Abstract
One of the few rRNA modifications conserved between bacteria and eukaryotes is the base dimethylation present at the 3′ end of the small subunit rRNA. In the yeast Saccharomyces cerevisiae, this modification is carried out by Dim1p. We previously reported that genetic depletion of Dim1p not only blocked this modification but also strongly inhibited the pre-rRNA processing steps that lead to the synthesis of 18S rRNA. This prevented the formation of mature but unmodified 18S rRNA. The processing steps inhibited were nucleolar, and consistent with this, Dim1p was shown to localize mostly to this cellular compartment. dim1-2 was isolated from a library of conditionally lethal alleles of DIM1. In dim1-2 strains, pre-rRNA processing was not affected at the permissive temperature for growth, but dimethylation was blocked, leading to strong accumulation of nondimethylated 18S rRNA. This demonstrates that the enzymatic function of Dim1p in dimethylation can be separated from its involvement in pre-rRNA processing. The growth rate of dim1-2 strains was not affected, showing the dimethylation to be dispensable in vivo. Extracts of dim1-2 strains, however, were incompetent for translation in vitro. This suggests that dimethylation is required under the suboptimal in vitro conditions but only fine-tunes ribosomal function in vivo. Unexpectedly, when transcription of pre-rRNA was driven by a polymerase II PGK promoter, its processing became insensitive to temperature-sensitive mutations in DIM1 or to depletion of Dim1p. This observation, which demonstrates that Dim1p is not directly required for pre-rRNA processing reactions, is consistent with the inhibition of pre-rRNA processing by an active repression system in the absence of Dim1p.
In most eukaryotes, three of the four rRNA species are excised from a single large RNA polymerase I (Pol I) transcript (35S pre-rRNA in yeast) which is processed in a complex pathway involving both endonucleolytic cleavages and exonucleolytic digestions (Fig. 1) (reviewed in references 24, 42, and 46). The fourth mature species, 5S rRNA, is transcribed and processed independently. During pre-rRNA processing, many specific nucleotides within the rRNAs are covalently modified. The major types of posttranscriptional modification are the isomerization of uracil to pseudouridine, 2′-O methylation of the ribose moieties, and base methylation. In eukaryotes, interactions between trans-acting guide small nucleolar RNAs (snoRNAs) and pre-rRNA identify nucleotides that are to be modified by 2′-O methylation or pseudouridylation (8, 12, 20, 31, 32). Surprisingly, all of the guide snoRNAs tested so far in yeast are nonessential for growth, demonstrating that the corresponding modifications are dispensable.
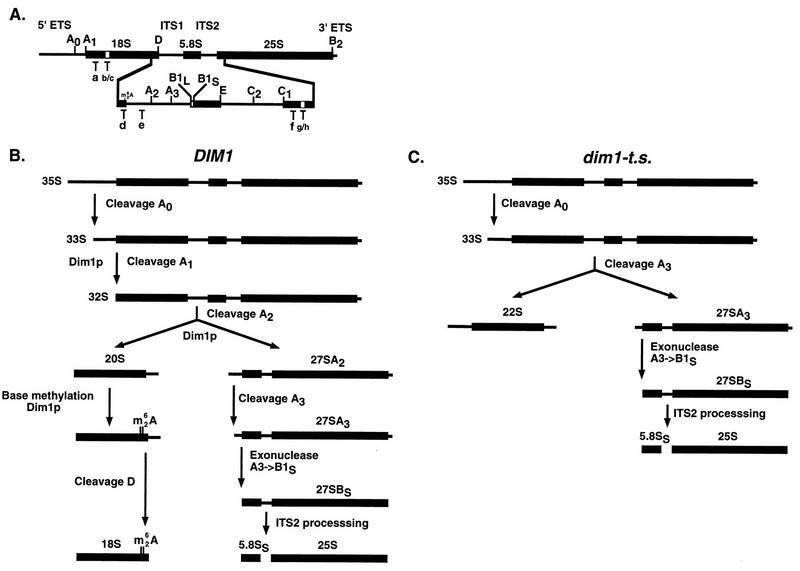
FIG. 1.
Structure of yeast 35S pre-rRNA and the pre-rRNA processing pathway. (A) 35S pre-rRNA. The sequences encoding the mature 18S, 5.8S, and 25S rRNAs are embedded in the 5′ and 3′ external transcribed spacers (ETS) and in ITS1 and ITS2. Sites of pre-rRNA processing are indicated with uppercase letters (A0 to E), and the oligonucleotides used for Northern hybridization and primer extension are indicated with lowercase letters (a to h). The site of dimethylation is denoted as m26A. (B) Pre-rRNA processing pathway. Processing of the 35S primary transcript starts at site A0 in the 5′ ETS. The resulting 33S pre-rRNA is processed at sites A1 and A2, giving rise successively to the 32S pre-rRNA and to the 20S and 27SA2 precursors. Cleavage at site A2 separates the pre-rRNAs destined for the small and large ribosomal subunits. The 20S precursor is dimethylated and endonucleolytically cleaved at site D to yield mature 18S rRNA. Cleavage of 27SA2 at site A3, by RNase MRP, is rapidly followed by exonucleolytic digestion to site B1S, generating the 27SBS precursor. Mature 25S rRNA and 7S pre-rRNA are released from 27SBS following cleavages at sites C1 and C2. 7S pre-rRNA undergoes a rapid 3′→5′ exonuclease digestion to site E, generating the mature 3′ end of 5.8S rRNA (not represented). For simplicity, only the major processing pathway, from 27SA2 to 5.8SS and 25S rRNA, is shown; an alternative pathway generates the minor 5.8SL rRNA, which is 7 to 9 nucleotides 5′ extended. The steps that require Dim1p are indicated. (C) Pre-rRNA processing in dim1 TS strains. 35S pre-rRNA is cleaved normally at site A0. 33S pre-rRNA accumulates and is cleaved at site A3, providing the 27SA3 that is normally processed to 5.8SS and 25S and the aberrant 22S pre-rRNA that is not dimethylated and not processed to 18S rRNA.
The only base modifications that have been studied in a eukaryote are the m26Am26A doublet present at the 3′ end of the small subunit (SSU) rRNA (18S rRNA). In the yeast Saccharomyces cerevisiae, these modifications are made by the Dim1p dimethylase. In contrast to 2′-O methylation and pseudouridylation, 18S dimethylation does not appear to involve a guide snoRNA cofactor. Moreover, expression of Dim1p in Escherichia coli is able to restore dimethylation to ksgA mutants that lack this activity, presumably showing that Dim1p is able to function in the absence of snoRNA cofactors (23).
Dim1p is essential for viability (23), but the requirement for the dimethylated nucleotides was unclear since cells depleted of Dim1p not only lacked dimethylation but were also strongly defective in the pre-rRNA cleavages at sites A1 and A2 that generate 20S pre-rRNA (Fig. 1) (26). 20S pre-rRNA is the immediate precursor to 18S rRNA, so this had the effect of preventing the synthesis of mature but nonmodified rRNA. Pre-rRNA molecules in which the two dimethylated adenosine residues are replaced by guanosine, which cannot be modified, were processed normally, showing that the m26Am26A nucleotides are not themselves required for cleavage (26). Expression of 18S rRNA with the double G substitution did not support growth, but it was not clear whether the lack of dimethylation or the substitution of the A residues, which are themselves universally conserved in evolution, was responsible for this defect.
In E. coli, ksgA strains that lack dimethylation are resistant to kasugamycin, an antibiotic belonging to the aminoglycoside family (14, 15). ksgA strains are only marginally affected for growth, but ksgA extracts show various defects in translation in vitro: (i) initiation requires a higher concentration of IF3 in the absence of IF1 (35), (ii) accuracy is affected (43), and (iii) the affinity between the subunits is decreased (34; reviewed in reference 45). This is consistent with the localization of the m26Am26A residues at the interface between the ribosomal subunits at a site where crucial interactions take place during translation (7, 29, 41). However, in vitro reconstitution experiments showed that neither dimethylation nor the twin adenosines are crucial for 30S subunit assembly and function (10, 22).
Previous analyses, therefore, had left two outstanding questions. What is the requirement for dimethylated nucleotides in the synthesis and function of eukaryotic ribosomes? What is the basis of the requirement for Dim1p in pre-rRNA processing reactions? Specifically, does a regulatory system monitor the association of Dim1p with pre-rRNA and inhibit processing in its absence, or does Dim1p have an additional function in pre-rRNA processing and/or ribosome assembly that is required for pre-rRNA cleavage? The approach that we adopted was to screen a library of conditionally lethal alleles of DIM1 in order to isolate mutations that uncouple the enzymatic function of Dim1p in methylation from its involvement in pre-rRNA processing.
MATERIALS AND METHODS
Media and plasmids.
Standard S. cerevisiae growth and handling techniques were used. 5-Fluoro-orotic acid (5-FOA) was used in minimal medium at 1 g/liter according to the recipe of Boeke et al. (4). The transformation procedure was as described by Gietz et al. (13).
Plasmid pDL31.42 (2μm URA3 DIM1) was isolated from a pFL44L (5)-based yeast genomic library as previously described (23). DIM1 was recovered from pDL31.42 as a 3,282-bp SmaI/XhoI fragment and subcloned in pFL36 (ARS CEN LEU2) (5) digested with PvuII/SmaI to give plasmid pTL6. A BamHI HIS3 cassette isolated from plasmid YDp-H (3) was filled and subcloned in the filled XbaI site of the terminator region of DIM1 on plasmids pTL6-dim1-2, pTL6-dim1-9 and pTL6-dim1-1 to give constructs pTL39, pTL42, and pTL43, respectively. Plasmid pTL17 is pJV12 (19) in which the LEU2 marker of pFL36 (5) was subcloned in SalI. In pTL29, the XhoI/SfiI ribosomal DNA (rDNA) fragment of pGAL::rDNA (16) was subcloned in pTL17. Plasmids pAT3 and pI12.34Δ33 and plasmid pHT4467 are gifts from T. Preiss (European Molecular Biology Laboratory [EMBL]) and H. Tekotte (EMBL), respectively.
Epitope-tagged versions of Dim1p were constructed in plasmid pTL6 by inserting three copies of the human c-Myc epitope at either the amino- or carboxy-terminal end of the protein (generating plasmids pTL18 and pTL25, respectively). For plasmid pTL18, an NcoI 3× Myc cassette was inserted at the ATG codon of DIM1 at the naturally occurring NcoI site. For the carboxy fusion, an in-frame SacI site was created at the end of the open reading frame (ORF) by site-directed mutagenesis and a SacI 3× Myc cassette was inserted. The 3× Myc cassettes were generated by PCR and have been described previously (25). All constructs were checked by sequencing.
Construction of a library of conditional alleles of DIM1.
The DIM1 ORF was mutagenized by PCR under the conditions described by Leung et al. (28). The primers used for amplification with pDL31.42 were 5′-TAAAATTATACCATGGGAAAGGCT-3′ and 5′-TGATAAGAGAGCTCATGAAAAATG-3′. The PCR product was subcloned as an NcoI/SacI fragment in plasmid pTL6, and the library was amplified in E. coli. Random sequencing revealed that the mutation rate was approximately 0.46%. The DIM1 ORF being ∼1 kb, this would give an average of 4 to 5 mutations per gene. Transitions, transversions, and insertions were detected.
Isolation of conditional alleles of DIM1.
The library was screened according to the cotransformation and plasmid-shuffling procedure described by Sikorski and Boeke (37). The shuffling strain was constructed as follows. A complete deletion of DIM1 was created in the diploid strain BMA38 by a one-step PCR strategy (2, 25). The oligonucleotides used for amplification with pRS313 (38) were 5′-GGTTATAAGATCGATAAATTAGGAACAGTGCTATTATACAGTCTC TTGGCCTCCTCTAG-3′ and 5′-TTTTTCTTATCTTAGGTAAATAGTATACAAGCACTTACATAATCGTTCAGAATGACACG-3′. The resulting strain, YDL303, was transformed with plasmid pDL31.42 and sporulated. Haploids containing the deletion rescued by pDL31.42 were identified (strains YDL304A and YDL304B [Table 1]).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or comments |
|---|---|---|
| D150 | a ura3-52 leu2-3,112 ade1-100 his4-519 GAL+ | BWG1-7A, L. Guarente |
| YDL302 | As D150 with URA3 GAL10::dim1 | 26 |
| BMA38 | a/α ade2-1/ade2-1 can1-100/can1-100 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 | B. Dujon |
| YDL303 | As BM38 with dim1Δ::HIS3/DIM1 | Full DIM1 deletion made by PCR in strain BMA38 |
| YDL304A | a ade2-1 can1-100 his3Δ200 leu2-3,112 trp1-1 ura3-1 dim1Δ::HIS3 + pDL31.42 (URA3 2μm DIM1) | Starting strain, segregant from YDL303 containing a URA3 2μm DIM1 plasmid |
| YDL304B | α ade2-1 can1-100 his3Δ200 leu2-3,112 trp1-1 ura3-1 dim1Δ::HIS3 + pDL31.42 (URA3 2μm DIM1) | YDL304A sister strain |
| YDL209 | a ade2-1 can1-100 his3Δ200 leu2-3,112 trp1-1 ura3-1 dim1Δ::HIS3 + pTL6 (LEU2 ARS/CEN DIM1) | YDL304A in which plasmid pDL31.42 has been shuffled with plasmid pTL6 |
| MBS | a ade2 his3 leu2 trp1 ura3 can1 L-0 M-0 | P. Sarnow (17) |
| YDL321 | As MBS with dim1-2 | dim1-2 integrated as a dim1-2/HIS3 cassette isolated from pTL39 |
| YDL324 | As MBS with dim1-1 | dim1-1 integrated as a dim1-1/HIS3 cassette isolated from pTL43 |
| CH1462 | α ade2 ade3 his3 leu2 ura3 can1 | 21 |
| YDL314 | As CH1462 with dim1-9 | dim1-9 integrated as a dim1-9/HIS3 cassette isolated from pTL42 |
| YDL315 | As CH1462 with dim1-1 | dim1-1 integrated as a dim1-1/HIS3 cassette isolated from pTL43 |
| YDL300 | a/α ura3-52/ura3-52 lys2-801amber/lys2-801amber ade2-101ochre/ade2-101ochre trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 dim1Δ::URA3/DIM1 | 23 |
| YDL100A | a; as YDL300 + pTL6 (LEU2 ARS/CEN DIM1) | dim1Δ::URA3 segregant from YDL300 |
| YDL101A | a; as YDL300 + pTL18 (LEU2 ARS/CEN 3× Myc-Dim1p) | dim1Δ::URA3 segregant from YDL300 |
| YDL102A | a; as YDL300 + pTL25 (LEU2 ARS/CEN Dim1p-3× Myc) | dim1Δ::URA3 segregant from YDL300 |
The library was transformed into strain YDL304A and plated on minimal medium lacking histidine and leucine (SD −His −Leu) at 30°C. One thousand eight hundred transformants were patched onto SD −His −Leu at 25°C. The resulting master plates were replica plated on 5-FOA at 25°C. After two rounds of selection on 5-FOA, the 5-FOA-resistant clones were tested for growth at various temperatures (18, 25, 30, and 37°C). Approximately 11% of the clones remained 5-FOA sensitive, probably due to nonconditional inactivation. Thirty-two thermosensitive (TS) clones were isolated, some of which were also slightly cryosensitive (CS). No clones showing only a CS phenotype were recovered. Ten conditional alleles, dim1-1 to dim1-10, were selected to be further characterized.
RNA extraction, Northern hybridization, and primer extension.
RNA extraction, Northern hybridization, and primer extension were as described previously (26). Oligonucleotides a, b, d, e, f, and g were named d, e, f, g, l, and m, respectively, in reference 26. Oligonucleotides c and h are TCTCTTCCAAAGGGTCG and GCACCGAAGGTACCAG, respectively. RNA analysis of dim1-1 to dim1-10 alleles were performed in strain YDL304A cured of plasmid pDL31.42 and bearing the corresponding pTL6-dim1-t.s. plasmid. The reference wild-type strain used was YDL209 (Table 1).
For the experiment presented in Fig. 7, dim1-1 HIS3 and dim1-9 HIS3 integrative cassettes were recovered from plasmids pTL43 and pTL42 by EcoRI/XhoI digestion and transformed in strain CH1462 (21).
FIG. 7.
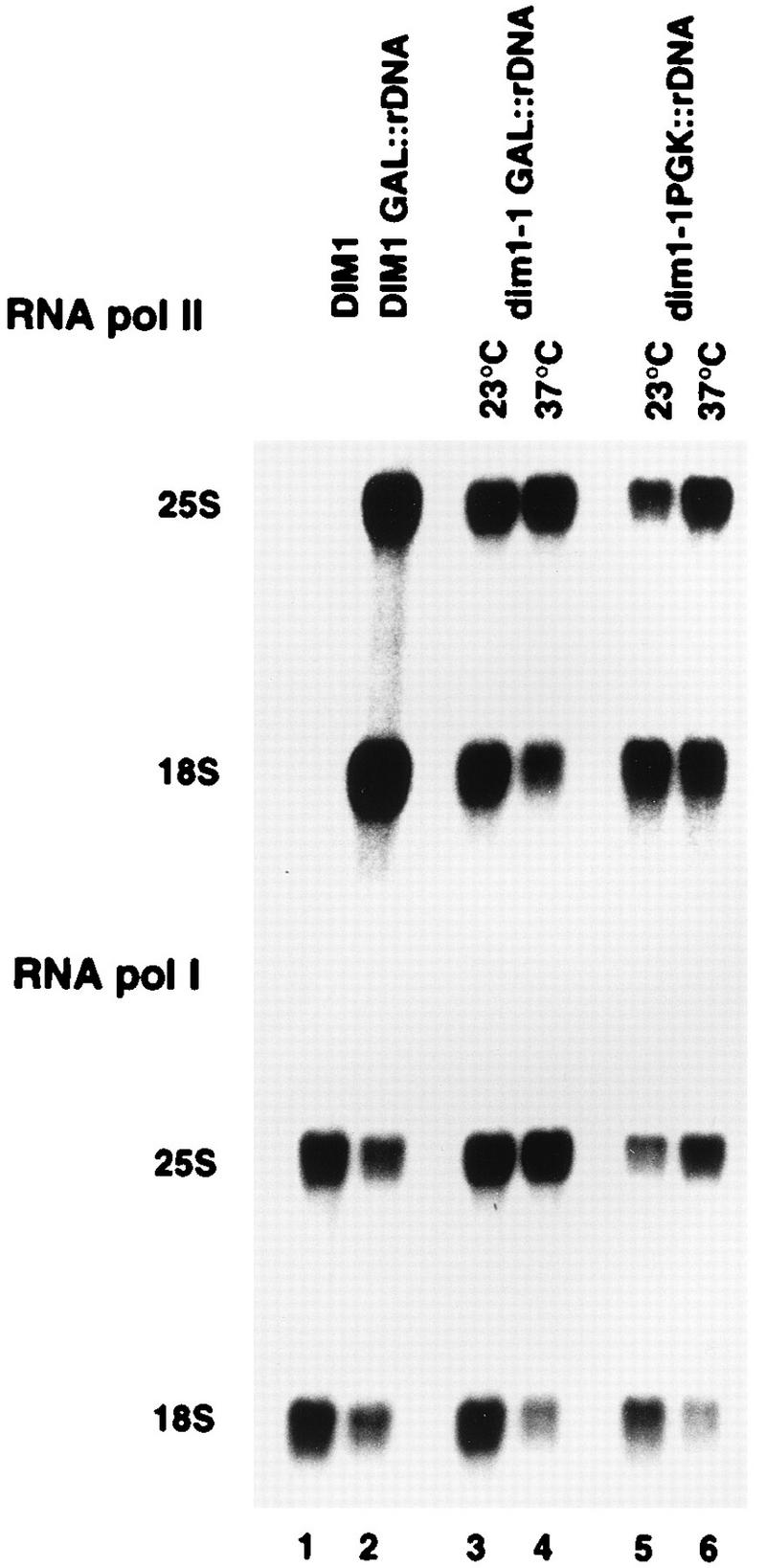
18S and 25S accumulation in dim1 TS strains expressing rDNA from different promoters. Lane 1, RNA extracted from a DIM1 wild-type strain; lane 2, RNA extracted from a DIM1 wild-type strain also expressing pre-rRNA from a GAL promoter; lanes 3 and 4, RNA extracted from a dim1-1 strain also expressing pre-rRNA from a GAL promoter; lanes 5 and 6, RNA extracted from a dim1-1 strain also expressing pre-rRNA from a PGK promoter. The same Northern filter was hybridized with probes complementary to the 25S and 18S rRNAs. The probes used are specific either for the tagged rRNAs synthesized from the RNA Pol II promoter or for the nontagged rRNAs transcribed from the chromosomal rDNA (RNA Pol I).
To allow HIS3 selection in strain CH1462 (ade2 ade3 [Table 1]), an ADE3 gene was expressed from plasmid pHT4467 (ADE3 URA3). The resulting strains, YDL314 (dim1-9) and YDL315 (dim1-1), were transformed with either pTL29 or pGAL::rDNA and grown at permissive temperature in glucose minimal medium lacking leucine or uracil, respectively, before being transferred to 37°C for 8 h. For GAL Dim1p depletion, strain YDL302 transformed with pTL29 was grown at 30°C in galactose minimal medium lacking uracil and leucine. Cells were harvested by centrifugation, washed, and resuspended in glucose minimal medium lacking uracil and leucine. During growth, cells were diluted with prewarmed medium and constantly maintained in early exponential phase.
In vitro translation.
Cytoplasmic S30 extracts of strains YDL321, YDL324, and the isogenic wild-type control strain MBS were prepared as described previously (17, 40). Strains YDL321 and YDL324 were constructed as follows: integrative cassettes (dim1 [TS] HIS3) were recovered from plasmids pTL39 (dim1-2) and pTL43 (dim1-1) by EcoRI/XhoI digestion and transformed into strain MBS. Poly(A) substrates were prepared as described previously (40) with constructs pAT3 (preprolactin) and pI12.34Δ33 (chloramphenicol acetyltransferase [CAT]). Both substrates were capped with the m7GpppG analog (New England Biolabs). Translation reactions (40) were for 60 min at 24°C with 0, 2, and 10 ng of preprolactin mRNA or 0, 20, and 100 ng of CAT mRNA. Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Antibiotic sensitivity assay.
As a preliminary experiment, equal amount of cells were plated on yeast extract-peptone-dextrose (YPD), and 5 and 10 μl of various antibiotic stock solutions were spotted on 5-mm sterile filter disks. Stock solutions were as follow: paromomycin (50 mg/ml), G418 (3 mg/ml), hygromycin B (3 mg/ml), cycloheximide (1 mg/ml), and neomycin B (20 mg/ml). Three prokaryote-specific antibiotics, streptomycin (100 mg/ml), erythromycin (50 mg/ml), and kasugamycin (100 mg/ml), were also tested. All antibiotics were purchased from Sigma. dim1 TS strains were found to be hypersensitive to paromomycin and neomycin B. All strains were also sensitive to cycloheximide but to the same extent as the wild-type isogenic control. The other antibiotics tested showed no effect. For the experiment presented in Fig. 10, paromomycin and neomycin B were used at 0, 250, 375, and 500 μg/μl and 0, 100, 400 and 800 μg/ml, respectively. Plates were incubated at the permissive temperature of 23°C.
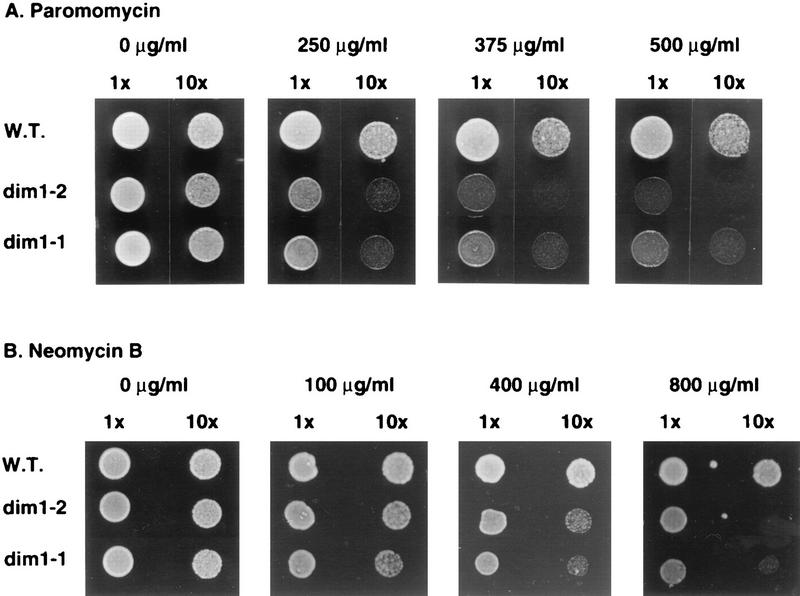
FIG. 10.
Antibiotic sensitivities of dim1 TS strains. Dilutions (1× and 10×) of dim1-1 and dim1-2 strains, along with the isogenic wild-type (W.T.) DIM1 control strain, were spotted on complete medium supplemented with paromomycin and neomycin B at the concentrations indicated. Plates were incubated at 23°C.
Indirect immunofluorescence.
Indirect immunofluorescence was performed on strains expressing 3× Myc-Dim1p or Dim1p-3× Myc fusion proteins (strains YDL101A and YDL102A, respectively [Table 1]). Strain YDL100A, expressing wild-type Dim1p, was used as a negative control. Cells were grown in YPD to an optical density at 600 nm of 0.5 to 1.0, fixed in 37% formaldehyde for 1 h, and permeabilized with Zymolyase in sorbitol buffer (1.2 M sorbitol, 20 mM KPI [pH 7.4]). Cells were incubated for 1 h with either anti-Nop1p antibody (monoclonal antibody 66, dilution 1/50; kindly provided by John Aris [1]) or anti-Myc antibody (9E10, dilution 1/50; BAbCO). Both antibodies are mouse monoclonal antibodies; therefore, cells were decorated independently. Detection with Texas red was for 1 h at a dilution of 1/50 (Texas red-conjugated affinipure F(ab′)2 goat anti-mouse immunoglobulin G [H+L]; Dianova). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Strains YDL100A, YDL101A, and YDL102A are haploids containing the dim1Δ::URA3 deletion of strain YDL300 (Table 1) rescued by the wild-type or epitope-tagged proteins expressed from plasmids pTL6 (Dim1p), pTL18 (3× Myc-Dim1p), and pTL25 (Dim1p-3× Myc), respectively. The growth rates of strains YDL100A, YDL101A, and YDL102A are identical, showing the fusion proteins to be fully functional.
RESULTS
Isolation of conditional alleles of DIM1.
A library of conditional alleles of DIM1 was created by mutagenic PCR and screened by plasmid exchange (37). A diploid strain containing a fully deleted dim1-Δ allele was transformed with a multicopy URA3 plasmid bearing a wild-type copy of DIM1. Haploid dim1-Δ progeny, rescued by the wild-type plasmid, were used as starting strains (strains YDL304A and YDL304B [Table 1]). The ORF of DIM1 was amplified under mutagenic PCR conditions and subcloned into an ARS/CEN LEU2 plasmid (see Materials and Methods). The resulting library was transformed into strain YDL304A. Transformants were replica plated on 5-FOA, which selects for the loss of the wild-type URA3 plasmid. 5-FOA-resistant clones, which have lost the wild-type DIM1 plasmid, were screened for growth defects at various temperatures. Among 1,600 5-FOA-resistant clones, 33 showed a conditional phenotype for growth. Most of them were TS, with some being also slightly CS (Fig. 2). No strain showed only a CS phenotype. Ten alleles, dim1-1 to dim1-10, were selected to be further characterized.
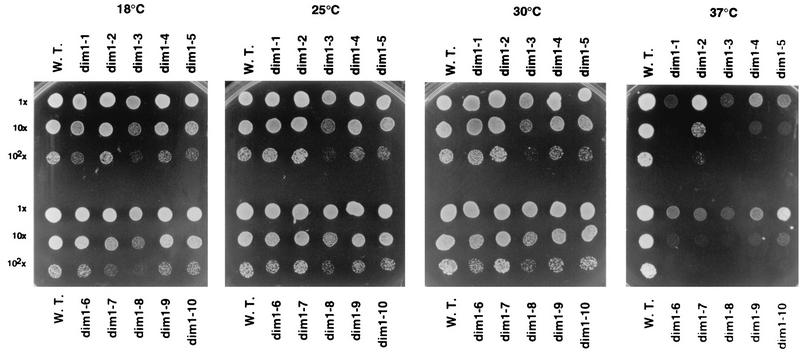
FIG. 2.
Conditional growth phenotypes of dim1 TS strains. Dilutions (1× to 102×) of strains dim1-1 to dim1-10, along with the wild-type isogenic DIM1 control (strain YDL209) (W.T.), were spotted on minimal plates at 18, 25, 30, and 37°C and incubated for 3 days. Most of the strains are very sensitive to elevated temperatures; some are also slightly CS (dim1-1 and dim1-7).
The coding sequences of dim1-1 to dim1-10 were fully sequenced; all contained from one to six substitution mutations (Fig. 3 and Table 2). It may be significant that mutations at four positions were selected twice (Table 2). Position 218 was mutated in both dim1-1 and dim1-4, and an identical substitution (S239P) is present in both dim1-6 and dim1-10. Notably, dim1-4 and dim1-5 have two identical mutations at identical positions (172 and 197), dim1-4 having an additional mutation at a position which is also altered in dim1-1 (position 218). Although mutations were spread over the whole length of the protein, we noticed a degree of clustering centered around positions 170 and 220; in the latter case, these fall in a region which is not conserved in E. coli KsgAp.
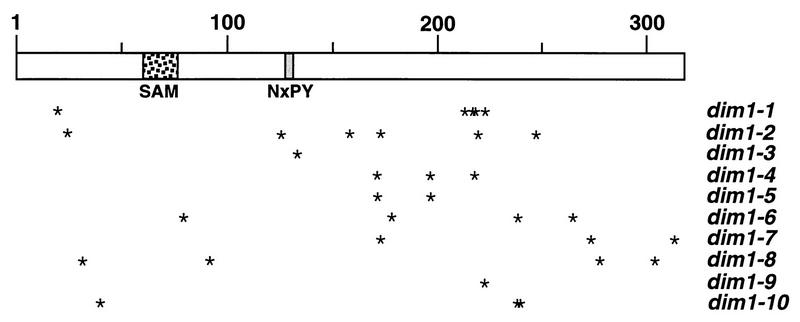
FIG. 3.
Mapping of dim1 TS mutations. Schematic representation of Dim1p (318 residues) drawn to scale. The S-adenosylmethionine binding domain (SAM), the putative catalytic residues (NxPY), and the 32 point mutations (asterisks) identified in the 10 dim1 TS alleles are represented.
TABLE 2.
Nature and positions of mutations in dim1 TS alleles
| Allele | Mutation(s) |
|---|---|
| dim1-1 | E21G; N213D; V218A; D219G; D224G |
| dim1-2 | S25G; I126V; F159S; R174S; Y220N; M273T |
| dim1-3 | I133V |
| dim1-4 | Y172D; R197S; V218E |
| dim1-5 | Y172D; R197S |
| dim1-6 | N80D; N178D; S239P; M265V |
| dim1-7 | C173R; H274R; V313A |
| dim1-8 | N31S; A92E; K278R; L304P |
| dim1-9 | W223R |
| dim1-10 | N41S; I238V; S239P |
None of the mutations fell into the predicted binding site for the cofactor S-adenosylmethionine (residues 60 to 76) (9, 18). The NXPY residues (positions 128 to 131) are conserved in all known SSU rRNA dimethylases, including human Dim1p (23a). These match the consensus sequence (N/D/S)PP(Y/F), which has been shown to be crucial for catalysis in the case of the N-6-adenine methylase EcoKI (48). Two mutations (I126V in dim1-2 and I133V in dim1-3) directly flank the NXPY residues, suggesting that they might directly affect catalysis.
dim1-2 uncouples the two functions of Dim1p.
RNA was extracted from the 10 dim1 TS strains and the isogenic wild-type strain (strain YDL209 [Table 1]) at the permissive temperature (23°C) and at 2, 8, and 23 h after transfer to 37°C.
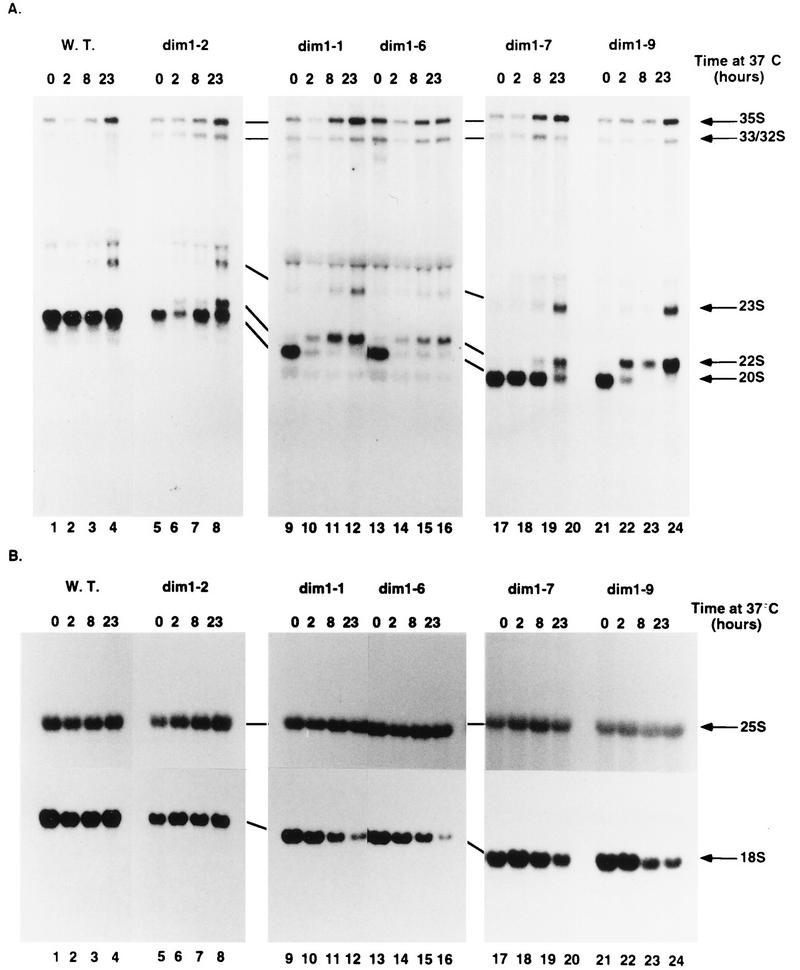
Northern hybridization revealed that at the nonpermissive temperature, all dim1 TS strains are inhibited in cleavage at sites A1 and A2, resulting in depletion of the 20S pre-rRNA and 18S rRNA (Fig. 1). These data are presented for five representative alleles (dim1-1, dim1-2, dim1-6, dim1-7, and dim1-9) in Fig. 4. In the dim1 mutants, there is seen an aberrant processing pathway in which the 35S pre-rRNA is processed normally at site A0 but the resulting 33S pre-rRNA is not processed at sites A1 and A2. Instead, cleavage at site A3 in internal transcribed spacer 1 (ITS1) generates the 22S RNA. Concomitant with the appearance of the 22S species, the 20S and 27SA2 pre-rRNAs are lost (Fig. 4A and data not shown). This is followed by depletion of the mature 18S rRNA (Fig. 4B), indicating that the aberrant 22S RNA cannot be processed to 18S rRNA. No alterations in the level of 27SB pre-rRNA (data not shown) or mature 25S rRNA (Fig. 4B) were observed, indicating that subsequent processing of the 3′ region of the pre-rRNA was unaffected.
FIG. 4.
Pre-rRNA processing in dim1 TS strains. (A) Probe against the 5′ region of ITS1 (oligonucleotide e [Fig. 1A]). (B) Probes against mature 18S and 25S rRNA (oligonucleotides a and f [Fig. 1A]). RNA was extracted from the DIM1 (strain YDL209) (W.T.) and dim1 TS strains following growth at 23°C (0-h lanes) and at intervals following transfer to 37°C (2-, 8-, and 23-h lanes) and separated on 1.2% agarose-formaldehyde gels. The 22S pre-rRNA extends from site A0 to site A3 and results from the inhibition of cleavages at sites A1 and A2 in the dim1 TS strains. At the late time point of transfer to 37°C (23 h), the normally minor and barely detectable 23S pre-rRNA that extends from the 5′ end of the primary transcript to site A3 accumulates to the same levels in the wild-type and dim1 TS strains. In this experiment, all samples, including the wild-type control, showed some accumulation of 35S pre-rRNA and 23S RNA after 23 h at 37°C; this was not observed in other experiments.
The pre-rRNA processing defects observed in the dim1 TS strains closely resemble those seen on depletion of Dim1p in GAL::dim1 strains (26). The timing of the appearance of the processing defects is strikingly different in the various dim1 TS strains. In the dim1-1, dim1-6, and dim-9 strains, processing is strongly inhibited 2 h after transfer to 37°C. In contrast, the 22S RNA is only weakly detected in the dim1-2 strain 2 h after transfer to 37°C, and the inhibition of processing is much stronger at later times. The 22S RNA is detected in the dim1-7 strain after 8 h. This may reflect a distinction between mutants in which Dim1p is rapidly inactivated by transfer to 37°C compared to mutations which do not allow synthesis of new, active Dim1p.
Although all strains accumulate 22S pre-RNA (Fig. 4 and data not shown), there are marked differences in 18S rRNA depletion. dim1-1, dim1-4, dim1-6, dim1-8, and dim1-10 strains all showed strong 18S rRNA depletion, while dim1-5 and dim1-9 strains showed only mild depletion and dim1-2, dim1-3, and dim1-7 strains were only slightly depleted (Fig. 4 and data not shown). No clear correlation could be established between the locations of the point mutations and the phenotypes, but it is notable that dim1-4, which contains only one additional mutation (at position 218) compared to dim1-5, shows stronger 18S rRNA depletion. A substitution at the same position is present in dim1-1, which shows a similar phenotype. dim1-6 and dim1-10 strains are also severely affected in 18S rRNA synthesis and share an identical mutation at position 239. No attempt was made to further separate the mutations.
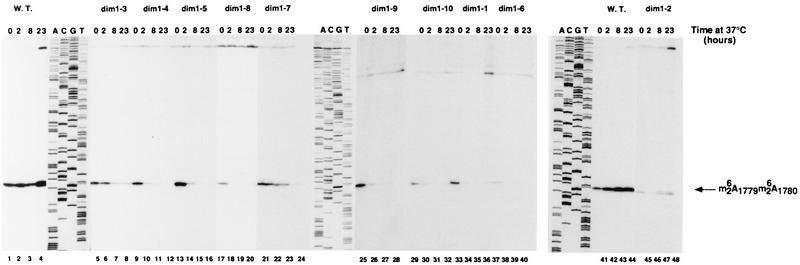
The level of pre-rRNA dimethylation was assessed by primer extension with an oligonucleotide complementary to a sequence in ITS1 located 218 nucleotides downstream of the site of modification (oligonucleotide e [Fig. 1A]). Following transfer to 37°C, the primer extension stop corresponding to dimethylation is strongly reduced in all dim1 TS strains (Fig. 5). Northern analysis data (Fig. 4) indicate that the overall levels of pre-rRNA species that contain the site of dimethylation are not strongly altered in most dim1 TS mutants. As with the pre-rRNA processing defects, dimethylation is inhibited to various extents and with different kinetics in the different dim1 TS alleles (Fig. 5).
FIG. 5.
Overall level of dimethylation in dim1 TS strains. RNA was extracted from the DIM1 (strain YDL209) (W.T.) and dim1 TS strains following growth at 23°C (0-h lanes) and at intervals following transfer to 37°C (2-, 8-, and 23-h lanes) and analyzed by primer extension with oligonucleotide e (Fig. 1A). The positions of primer extension stops due to the presence of the modifications are indicated. A DNA sequence made with the same primer is shown as a size marker.
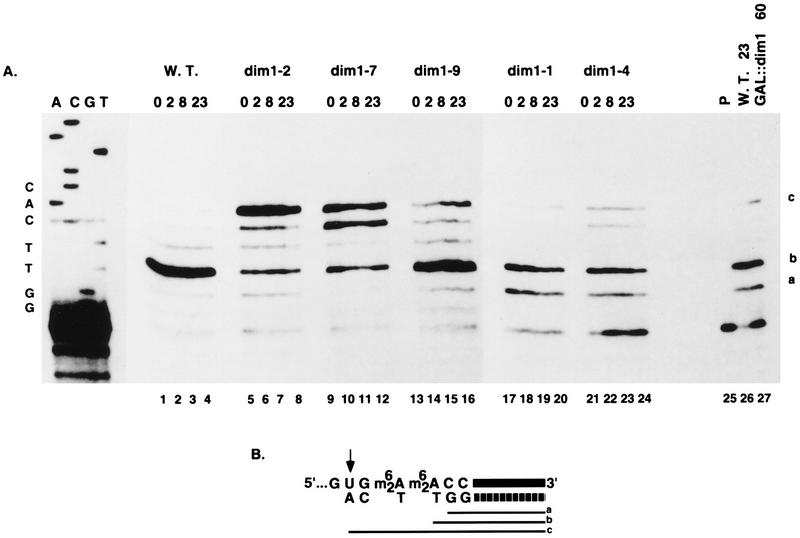
The accumulation of nondimethylated 18S rRNA can be monitored by primer extension with an oligonucleotide complementary to the very 3′ end of the mature rRNA (oligonucleotide d [Fig. 1A]), since the m26A modification blocks reverse transcription (see the legend to Fig. 6). The level of nondimethylated rRNA and pre-rRNA is indicated by the stop at position c, which represents reverse transcriptase molecules that were able to read through the unmodified site of dimethylation (26). The very faint band visible at this position in the wild-type strain (Fig. 6, lanes 1 to 4) represents primer extension on the unmodified pre-rRNA, which is very much less abundant than the mature rRNA (see reference 26). The GAL::dim1 strain (Fig. 6, lane 27) does not accumulate nondimethylated 18S rRNA; this is as previously reported (26). In contrast, several dim1 TS strains accumulate unmodified 18S rRNA. Strong accumulation is seen in the dim1-2 and dim1-7 strains (Fig. 6, lanes 5 to 12), even at the permissive temperature, and some accumulation is seen in the dim1-9 strain at 37°C (Fig. 6, lanes 13 to 16). Little nondimethylated 18S was detected in the dim1-1 or dim1-4 strains (Fig. 6, lanes 17 to 24).
FIG. 6.
Levels of nondimethylated 18S rRNA in dim1 TS strains. (A) RNA was extracted from the DIM1 (strain YDL209) (W.T.) and dim1 TS strains following growth at 23°C (0-h lanes) and at intervals following transfer to 37°C (2-, 8-, and 23-h lanes) and analyzed by primer extension with oligonucleotide d, which is complementary to the very 3′ end of 18S rRNA (Fig. 1A). The reactions were performed with dideoxyadenosine nucleotides in place of deoxyadenosine. The site of priming is 3 nucleotides 3′ to A1780, and no A residues are incorporated before the site of modification (see panel B). Dimethylation of A1779 A1780 blocks primer extension. Extensions carried out on nondimethylated rRNA extend through the A1779 A1780 site but are blocked 2 nucleotides 5′ to A1779 (position U1777). The positions of primer extension stops due to the presence of the modifications are indicated (a, b and c). A DNA sequence made with the same primer is shown as a size marker. Lanes 25 to 27, control lanes (lane 25, no RNA [P denotes primer alone]; lane 26, same as lane 4; lane 27, RNA extracted from the GAL::dim1 strain (strain YDL302) following transfer to glucose for 60 h). (B) Schematic representation of the 3′ end of 18S rRNA. Upper line, rRNA strand (the thick line represents the last 16 nucleotides). Lower line, complementary cDNA strand (the thick broken line represents oligonucleotide d). The three potential extended products are represented by thin lines (a, b, and c). The positions of the primer extension stops due to the presence of the modifications are indicated as a and b; the position of the primer extension stop due to read-through of the dimethylation site is indicated as c.
Thus, at the nonpermissive temperature, all of the dim1 TS strains analyzed are impaired both in cleavage of pre-rRNA at sites A1 and A2 and in dimethylation of pre-rRNA, although the kinetics and severity of these phenotypes vary between mutants. Some of the dim1 TS strains accumulate nondimethylated 18S rRNA, showing that the pre-rRNA methylation defect can be uncoupled from the pre-rRNA processing defect. This is particularly true in the dim1-2 strain, which shows no clear pre-rRNA processing defect at the permissive temperature but has very low levels of pre-rRNA modification.
It is notable that no dim1 TS strain defective only in pre-rRNA processing was isolated (Fig. 4 and 5). Dim1p must bind its substrate to modify the two adenosines, and this interaction with the pre-rRNA may be both necessary and sufficient to fulfill the requirement for Dim1p in pre-rRNA processing. Since dimethylation is dispensable in vivo (see below), the TS screen would not have isolated strains defective only in methylation without a pre-rRNA processing defect.
Pre-rRNAs transcribed from the PGK promoter do not require Dim1p for processing.
The studies on pre-rRNA processing described above analyzed pre-rRNA transcribed from the chromosomal rDNA by RNA Pol I. Unexpectedly, the processing of pre-rRNA transcribed by the constitutive RNA Pol II PGK promoter is much less sensitive to the dim1-1 and dim1-9 mutations than is processing of the Pol I-transcribed pre-rRNA (shown for dim1-1 in Fig. 7). The Pol II-driven rDNA repeat contains neutral tags within the mature rRNA sequences; these allow the use of hybridization probes that are specific for either the chromosomal or Pol II-transcribed rRNAs. Transfer of the dim1-1 strain to 37°C for 20 h resulted in underaccumulation of the 18S rRNA synthesized from pre-rRNA transcribed from chromosomal rDNA (Fig. 7, lower panel [compare lane 3 with 4 and lane 5 with 6]) as described above. In contrast, the level of 18S rRNA synthesized from pre-rRNA transcribed from the PGK promoter carried on plasmid pTL29 (see Materials and Methods) was reduced to a lesser extent (Fig. 7, upper panel [compare lanes 5 and 6]). Accumulation of 18S rRNA transcribed from a different RNA Pol II promoter, GAL10 (Fig. 7, upper panel [compare lanes 3 and 4]), was reduced to an extent similar to the rRNA transcribed by Pol I. The same effects were seen in the dim1-9 strain (data not shown); other dim1 alleles were not tested.
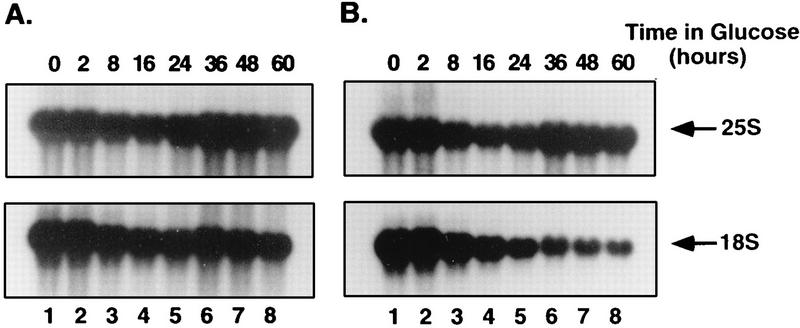
Similar effects were observed in the GAL::dim1 strain (YDL302 [Table 1]) following depletion of Dim1p. Following transfer to glucose, the level of 18S rRNA synthesized by Pol I from chromosomal rDNA (Fig. 8B) is strongly depleted. In contrast, little depletion of the 18S rRNA synthesized from the pPGK::rDNA construct is observed (Fig. 8A). The effects of genetic depletion of two other components required for pre-rRNA processing at sites A1 and A2 were also tested. On depletion of snR30 (30) or Rrp5p (47), the processing of the PGK-driven pre-rRNAs was inhibited to the same extent as processing of the Pol I-transcribed pre-rRNA (33a).
FIG. 8.
pPGK::rDNA transcripts are insensitive to Dim1p depletion. (A) Probes specific to mature 25S and 18S rRNA produced from the pPGK::rDNA construct (oligonucleotides b and g [Fig. 1A]). (B) Probes specific to the mature 25S and 18S rRNA produced from the chromosomal rDNA units (oligonucleotides c and h [Fig. 1A]). RNA was extracted from the GAL::dim1 strain transformed with the pPGK::rDNA construct following growth in galactose (0-h lanes) and at intervals following transfer to glucose (2- to 60-h lanes) and separated on a 1.2% agarose-formaldehyde gel.
The processing of the pre-rRNAs transcribed from the PGK-driven rDNA cannot readily be assessed in these strains, since Pol I transcription is much stronger than PGK-driven transcription. However, we interpret these data as showing that the cleavage of sites A1 and A2 is specifically resistant to mutations in DIM1 or depletion of Dim1p in pre-rRNA molecules that have been transcribed from the PGK promoter. The mechanism that relieves the Dim1p dependence may be related to the packaging of the pre-rRNA transcripts with different sets of proteins. Whatever the mechanism, this observation demonstrates that Dim1p is not directly required for pre-rRNA processing. We interpret this as strong evidence for the existence of a system that represses pre-rRNA processing in the absence of Dim1p.
m26A1779m26A1780 is not essential in vivo but is required for translation in vitro.
At the permissive temperature, the dim1-2 strain showed no pre-rRNA processing defect but predominantly contained nondimethylated 18S rRNA. The dim1-2 strain showed no clear growth inhibition compared to the otherwise isogenic DIM1 strain in liquid or solid minimal medium or complete medium at 23°C (data not shown). This demonstrates that 18S rRNA dimethylation is dispensable for translation in vivo.
In order to test whether dimethylation affected ribosome function at a more subtle level, extracts prepared from wild-type and dim1-2 strains grown at the permissive temperature were tested for the ability to translate exogenous mRNA in vitro. As a control, extracts from a dim1-1 strain that was not impaired in rRNA methylation at the permissive temperature were also tested.
The dim1 alleles were integrated into strain MBS (see Materials and Methods and Table 1), which is particularly suitable for such analysis (17). Standardized amounts of extracts prepared from strains YDL321 (dim1-2), YDL324 (dim1-1), and the isogenic wild-type control strain MBS were incubated with various amounts of either CAT mRNA (Fig. 9B and D) or preprolactin mRNA (Fig. 9A). Figures 9A and B present the analyses of two independently isolated dim1-2 strains (YDL321-1 and YDL321-2). Translation products were easily detected at the expected lengths when wild-type extracts were incubated with either of the two mRNAs (Fig. 9A, B, and D, lanes 2 and 3). No product was detected for either mRNA substrate when the dim1-2 extracts were used (Fig. 9A and B, lanes 4 to 9, and Fig. 9D, lanes 4 to 6). Extracts from the dim1-1 strain were less competent for translation than was the wild-type control (Fig. 9D, lanes 8 and 9) but consistently exhibited substantially greater translation than did the dim1-2 strain. The reduced translation may be related to the mild processing defect present in the dim1-1 strain, which accumulated low levels of the 22S pre-rRNA even at the permissive temperature (Fig. 4A, lane 9). Figure 9C shows the analysis of the level of 18S rRNA dimethylation in the cell extracts used for Fig. 9A and B. We concluded that in the dim1-2 strains, the 40S subunits contain nondimethylated 18S rRNA and are not competent for translation in vitro.
FIG. 9.
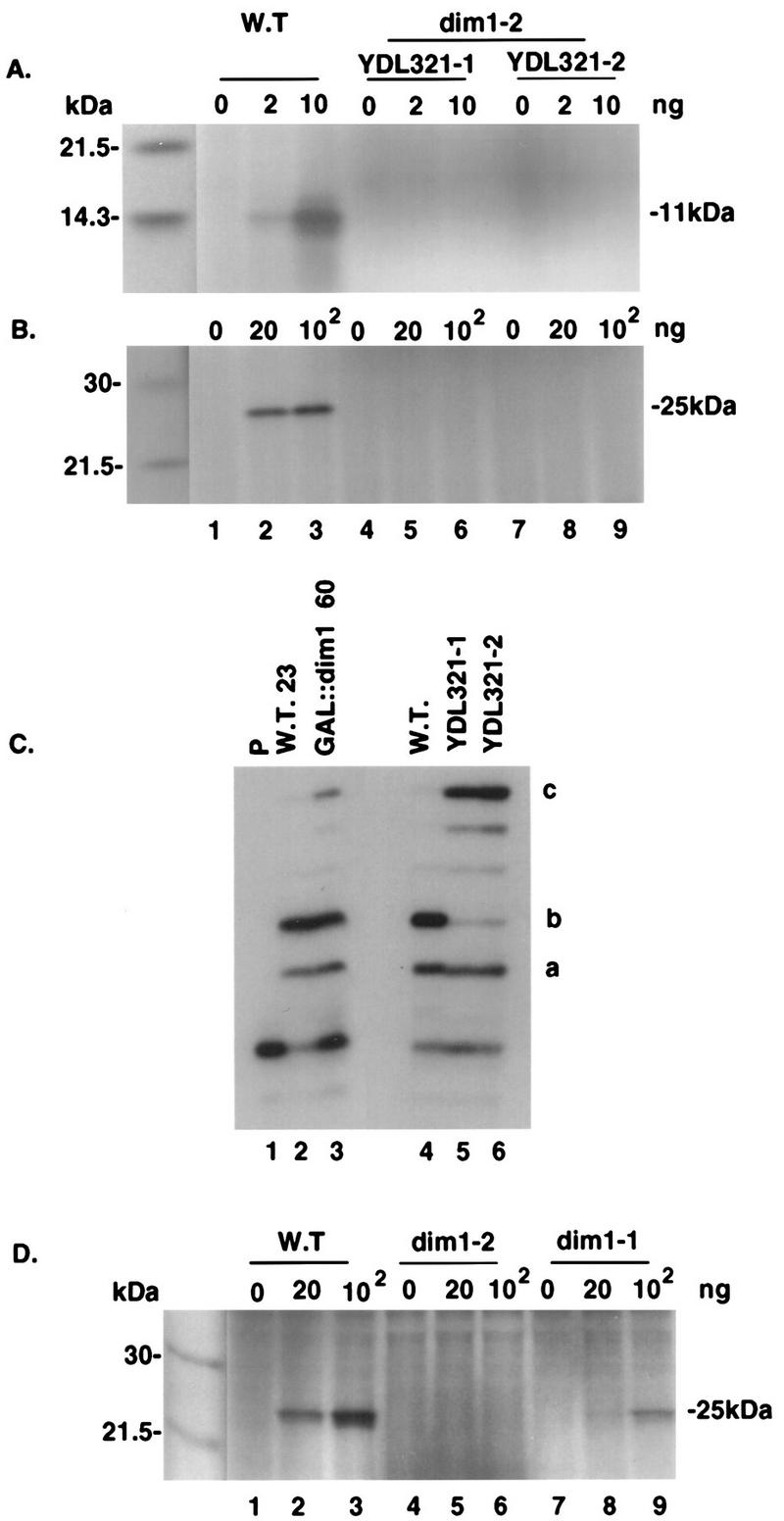
In vitro translation analysis of dim1 TS strains. Cytoplasmic S30 extracts of dim1-2 strains (YDL321-1 and YDL321-2), a dim1-1 strain (YDL324), and the wild-type (W.T.) isogenic control (strain MBS) were prepared following growth at 23°C. Standardized amount of extracts were incubated with 0, 2, and 10 ng of preprolactin mRNA (A) or 0, 20, and 100 ng of CAT mRNA (B and D). YDL321-1 and YDL321-2 are two independently isolated integrants of the dim1-2 allele in the MBS strain (see Materials and Methods and Table 1). The dim1-2 strain used in panel D is YDL321-2. Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and are indicated at their expected lengths (11 and 25 kDa). (C) 18S rRNA dimethylation in cell extracts, analyzed as described in the legend to Fig. 6. RNA was extracted from strains YDL321-1, YDL321-2, and the wild-type strain (strain MBS) following growth at 23°C. Lanes 1 to 3, control lanes (lane 1, no RNA [P denotes primer alone]; lane 2, RNA extracted from strain MBS following transfer to 37°C for 23 h; lane 3, RNA extracted from the GAL::dim1 strain [strain YDL302] following transfer to glucose for 60 h).
dim1 TS strains are hypersensitive to aminoglycoside antibiotics.
To identify more subtle effects on translation in vivo, representative dim1 TS strains were tested for their sensitivities towards a range of antibiotics at the permissive temperature (see Materials and Methods). Strains carrying dim1-1, dim1-2, and dim1-9 were hypersensitive to paromomycin and neomycin B to approximately equal extents (Fig. 10 and data not shown). These two antibiotics belong to the aminoglycoside family and are known to induce misreading and suppression of nonsense mutations in yeast (33, 39). This observation suggested that each of the dim1 TS strains analyzed had an additional assembly defect which resulted in antibiotic sensitivity. This appears to be unrelated to the degree of rRNA dimethylation.
Dim1p is localized to the nucleus with nucleolar enrichment.
To determine the subcellular location of Dim1p, the protein was tagged with three copies of the human c-Myc epitope at either the amino- or carboxy-terminal end (see Materials and Methods). Both fusion proteins were expressed in a dim1-Δ deleted background and were shown to be fully functional (strains YDL101A and YDL102A [Table 1]).
The fusion proteins were localized by indirect immunofluorescence. Figure 11D presents the results of the carboxy-terminal fusion (strain YDL102A). The amino-terminal fusion gave an identical signal (data not shown). Comparison to DAPI staining of the DNA (Fig. 11C) and the localization of the nucleolar protein Nop1p (Fig. 11B) reveals that the Dim1p fusion protein is localized to the nucleus with enrichment in the nucleolus, which forms a cap-like structure slightly displaced from the DAPI-stained region. Little cytoplasmic staining was detected.
FIG. 11.
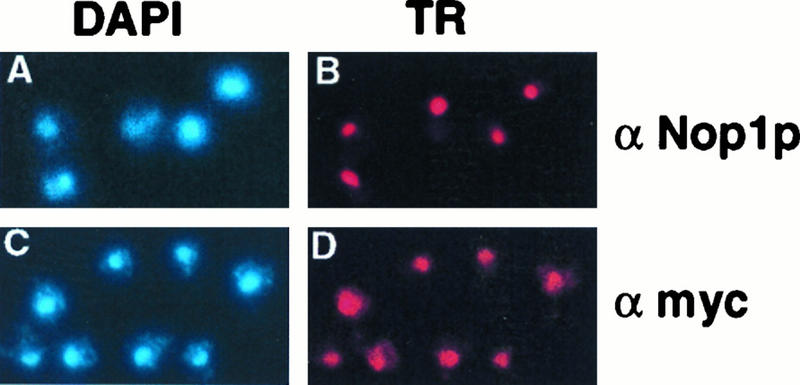
Subcellular localization of Dim1p. Indirect immunofluorescence with strain YDL102A (Dim1p-3× Myc). Cells were incubated with either anti-Nop1p (α-Nop1p) or anti-Myc (α-Myc) antibody followed by Texas red (TR) and DAPI staining. Both primary antibodies are mouse monoclonal antibodies; therefore, cells were labelled independently.
DISCUSSION
We report here that mutations in DIM1 can uncouple the requirements for Dim1p in pre-rRNA modification and processing. All of the dim1 alleles that were isolated from a temperature-sensitive library were found to block pre-rRNA processing at the nonpermissive temperature (37°C), although the kinetics of inhibition varied. In addition, the dim1-2 allele blocks rRNA dimethylation at the permissive temperature (23°C) but shows no pre-rRNA processing defect at this temperature. This resulted in the accumulation of high levels of nondimethylated 18S rRNA. The dim1-2 strains had no detectable growth defect at 23°C, showing the dimethylation to be dispensable for ribosome function in vivo.
Pre-rRNA molecules in which the two adenosine residues are replaced by guanosines that cannot be modified are processed normally but do not support growth (26). Additionally, these substitutions give rise to a reduced level of 18S rRNA at low temperatures, possibly reflecting an assembly defect, which is often associated with cold sensitivity. From the dim1-2 analysis, it is clear that the lethality of the G1779 G1780 cis mutation and the cold-sensitive processing phenotype are due to the substitution of the adenosines at these two universally conserved positions. In E. coli, both dimethylation and the twin adenosines were shown to be dispensable for function and assembly in vitro (10, 22), but their importance in vivo has not been assessed.
Strikingly, extracts prepared from dim1-2 strains grown at 23°C lacked detectable activity for translation in vitro. Extracts prepared from dim1-1 strains, which have normal levels of rRNA dimethylation, were competent for in vitro translation, although with lower activity than in the wild-type extract. While we cannot exclude the possibility that ribosomes synthesized in the dim1-2 mutant have some additional defect, it seems likely that dimethylation is required for in vitro translation in yeast extracts. We speculate that the dimethylation fine-tunes the function of the 40S ribosomal subunit in vivo but is essential under the suboptimal in vitro conditions.
Other rRNA modifications, 2′-O methylation and pseudouridine formation, are directed by guide snoRNAs (8, 12, 20, 31, 32). In all cases tested, the guide functions of the snoRNAs were completely dispensable for growth, indicating that these modifications are, like Dim1p dimethylation, dispensable for ribosome function in vivo. The requirements for the 2′-O-methyl and pseudouridine modifications, however, have not been tested in vitro.
Prokaryotic ksgA strains lack rRNA dimethylase activity and are resistant to the aminoglycoside antibiotic kasugamycin (14, 15, 44). Hypersensitivity toward the aminoglycoside antibiotics paromomycin and neomycin B was observed in dim1 TS strains. This could not be correlated, however, with dimethylation; dim1-1 and dim1-9 strains, which do not accumulate nondimethylated SSU rRNA, are as sensitive to aminoglycosides as are dim1-2 strains. We speculate that the dim1 TS strains analyzed bear an additional assembly defect responsible for antibiotic sensitivity. Hypersensitivity to paromomycin and neomycin B has previously been reported in yeast strains carrying mutations in NSR1 and RRP1, which are also required for normal pre-rRNA processing (11, 27). Neither of these proteins were reported to be involved in rRNA modification, and in both cases an assembly defect is likely to underlie the antibiotic sensitivity observed.
Model for a regulatory mechanism in ribosome synthesis.
We reported previously that cells depleted of Dim1p are inhibited in cleavage of the 33S pre-rRNA at sites A1 and A2 (26), and this was also the case in all of the dim1 TS strains at the nonpermissive temperature. However, dimethylation was found to occur on the 20S pre-rRNA, which is the product of cleavage at sites A1 and A2 (Fig. 1), consistent with early reports that the formation of m26Am26A is a late event in ribosome synthesis (6, 36). Since the cleavages at sites A1 and A2 occur before dimethylation, they cannot be directly dependent on the presence of the modification. This conclusion was supported by the observation that pre-rRNAs containing the G1779 G1780 mutations, which cannot be dimethylated, can be processed at sites A1 and A2. We concluded that the Dim1p protein, rather than the modification itself, was required for pre-rRNA processing (26).
Our interpretation was that a quality control system probably inhibited the processing of the pre-rRNA in the absence of Dim1p (26). However, the data left open the alternative possibility that Dim1p had an additional function and was directly required for pre-rRNA processing. During the course of this work, this question was unexpectedly resolved. When transcription of an rDNA unit is driven by an RNA Pol II PGK promoter, the pre-rRNAs produced are largely insensitive to dim1-1 and dim1-9 mutations that strongly inhibit processing of pre-rRNAs transcribed from the chromosomal rDNA driven by RNA Pol I. This phenomenon is not allele specific: the PGK-transcribed pre-rRNAs are also resistant to genetic depletion of Dim1p. Moreover, the otherwise identical pre-rRNA transcribed from a GAL promoter was not resistant to the dim1-1 or dim1-9 mutations. These observations demonstrate that Dim1p does not have a direct role in pre-rRNA processing.
We speculate that a repression system blocks pre-rRNA processing in the absence of the binding of Dim1p to pre-rRNA. According to this model, Dim1p normally binds to pre-rRNA in the nucleolus at an early stage in ribosome synthesis, and a component of the processing machinery senses this interaction. This could occur through direct interaction with Dim1p or through an interaction with preribosomal particles (a conformational change could be monitored). Cleavages at sites A1 and A2 occur in the nucleolus; consistent with this, Dim1p was shown to localize mostly to this cellular compartment. If Dim1p has bound to pre-rRNA, processing at sites A1 and A2 proceeds; otherwise, processing is blocked. In mutant strains that lack Dim1p, this leads to the synthesis of a dead-end intermediate, the 22S pre-rRNA, and prevents synthesis of the 18S rRNA. In wild-type strains, pre-rRNA processing is presumably only delayed until Dim1p finds and binds to its target, preventing the formation of mature but nonmodified 18S rRNA. This is desirable since, as shown here, the unmodified ribosomal subunits are impaired in function.
The pre-rRNAs that are transcribed from the PGK promoter are very likely to be associated with a different set of hnRNP proteins than are the Pol I transcripts. We speculate that one of these occupies the Dim1p binding site and is detected by the pre-rRNA processing machinery as Dim1p, thus alleviating the need for authentic Dim1p.
The m26Am26A modification is highly conserved and is present in both bacteria and eukaryotes. In ksgA mutant strains of E. coli that lack dimethylation, growth is mildly impaired and the nondimethylated ribosomes show defects in translation in vitro (reviewed in reference 45). Expression of Dim1p in E. coli restores dimethylation, and E. coli KsgAp is highly homologous to Dim1p. However, while bacteria lacking the KsgAp methyltransferase synthesize the unmodified rRNA, eukaryotes have evolved a regulatory system to prevent this. We anticipate that many such quality control mechanisms act to coordinate the numerous steps of eukaryotic pre-rRNA processing, rRNA modification, and ribosome assembly.
ACKNOWLEDGMENTS
We thank E. Petfalski for analysis of the GAL::RRP5 and GAL::snr30 strains, Jaap Venema for supplying the GAL::RRP5 strain, B. Séraphin and M. Hentze (EMBL) for fruitful discussions, G. Berben (Station de Chimie, Gembloux, Belgium) for making the YDp plasmids available, H. Tekotte (EMBL) for supplying plasmid pHT4467, and the EMBL sequencing service.
This work was partially supported by the Wellcome Trust. During the course of this work, D. L. J. Lafontaine was the recipient of an EMBO long-term fellowship.
REFERENCES
- 1.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 5.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 6.Brand R C, Klootwijk J, van Steenbergen T J M, de Kok A J, Planta R J. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 7.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkarriov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 8.Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X, Kumar S, Posfai J, Pflugrath J W, Roberts R J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham P R, Weitzmann C J, Nurse K, Masurel R, van Knippenberg P H, Ofengand J. Site-specific mutation of the conserved m26Am26A residues of E. coli 16S ribosomal RNA. Effects on ribosome function and activity of the ksgA methyltransferase. Biochim Biophys Acta. 1990;1050:18–26. doi: 10.1016/0167-4781(90)90135-o. [DOI] [PubMed] [Google Scholar]
- 11.Fabian G R, Hopper A K. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J Bacteriol. 1987;169:1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 13.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helser T L, Davies J E, Dahlberg J E. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 15.Helser T L, Davies J E, Dahlberg J E. Mechanism of kasugamycin resistance in Escherichia coli. Nature New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 16.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingrosso D, Fowler A V, Bleibaum J, Clarke S. Sequence of the d-aspartyl/l-isoaspartyl protein methyltransferase from human erythrocytes. J Biol Chem. 1989;264:20131–20139. [PubMed] [Google Scholar]
- 19.Jeeninga R E, Van Delft Y, de Graaff-Vincent M, Dirks-Mulder A, Venema J, Raué H A. Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA. 1997;3:476–488. [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 21.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke C W, Agris P F, Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into functional 30S ribosomes. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 23.Lafontaine D, Delcour J, Glasser A-L, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m26Am26A dimethylation in the 3′ terminal loop of 18S rRNA is essential in yeast. J Mol Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 23a.Lafontaine, D., and D. Tollervey. Unpublished data.
- 24.Lafontaine D, Tollervey D. Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem Cell Biol. 1995;73:803–812. doi: 10.1139/o95-088. [DOI] [PubMed] [Google Scholar]
- 25.Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 27.Lee W C, Zabetakis D, Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 29.Maden B E H. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 30.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 32.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 33.Palmer E, Wilhelm J M, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 33a.Petfalski, E., and D. Tollervey. Unpublished data.
- 34.Poldermans B, Bakker H, van Knippenberg P H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′-end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methyl groups on ribosomal subunit interaction. Nucleic Acids Res. 1980;8:143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poldermans B, van Buul C P J J, van Knippenberg P H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′-end of 16S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979;254:9090–9094. [PubMed] [Google Scholar]
- 36.Salim M, Maden B E H. Early and late methylations in HeLa cell ribosome maturation. Nature. 1973;244:334–336. doi: 10.1038/244334a0. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 40.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 41.Thamana P, Cantor C R. Studies on ribosome structure and interactions near the m26Am26A sequence. Nucleic Acids Res. 1978;5:805–823. doi: 10.1093/nar/5.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tollervey D. Trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 43.van Buul C P J J, Visser W, van Knippenberg P H. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 1984;177:119–124. doi: 10.1016/0014-5793(84)80994-1. [DOI] [PubMed] [Google Scholar]
- 44.van Buul C P J J, Damn J B L, van Knippenberg P H. Kasugamycin resistant mutants of Bacillus stearothermophilus lacking the enzyme for the methylation of two adjacent adenosines in 16S ribosomal RNA. Mol Gen Genet. 1983;189:475–478. doi: 10.1007/BF00325912. [DOI] [PubMed] [Google Scholar]
- 45.van Knippenberg P H. Structural and functional aspects of the N6,N6-dimethyladenosines in 16S ribosomal RNA. In: Hardesty B, Kramer G, editors. Structure, function and genetics of ribosomes. New York, N.Y: Springer-Verlag Inc.; 1986. pp. 412–424. [Google Scholar]
- 46.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 47.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 48.Willcock D F, Dryden D T F, Murray N E. A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J. 1994;13:3902–3908. doi: 10.1002/j.1460-2075.1994.tb06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]