missense substitutions caused by our five different MRP51 suppressor alleles are clustered within a 45-amino-acid region of the protein, which could be involved in mRNA interactions.
The mechanism by which yeast (and other) mitochondrial ribosomes identify translation initiation sites is not clear, largely owing to the lack of suitable in vitro systems (20). However, it does not involve either a classical Shine-Dalgarno interaction or a simple scanning mechanism (27). AUG codons clearly play a role in start site selection, but additional information is also used (26, 47). Genetic studies have strongly supported a model in which mRNA-specific translational activators mediate the mRNA-ribosome interaction leading to initiation and possibly influence start site selection (reviewed in reference 27).
Our present data demonstrate that, in conjunction with mRNA-specific activators, the yeast mitochondrial ribosome itself plays an active role in recognizing translatable mRNAs. The pattern of suppression observed suggests that the ribosomes may recognize a feature common to all yeast mitochondrial mRNA 5′-UTLs. A candidate for such a common feature, the octanucleotide sequence UAUAAAUA, has recently been identified based on a functional analysis of the COX2 mRNA 5′-UTL and comparisons with other 5′-UTLs (22). While this sequence is not directly altered in the suppressible alleles studied here, cox2-11 and cox3-15, it is within 10 bases upstream of both mutations. This octanucleotide is complementary to several sites in the mitochondrial small-subunit rRNA and could thus be involved in mRNA-rRNA base pairing. Clearly, Mrp21p and Mrp51p could play a role in establishing such an mRNA-rRNA interaction. However, this putative interaction would not closely resemble the Shine-Dalgarno mechanism (55, 61), since the octanucleotide does not occur at fixed distances from translation initiation codons of mRNAs and its complement is not located at the 3′ end of the rRNA. Alternatively, yeast mitochondrial ribosomes could interact with mRNAs purely through protein-mRNA contacts, possibly involving Mrp21p and Mrp51p directly.
ACKNOWLEDGMENTS
We thank C. Shamu and J. Nunnari for providing us with the plasmid pCS124, R. Zitomer for providing plasmids carrying the ROX3 region, T. L. Mason for gifts of antisera, and P. Nagley for the gift of strain h45.
This work was supported by National Institutes of Health research grant GM29362. N.S.G.-W. was supported by National Institutes of Health predoctoral training grant GM07617.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anderson M S, Muehlbacher M, Street I P, Proffitt J, Poulter C D. Isopentenyl diphosphate:dimethylallyl diphosphate isomerase. An improved purification of the enzyme and isolation of the gene from Saccharomyces cerevisiae. J Biol Chem. 1989;264:19169–19175. [PubMed] [Google Scholar]
- 4.Boguta M, Dmochowska A, Borsuk P, Wrobel K, Gargouri A, Lazowska J, Slonimski P P, Szczesniak B, Kruszewska A. NAM9 nuclear suppressor of mitochondrial ochre mutations in Saccharomyces cerevisiae codes for a protein homologous to S4 ribosomal proteins from chloroplasts, bacteria, and eucaryotes. Mol Cell Biol. 1992;12:402–412. doi: 10.1128/mcb.12.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botstein D, Falco S C, Stewart S E, Brennan M, Scherer S, Stinchcomb D T, Struhl K, Davis R W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- 6.Brown N G, Costanzo M C, Fox T D. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Glake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Capel M S, Kjeldgaard M, Engelman D M, Moore P B. Positions of S2, S13, S16, S19, and S21 in the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1988;200:65–87. doi: 10.1016/0022-2836(88)90334-8. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo M C, Fox T D. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986;6:3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo M C, Fox T D. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci USA. 1988;85:2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo M C, Fox T D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo M C, Fox T D. Suppression of a defect in the 5′-untranslated leader of the mitochondrial COX3 mRNA by a mutation affecting an mRNA-specific translational activator protein. Mol Cell Biol. 1993;13:4806–4813. doi: 10.1128/mcb.13.8.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo M C, Fox T D. A point mutation in the 5′-untranslated leader that affects translation of the mitochondrial COX3 mRNA. Curr Genet. 1995;28:60–66. doi: 10.1007/BF00311882. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo M C, Seaver E C, Fox T D. At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 1986;5:3637–3641. doi: 10.1002/j.1460-2075.1986.tb04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czworkowski J, Odom O W, Hardesty B. Fluorescence study of the topology of messenger RNA bound to the 30S ribosomal subunit of Escherichia coli. Biochemistry. 1991;30:4821–4830. doi: 10.1021/bi00233a026. [DOI] [PubMed] [Google Scholar]
- 16.Dabbs E R. The gene for ribosomal protein S21, rpsU, maps close to dnaG at 66.5 min on the Escherichia coli chromosomal linkage map. J Bacteriol. 1980;144:603–607. doi: 10.1128/jb.144.2.603-607.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dake E, Hofmann T J, McIntire S, Hudson A, Zassenhaus H P. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988;263:7691–7702. [PubMed] [Google Scholar]
- 18.Dang H, Ellis S R. Structural and functional analyses of a yeast mitochondrial ribosomal protein homologous to ribosomal protein-S15 of Escherichia coli. Nucleic Acids Res. 1990;18:6895–6901. doi: 10.1093/nar/18.23.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis S C, Tzagoloff A, Ellis S R. Characterization of a yeast mitochondrial ribosomal protein structurally related to the mammalian 68-kDa high affinity laminin receptor. J Biol Chem. 1992;267:5508–5514. [PubMed] [Google Scholar]
- 20.Dekker P J T, Papadopoulou B, Grivell L A. In-vitro translation of mitochondrial mRNAs by yeast mitochondrial ribosomes is hampered by the lack of start-codon recognition. Curr Genet. 1993;23:22–27. doi: 10.1007/BF00336745. [DOI] [PubMed] [Google Scholar]
- 21.Dontsova O, Kopylov A, Brimacombe R. The location of mRNA in the ribosomal 30S initiation complex: site-directed cross-linking of mRNA analogues carrying several photoreactive labels simultaneously on either side of the AUG start codon. EMBO J. 1991;10:2613–2620. doi: 10.1002/j.1460-2075.1991.tb07803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunstan H M, Green-Willms N S, Fox T D. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faye G, Sor F. Analysis of mitochondrial ribosomal proteins of Saccharomyces cerevisiae by two dimensional polyacrylamide gel electrophoresis. Mol Gen Genet. 1977;155:27–34. doi: 10.1007/BF00268557. [DOI] [PubMed] [Google Scholar]
- 24.Fearon K, Mason T L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol Cell Biol. 1988;8:3636–3646. doi: 10.1128/mcb.8.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Field J, Nikawa J-I, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folley L S, Fox T D. Site-directed mutagenesis of a Saccharomyces cerevisiae mitochondrial translation initiation codon. Genetics. 1991;129:659–668. doi: 10.1093/genetics/129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox T D. Genetics of mitochondrial translation. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 733–758. [Google Scholar]
- 28.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 29.Glick B S, Pon L A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 30.Gray M W. Origin and evolution of organelle genomes. Curr Opin Genet Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 31.Grivell L A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989;182:477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- 32.Grohmann L, Kitakawa M, Isono K, Goldschmidt-Reisin S, Graack H-R. The yeast nuclear gene MRP-L13 codes for a protein of the large subunit of the mitochondrial ribosome. Curr Genet. 1994;26:8–14. doi: 10.1007/BF00326298. [DOI] [PubMed] [Google Scholar]
- 33.Haffter P, Fox T D. Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and PET127. Mol Gen Genet. 1992;235:64–73. doi: 10.1007/BF00286182. [DOI] [PubMed] [Google Scholar]
- 34.Haffter P, McMullin T W, Fox T D. A genetic link between an mRNA-specific translational activator and the translation system in yeast mitochondria. Genetics. 1990;125:495–503. doi: 10.1093/genetics/125.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haffter P, McMullin T W, Fox T D. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991;127:319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 37.Hochuli E. Purification of recombinant proteins with metal chelate adsorbent. In: Setlow J K, editor. Genetic engineering, principle and methods. Vol. 12. New York, N.Y: Plenum Press; 1990. pp. 87–98. [DOI] [PubMed] [Google Scholar]
- 38.Kloeckener-Gruissem B, McEwen J E, Poyton R O. Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene in Saccharomyces cerevisiae. J Bacteriol. 1988;170:1399–1402. doi: 10.1128/jb.170.3.1399-1402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotter P, Entian K D. Cloning and analysis of the nuclear gene MRP-S9 encoding mitochondrial ribosomal protein S9 of Saccharomyces cerevisiae. Curr Genet. 1995;28:26–31. doi: 10.1007/BF00311878. [DOI] [PubMed] [Google Scholar]
- 40.Lupski J R, Smiley B L, Godson G N. Regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon and the initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1983;189:48–57. doi: 10.1007/BF00326054. [DOI] [PubMed] [Google Scholar]
- 41.Marykwas D L, Fox T D. Control of the Saccharomyces cerevisiae regulatory gene PET494: transcriptional repression by glucose and translational induction by oxygen. Mol Cell Biol. 1989;9:484–491. doi: 10.1128/mcb.9.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMullin T W, Haffter P, Fox T D. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990;10:4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mieszczak M, Kozlowski M, Zagorski W. Protein composition of Saccharomyces cerevisiae mitochondrial ribosomes. Acta Biochim Pol. 1988;35:105–118. [PubMed] [Google Scholar]
- 44.Mulero J J. Studies on the translation of the yeast mitochondrial COX2 mRNA with emphasis on its translational activator PET111. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1993. [Google Scholar]
- 45.Mulero J J, Fox T D. Alteration of the Saccharomyces cerevisiae COX2 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulero J J, Fox T D. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulero J J, Fox T D. Reduced but accurate translation from a mutant AUA initiation codon in the mitochondrial COX2 mRNA of Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:383–390. doi: 10.1007/BF00281787. [DOI] [PubMed] [Google Scholar]
- 48.Muralikrishna P, Cooperman B S. A photolabile oligodeoxyribonucleotide probe of the decoding site in the small subunit of the Escherichia coli ribosome: identification of neighboring ribosomal components. Biochemistry. 1994;33:1392–1398. doi: 10.1021/bi00172a015. [DOI] [PubMed] [Google Scholar]
- 49.Myers A M, Crivellone M D, Tzagoloff A. Assembly of the mitochondrial membrane system: MRP1 and MRP2, two yeast nuclear genes coding for mitochondrial ribosomal proteins. J Biol Chem. 1987;262:3388–3397. [PubMed] [Google Scholar]
- 50.Myers A M, Pape L K, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ooi B G, Lukins H B, Linnane A W, Nagley P. Biogenesis of mitochondria: a mutation in the 5′-untranslated region of yeast mitochondrial oli1 mRNA leading to impairment in translation of subunit 9 of the mitochondrial ATPase complex. Nucleic Acids Res. 1987;15:1965–1977. doi: 10.1093/nar/15.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orr-Weaver T L, Szostak J W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Partaledis J A, Mason T L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol Cell Biol. 1988;8:3647–3660. doi: 10.1128/mcb.8.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poutre C G, Fox T D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G D, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 56.Rose M D, Broach J R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 57.Rosenblum-Vos L S, Rhodes L, Evangelista C C, Jr, Boayke K A, Zitomer R S. The ROX3 gene encodes an essential nuclear protein involved in CYC7 gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5639–5647. doi: 10.1128/mcb.11.11.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Shamu, C., and J. Nunnari. Personal communication.
- 60.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 61.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steele D F, Butler C A, Fox T D. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinstock K G, Strathern J N. Molecular genetics in Saccharomyces kluyveri: the HIS3 homolog and its use as a selectable marker gene in S. kluyveri and Saccharomyces cerevisiae. Yeast. 1993;9:351–361. doi: 10.1002/yea.320090405. [DOI] [PubMed] [Google Scholar]
- 67.Wiesenberger G, Costanzo M C, Fox T D. Analysis of the Saccharomyces cerevisiae mitochondrial COX3 mRNA 5′-untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995;15:3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wool I G, Chan Y-L, Glück A. Mammalian ribosomes: the structure and the evolution of the proteins. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 685–732. [Google Scholar]
- 69.Yang D, Oyaizu Y, Oyaizu H, Olsen G J, Woese C R. Mitochondrial origins. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]