Abstract
It has been suggested that DNA contains sacrificial nucleobase sequences that protect sensitive regions of the genome from oxidative damage. Oxidation of DNA by loss of an electron generates a radical cation that can migrate long distances by hopping. The radical cation can be trapped irreversibly at certain sites (GG steps) by reaction with H2O or O2 leading to the formation of lesions (oxidative damage). A series of DNA oligomers that contain regularly spaced GG steps and an 8-oxo-7,8-dihydroguanine (8-oxoG), which serves as a proxy for possibly sacrificial protective low oxidation potential sites, was prepared and analyzed. We find that in certain special sequences of DNA nucleobases that 8-oxoG protects remote GG steps from oxidative damage but that this is not a general phenomenon extending to normal mixed sequence DNA. This is a consequence of the change in the relative rate of charge hopping compared with trapping of the radical cation. When hopping is relatively slow, 8-oxoG exerts no protective effect. Thus, it seems unlikely that low oxidation potential sequences play a meaningful part in protecting mixed sequence DNA from damage.
INTRODUCTION
It is generally accepted that oxidative damage to DNA is a major cause of mutations that lead to disease and aging (1). Consequently, it is critically important that living systems maintain the fidelity of their genetic information in the face of an onslaught of oxidative attacks by ionizing radiation, reactive oxygen species and photooxidation. One approach has been the evolution of sophisticated repair systems that detect and correct the lesions in DNA that result from oxidative reactions. Not surprisingly, repair system malfunctions lead to the rapid build up of mutations with disastrous consequences (2). Correspondingly, it is suspected that certain intracellular antioxidants help prevent damage to DNA in the first place (3,4). Of special significance to the work reported here is the suggestion that DNA itself contains sacrificial nucleobases sequences that protect sensitive regions of the genome from oxidative damage (5–7). This proposal is based on the finding that one-electron oxidation of DNA introduces a radical cation that may migrate long distances by hopping through the duplex before being trapped by reaction with water or O2 at a relatively low oxidation potential (EOX) site to generate a lesion (8–12). It has been suggested that concentration of these reactive sites in non-critical regions of the DNA (e.g. introns or intergenic domains) will reduce the oxidative load on important genomic domains and consume a radical cation without the concomitant formation of a deleterious mutation (7). The general validity of this proposal relies on efficient migration of the radical cation to a remote trap through oligomers containing nucleobases of various sequences. We carried out experiments designed to assess this aspect of the proposal.
Lesions in DNA formed from the trapping of a radical cation are generally found at sites of low EOX such as (GG)n (n = 2 or 3) sequences (13). 8-Oxo-7,8-dihydroguanine (8-oxoG), a product of guanine oxidation (14,15) has an EOX that is ∼0.4 V below guanine (16). Consequently, 8-oxoG has been observed to be far more susceptible to oxidative reactions than the normal DNA bases (17), and this observation has led to the suggestion that it plays a protective role in genomic DNA (18). In this work, 8-oxoG is used as a proxy for sacrificial low oxidation potential sites in DNA. A series of oligonucleotide constructs that contain GG steps separated by bridges containing A/T base pairs and a strategically located 8-oxoG was prepared and investigated. For certain nucleobase sequences, a remote 8-oxoG protects all of the GG steps in the oligomer from oxidative damage, for others this protective effect is greatly diminished.
MATERIALS AND METHODS
Terminal dinucleotide terminase (TdT) enzyme and [α-32P]ATP were purchased from GE Health Sciences. T4 polynucleotide kinase (T4 PNK) was purchased from New England Biolabs. DNA oligomers and anthraquinone (AQ) containing oligomers were synthesized on an automated Expedite 8909 DNA synthesizer. 8-Oxo-dG phosphoramidite and other nucleotide phosphoramidites were obtained from Glen Research and used as received. The sequences containing 8-oxoG were deprotected in a solution of 2-mercaptoethanol in ammonium hydroxide (0.25 M) for 17 h at 55°C to avoid oxidative degradation. The oligonucleotides were purified by means of reversed phase high-performance liquid chromatography (HPLC) on a Hitachi preparative HPLC system using a Dynamax C18 column. Purified oligomers were desalted and characterized by mass spectroscopy.
Preparation of radiolabeled DNA
The oligomers were radiolabeled at the 3′ end using [α-32P] ATP and TdT enzyme. A 5 µl sample of desired single-stranded DNA was incubated with that of [α-32P]ATP and TdT enzyme at 37°C for ∼45 min. After incubation, the DNA sample was suspended in a denaturing loading dye and was purified on a 20% non-denaturing polyacrylamide gel. The desired DNA band was excised from the gel and eluted at 37°C for 12 h. The DNA was precipitated from the supernatant by addition of cold ethanol and glycogen. The mixture was vortexed, placed on dry ice for a short duration and centrifuged at 12 000 g for 30 min. The supernatant was removed and the residual DNA was washed with ethanol and air-dried. Suitable volumes of water were added for further experimentation.
UV irradiation and cleavage analysis
Samples for irradiation were prepared by hybridizing a mixture of unlabeled (5.0 µM) and radiolabeled (10 000 c.p.m.) oligonucleotides with complementary DNA in a pH ∼7.0 sodium phosphate buffer solution. Hybridization was achieved by heating the samples at 90°C for 10 min, followed by slow cooling to room temperature. Samples were irradiated at ∼30°C in microcentrifuge tubes in a Rayonet Photoreactor (Southern New England Ultraviolet Co., Barnsford, CT) equipped with four 350 nm lamps. After irradiation, the samples were precipitated once with cold ethanol and glycogen. The precipitated samples were washed twice with 80% ethanol, dried and treated with piperidine at 90°C for 30 min. After evaporation of the piperidine and co-evaporation with water, the samples were dissolved in denaturing loading dye and analyzed on a 20% 19:1 acrylamide:bisacrylamide gel. The gels were dried and the cleavage sites were visualized by autoradiography.
RESULTS
Design of oligonucleotides
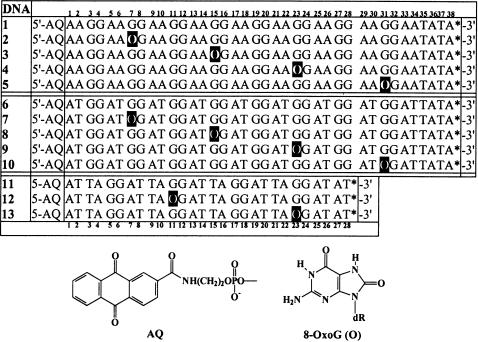
The series of duplex DNA oligomers used in this work is shown in Figure 1. In the first group, DNA(1) through DNA(5), one strand has the sequence d[(AAXG)8AAATAT] where X is guanine or 8-oxoG (abbreviated O in Figure 1) and the second strand, d[(TTCC)8TTTATA], is fully complementary to the first. An AQ group is linked covalently to the 5′ end of the first strand and a 32P-radiolabel (indicated by asterisk on Figure 1) is incorporated at its 3′ end. Similarly, oligomers in the second group, DNA(6) through DNA(10), have the structure d[(ATXG)8ATAT]/d[ATAT(CCAT)8], and the third group, DNA(11) through DNA(13), has the structure d[(ATTAXG)4ATAT/d[ATAT(CCTAAT)4]. Thus, DNA oligomers 1, 6 and 11 contain a succession of GG steps separated by AA, AT or ATTA sequences, respectively. All of the other oligomers shown in Figure 1 have one 8-oxoG in place of a 5′-G in the succession of GG steps. The position of the 8-oxoG is varied systematically in these series. For example, in DNA(2) it is at a position 7 bp (O7) from the 5′ end that is linked to the AQ.
Figure 1.
Structures of DNA oligomers. One strand is shown; the second strand is fully complementary with C opposite O.
The UV irradiation of an AQ group covalently-linked to DNA causes the one-electron oxidation of an adjacent base, and thus the radical cation that results must reside temporarily on intervening nucleobases before it encounters an 8-oxoG. In DNA(5), the 8-oxoG is 31 bp from the AQ and the intervening sequence contains seven GG steps that are potential traps for the radical cation. In the other oligomers of the first group, the 8-oxoG is located at intermediate positions between the AQ and the last GG step. By this construction, we systematically probed the distance dependence of the remote 8-oxoG oxidative protective effect on GG steps separated by AA bridges. Similarly, in the oligomers of second and third groups the effect of AT and ATTA sequences bridging the GG steps was investigated. The results show that the nature of the bridging groups significantly affects the 8-oxoG protective effect.
Characterization: melting temperature (Tm) and circular dichroism (CD) studies
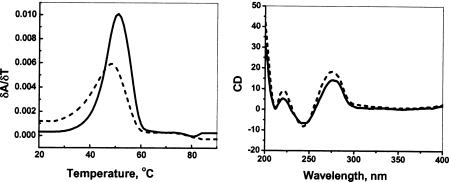
It is known that 8-oxoG in duplex DNA forms normal Watson–Crick base pairs with cytosine (19). The duplex oligomers prepared here that contain 8-oxoG in place of G were characterized by their Tm and by CD spectroscopy to confirm that this substitution does not cause an unanticipated change in oligomer stability or to its gross structure. The melting data show no significant changes in Tm when 8-oxoG is incorporated. For example, DNA(1) is observed to melt cooperatively at 47°C and DNA(2), where there is a substitution for G by 8-oxoG, has a Tm of 46°C. Similarly, the CD spectra (20) of each of the duplexes examined are indistinguishable and show that these oligomers adopt the expected B-form structure in solution (Figure 2).
Figure 2.
Tm and CD spectral data of DNA(1) (solid line) and DNA(3) (dashed line).
Irradiation of the DNA oligomers: one-electron oxidation and strand cleavage
Irradiation of an AQ-linked DNA oligomer at 350 nm causes the electronic excitation of the AQ group and initiates a charge transfer reaction that generates a base radical cation and the AQ radical anion. The AQ radical anion is consumed rapidly by reaction with O2 to form superoxide, and the radical cation migrates through the DNA until it is consumed irreversibly by reaction at a low oxidation potential site. Treatment of the irradiated sample with piperidine results in strand cleavage that is detected by PAGE and autoradiography for oligomers labeled with [α-32P]ATP at the 3′-terminus of the strand that contains the GG steps. Reaction of the guanine radical cation gives 8-oxoG, which reacts slowly with piperidine, and other products that give rapid strand cleavage under these conditions (21–23). These reactions are carried out to low conversion (single-hit conditions) where each DNA molecule reacts once or not at all. This condition was confirmed by comparing reactions carried to different extents, the data are shown in the Supplementary Data to this paper. Under these conditions, the observed strand cleavage pattern reflects the reactive distribution of the radical cation in the DNA oligomer. That is, the greatest amount of strand cleavage is detected at those sites where the likelihood of an irreversible trapping reaction of the radical cation is highest (24).
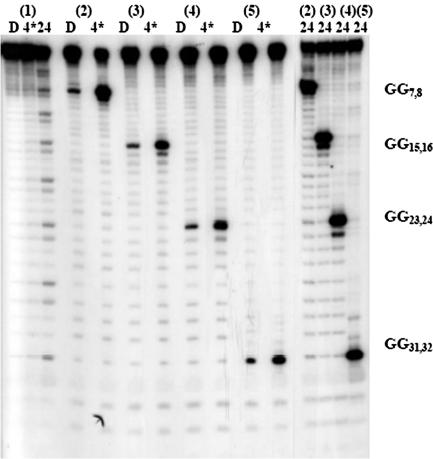
DNA(1) contains eight regularly spaced GG steps, the closest (G7) is ∼7.2 Å from the AQ group and the farthest (G32) is >113 Å from that point. However, the amount of strand cleavage at each GG step resulting from irradiation of the AQ group is comparable (Figure 3), which means that migration of the radical cation through this oligomer occurs very efficiently (24). In contrast, piperidine treatment of UV-irradiated DNA(2) results in meaningful strand cleavage only at the 8-oxoG at O7. Oxidation of 8-oxoG forms compounds that are piperidine labile (22). No significant strand cleavage is seen at the GG steps in DNA(2) that precede (G4) or follow (G12,20,24,28,32) the 8-oxoG even after 24 min of irradiation, which is beyond single-hit conditions. Similarly for DNA(3), DNA(4) and DNA(5), the presence of an 8-oxoG protects each of the GG steps from oxidative damage. These experiments show that for the DNA oligomers of this group, where the GG steps are separated by AA bridges, charge migration through the (AAGG)n segment is much faster than its consumption at a GG step. However, the radical cation is irreversibly consumed the first time it encounters an 8-oxoG and thus no reaction is detected at any other nucleobase in the oligomer.
Figure 3.
Autoradiogram from the irradiation of DNA(1)–DNA(5). D is dark control (no irradiation), 4* is 4 min of irradiation (4 lamps), 24 is 24 min of irradiation (8 lamps). DNA(7)–DNA(10) contain 8-oxoG at various positions. All the irradiated samples have been treated with 1 M piperidine before gel electrophoresis, which causes rapid strand cleavage at some oxidized guanine products (22) but slow reaction at 8-oxoG (25). The minor amount of strand cleavage at the 8-oxoG site in the dark control lanes is presumably due to oxidation of 8-oxoG during reaction (22).
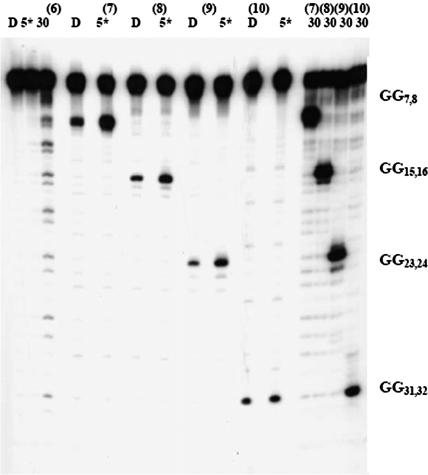
The results of photooxidation of the second group of oligomers, DNA(6–10), studied are somewhat different. In the case of DNA(6), there are eight GG steps separated by AT bridges. Its irradiation leads to reaction at each GG step, but there is somewhat more strand cleavage at those that are closer to the AQ than those that are farther away. This indicates that the rates for charge migration and trapping in the (ATGG)8 segment of this compound are of comparable magnitude (24). However, just as was observed for the first group, an 8-oxoG protects all of the GG steps that precede or follow it even after extensive irradiation (30 min, Figure 4).
Figure 4.
Autoradiogram from the irradiation of DNA(6)–DNA(10). D is dark control, 5* is 5 min of irradiation with 4 lamps and 30 is 30 min of irradiation with 8 lamps. DNA(6)–DNA(10) contains 8-oxoG at various positions. All the irradiated samples have been treated with 1 M piperidine before gel electrophoresis.
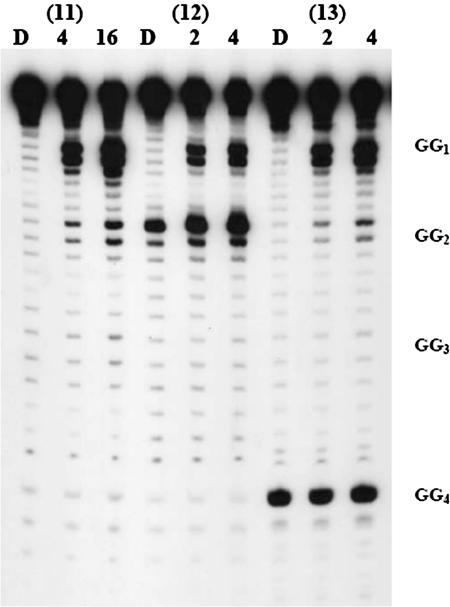
The oligomers studied in the final group, DNA(11–13), contain four GG steps separated by ATTA sequences. In contrast to the result of irradiation of DNA(1) and DNA(6), irradiation of DNA(11) gives an unequal distribution of strand cleavage efficiencies (Figure 5). The GG step closest to the AQ is far more reactive than those that are farther away, and the amount of strand cleavage falls off approximately exponentially with distance from the AQ group. The amount of reaction at the farthest position, G23, is not detectable above background. This behavior indicates that the rate of radical cation hopping is slower than its trapping (24). Most significantly, examination of DNA(12) and DNA(13) show that in these cases the presence of an 8-oxoG does not protect GG steps that precede it from oxidative damage. The GG at position 6 of DNA(12) is damaged by irradiation of the AQ even though this compound contains an 8-oxoG at position 11. Similarly, for DNA(13) the GG steps at positions 6, 12 and 18 are not protected by the 8-oxoG at position 23. These findings show that the protective effect of an 8-oxoG is strongly dependent on the specific sequence of nucleobases in its immediate environment.
Figure 5.
Autoradiogram from the irradiation of DNA(11)–DNA(13). 8-OxoG is incorporated at GG2 or GG4. D is dark control, 4 and 16 are times of irradiation (in min; 2 lamps). All the samples were treated with 1 M piperidine after irradiation.
DISCUSSION
Effect of 8-oxoG on strand cleavage
Irradiation of an AQ-linked duplex DNA at 350 nm, where only the AQ absorbs, injects a radical cation that migrates through the oligomer (21,23,24). Under these conditions the radical cation will be trapped irreversibly by reaction with H2O or O2 to create a ‘damaged’ DNA nucleobase. Subsequent treatment of the irradiated oligomer with piperidine results in strand cleavage at the site of damage (22). The amount of strand cleavage at a particular base at low conversion reveals the reactive distribution of the radical cation among the sites of the oligomer. If the rate of radical cation hopping (khop) is much faster than the rate of irreversible trapping (ktrap), then the radical cation will be distributed equally among all equivalent sites. If ktrap is comparable with or faster than khop, then reaction at the GG steps closer to the AQ predominates (24).
Irradiation of DNA(1) and subsequent piperidine treatment results in strand cleavage at each of the eight GG steps in this compound. Within experimental error, the amount of cleavage is the same at each GG step, Figure 3, which indicates that khop from GG to GG across the AA bridges is much faster than ktrap at the GG step. In contrast, no significant strand cleavage at any GG step is detected from irradiation of DNA(2); instead, reaction occurs exclusively at the 8-oxoG located 7 bp (O7) from the AQ. Similarly, strand cleavage is detected only at O31 of DNA(5), which is ∼107 Å from the AQ and is preceded by seven GG steps. Related results are obtained from irradiation of DNA oligomers 3 and 4: strand cleavage occurs exclusively at the 8-oxoG and not at any of the GG steps that precede or follow it. Clearly, 8-oxoG protects guanines from oxidative reaction in DNA oligomers comprised of repeating AAGG sequences. These findings confirm that hopping to an 8-oxoG in these cases is much faster than irreversible reaction at a GG step and that 8-oxoG is a deep trap from which the radical cation cannot escape before it is consumed by a reaction that leads to strand cleavage. Evidently, for DNA comprised of nucleotides in a repeating (AAGG) sequence, 8-oxoG, a proxy for low oxidation potential sites generally, protects the other guanines in the oligomer from oxidative damage by collecting and trapping all of the radical cations. This protective role of 8-oxoG depends on the nucleobase composition of the oligomer because that controls the ratio of khop and ktrap. We explored DNA oligomers 6 through 13 to determine the result of base composition on the protective effect.
The results from irradiation of DNA(6–10), which contains an (ATGG)8 sequence, are similar to those obtained from DNA(1–5). For DNA(6), which does not contain an 8-oxoG, all eight GG steps react but there is a slight distance dependence, which indicates that khop and ktrap are of similar magnitude. However, when an 8-oxoG is substituted for a 5′-G in a GG step of DNA(6), and DNA oligomers 7 through 10, there is no detectable reaction at the GG steps. Thus, hopping through the AT bridges is sufficiently fast so that irreversible capture of the radical cation by the 8-oxoG provides significant protection for preceding and following GG steps.
A different result is obtained from irradiation of DNA oligomers 11 through 13 where the GG steps are separated by ATTA sequences. For DNA(11), which does not contain an 8-oxoG, the amount of strand cleavage observed at the GG steps falls off rapidly with distance from the AQ. This is the expected behavior when khop is significantly less than ktrap (24). In DNA(12), an 8-oxoG replaces the 5′-G in the second GG step of DNA(11). In this case, the 8-oxoG does not protect the preceding GG step from reaction. Similarly, for DNA oligomer 13, replacement of a G by 8-oxoG provides no detectable protective effect for any preceding guanine. Quite clearly, the ability of a low oxidation potential site such as 8-oxoG to protect remote guanines from oxidation is highly dependent on the local sequence of nucleobases.
Protection of DNA: 8-oxoG as a proxy for low oxidation potential sites
The experiments reported here were designed as an analysis of the idea that long-distance radical cation migration in DNA has been adopted by nature as a mechanism to protect sensitive genomic regions from oxidative damage (5–7). Transfer of a radical cation through intervening DNA to low oxidation potential sites (cathodic protection) could channel it away from regions where lesions may do damage. To test this hypothesis, we used 8-oxoG as a generic proxy for low oxidation potential sites, and assessed the influence of the intervening DNA sequence on the effectiveness of the remote protective effect.
For certain sequences, where charge hopping is faster than trapping, the 8-oxoG is able to exert a powerful protective effect. In essence, its presence in these oligonucleotides renders damage at GG steps undetectable. However, in other circumstances the 8-oxoG is completely ineffective in protecting GG steps from oxidative damage. This is a consequence of the change in the relative rate of charge hopping compared with trapping of the radical cation by reaction. In particular, when charge hopping is slowed, as it is for example in DNA(11) by the imposition of an ATTA sequence between GG steps, the 8-oxoG exerts no protective effect.
It is clear from the results reported here and from related studies (24,26) that the specific sequence of nucleic acid bases in an oligomer determines the distance dependence and the efficiency of radical cation migration. For certain sequences such as (AAGG)n where khop is much greater than ktrap, low EOX sites can exert a protective effect on guanine oxidation over a significant distance. However for other sequences where khop is not much greater than ktrap, the protective effect of a low EOX site will be extremely localized. Normal DNA is mainly comprised of sequences where khop is less than ktrap, thus sacrificial low oxidation potential sites, such as 8-oxoG, can only be locally protective. From these results and recent findings from investigation of radiation damage in cells (27), it is unlikely that sacrificial sites play a meaningful part in protecting DNA from oxidative damage in mixed sequence oligomers. In genomic DNA, radical cation hopping from strand to strand may provide a means to circumvent this restriction (28). We are currently exploring this possibility.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation and by The Vassar Woolley Foundation, for which we are grateful. Funding to pay the Open Access publication charges for this article was provided by The Vassar Woolley Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Poulsen H.E., Prieme H., Loft S. Role of oxidative DNA damage in cancer initiation and promotion. Eur. J. Cancer. Prev. 1998;7:9–16. [PubMed] [Google Scholar]
- 2.Boiteux S., Gellon L., Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 2002;15:1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 3.Malins D.C., Hellstrom K.E., Anderson K.M., Johnson P.M., Vinson M.A. Antioxidant-induced changes in oxidized DNA. Proc. Natl Acad. Sci. USA. 2002;99:5937–5941. doi: 10.1073/pnas.082111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanvah S., Schuster G.B. Long-range oxidative damage to DNA: protection of guanines by a nonspecifically bound disulfide. J. Am. Chem. Soc. 2002;124:11286–11287. doi: 10.1021/ja027259x. [DOI] [PubMed] [Google Scholar]
- 5.Heller A. On the hypothesis of cathodic protection of genes. Faraday Discuss. 2000;116:1–13. doi: 10.1039/b006196o. [DOI] [PubMed] [Google Scholar]
- 6.Friedman K.A., Heller A. On the non-uniform distribution of guanine in introns of human genes: possible protection of exons against oxidation by proximal intron poly-G sequences. J. Phys. Chem. B. 2001;105:11859–11865. [Google Scholar]
- 7.Friedman K.A., Heller A. Guanosine distribution and oxidation resistance in eight eukaryotic genomes. J. Am. Chem. Soc. 2004;126:2368–2371. doi: 10.1021/ja038217r. [DOI] [PubMed] [Google Scholar]
- 8.Ly D., Kan Y., Armitage B., Schuster G.B. Cleavage of DNA by irradiation of substituted anthraquinones: intercalation promotes electron transfer and efficient reaction at GG steps. J. Am. Chem. Soc. 1996;118:8747–8748. [Google Scholar]
- 9.Henderson P.T., Jones D., Hampikian G., Kan Y., Schuster G.B. Long-distance charge transport in duplex DNA: the phonon-assisted polaron-like hopping mechanism. Proc. Natl Acad. Sci. USA. 1999;96:8353–8358. doi: 10.1073/pnas.96.15.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giese B. Long-distance electron transfer through DNA. Annu. Rev. Biochem. 2002;71:51–70. doi: 10.1146/annurev.biochem.71.083101.134037. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill M.A., Barton J.K. DNA charge transport: conformationally gated hopping through stacked domains. J. Am. Chem. Soc. 2004;126:11471–11483. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 12.Schuster G.B., editor. Topics in Current Chemistry. Vol. 236. Heidelberg: Springer-Verlag; 2004. Long-range charge transfer in DNA I; p. 237. [Google Scholar]
- 13.Lewis F.D., Liu X., Liu J., Hayes R.T., Waielswski M.R. Dynamics and equilibria for oxidation of G, GG, and GGG sequences in DNA hairpins. J. Am. Chem. Soc. 2000;123:12037–12038. [Google Scholar]
- 14.Gasper S.M., Schuster G.B. Intramolecular photo-induced electron transfer to anthraquinones linked to duplex DNA: the effect of gaps and traps on long range radical cation migration. J. Am. Chem. Soc. 1997;119:12762–12771. [Google Scholar]
- 15.Cai Z., Sevilla M.D. Electron and hole transfer from DNA base radicals to oxidixed products of guanine in DNA. Radiat. Res. 2003;159:411–419. doi: 10.1667/0033-7587(2003)159[0411:eahtfd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Sheu C., Foote C.S. Solvent and electronic effects on the reaction of guanosine derivatives with singlet oxygen. J. Org. Chem. 1995;60:4498–4503. [Google Scholar]
- 17.Ravanat J.-L., Saint-Pierre C., Cadet J. One-electron oxidation of the guanine moiety of 2′-deoxyguanosine: influence of 8-oxo-7,8-dihydro-2′-deoxyguanosine. J. Am. Chem. Soc. 2003;125:2030–2031. doi: 10.1021/ja028608q. [DOI] [PubMed] [Google Scholar]
- 18.Kim J.E., Choi S., Yoo J.A., Chung M.H. 8-Oxoguanine induces intramolecular DNA damage but free 8-oxoguanine protects intermolecular DNA from oxidative stress. FEBS Lett. 2004;556:104–110. doi: 10.1016/s0014-5793(03)01385-1. [DOI] [PubMed] [Google Scholar]
- 19.Lipscomb L.A., Peek M.E., Morningstar M.L., Verghis S.M., Miller E.M., Rich A., Essigmann J.M., Williams L.D. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray D.M., Hung S.-H., Johnson K.H. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–36. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 21.Schuster G.B. Long-range charge transfer in DNA: transient structural distortions control the distance dependence. Acc. Chem. Res. 2000;33:253–260. doi: 10.1021/ar980059z. [DOI] [PubMed] [Google Scholar]
- 22.Burrows C.J., Muller J.G. Oxidative nucleobase modifications leading to strand cleavage. Chem. Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 23.Henderson P.T., Boone E., Schuster G.B. Bulged guanine is uniquely sensitive to damage caused by visible light irradiation of ethidium bound to DNA: a possible role in mutagenesis. Helv. Chim. Acta. 2002;85:135–151. [Google Scholar]
- 24.Liu C.-S., Hernandez R., Schuster G.B. The mechanism of radical cation transport in DNA. J. Am. Chem. Soc. 2004;126:2877–2884. doi: 10.1021/ja0378254. [DOI] [PubMed] [Google Scholar]
- 25.Cullis P.M., Malone M.E., Merson-Davies L.A. Guanine radical cations are precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J. Am. Chem. Soc. 1996;118:2775–2781. [Google Scholar]
- 26.Misiaszek M., Crean C., Joffe A., Geacintov N.E., Shafirovich V. Oxidative DNA damage associated with combination of guanine and superoxide radicals and repair mechanisms via radical trapping. J. Biol. Chem. 2004;279:32106–32115. doi: 10.1074/jbc.M313904200. [DOI] [PubMed] [Google Scholar]
- 27.Pouget J.P., Frelon S., Ravant J.-L., Testard I., Odin F., Cadet J. Formation of modified DNA bases in cells exposed to either gamma radiation or to high –LET particles. Radiat. Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Shukla L.I., Adhikary A., Pazdro R., Becker D., Sevilla M.D. Formation of 8-oxo-7,8-dihydroguanine-radicals in gamma-irradiated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004;32:6565–6574. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.