Abstract
Background
People living with HIV are vulnerable to antibiotic-resistant bacterial infections because of frequent healthcare visits, the consumption of many antimicrobials, and the weakened immune system to fight infections. Our objective was to provide comprehensive data about ESBL-producing Enterobacterales among HIV-positive individuals across the globe.
Methods
This meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. To select eligible articles published between January 1, 2010, and May 12, 2024, a literature search was performed on available electronic databases such as PubMed, Hinari, Google Scholar, and Scopus. The quality of the included studies was assessed via the Joanna Briggs Institute critical appraisal tool. The data were extracted from the eligible studies via Microsoft Excel 2019 and analyzed via STATA version 17. A random effects model was constructed via the DerSimonian and Laird method. The heterogeneity was checked through the Cochrane Q statistic, and the magnitude was quantitatively measured via I2 statistics. To determine the possible sources of heterogeneity, a subgroup analysis was performed. Additionally, a sensitivity analysis was conducted, and publication bias was checked via funnel plots and Egger’s regression tests. A p value of less than 0.05 was considered evidence of heterogeneity and small study effects according to the Cochrane Q statistic and Egger’s test, respectively. The protocol was registered previously (PROSPERO ID: CRD42024557981).
Results
A total of 5305 HIV-positive individuals from 20 studies were included in our meta-analysis. The overall pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals was 20.30% (931/5305; 95% CI: 15.13–25.47%, P < 0.001), with a high level of heterogeneity (I2 = 97.82%, P < 0.001). The predominant ESBL producers were K. pneumoniae, with a pooled prevalence of 40.84% (76/217; 95% CI: 26.87–54.81%), followed closely by E. coli at 40.14% (348/985; 95% CI: 27.83–52.45%). In the subgroup analysis, the highest magnitude of ESBL-producing pathogens was observed in Asia (195/715; 28.55%), followed by Africa (666/3981; 19.12%). Additionally, the highest pooled prevalence of ESBL-producing pathogens among HIV-positive individuals was reported to be colonization 23.78% (613/2455; 95% CI: 15.36–32.19, I² = 96.78%, p < 0.001), followed by infection 15.77% (318/2850; 95% CI: 10.06–21.49, I² = 97.45%, p < 0.001). Among the different types of ESBL enzyme-encoding genes, blaCTX-M was the most common (73 out of 150 isolates, 48.7%), followed by blaTEM (49 out of 150, 32.7%).
Conclusion and recommendations
This study demonstrated that HIV-positive individuals are commonly colonized and infected with ESBL-producing Enterobacterales. The highest prevalence of these pathogens was reported in Asia and Africa. To reduce mortality from severe bacterial infections in HIV patients, resources should be distributed equitably across all regions. Particular attention should be given to high-prevalence areas, where early detection of colonization and infection with antibiotic-resistant pathogens is critical. Enhanced surveillance of ESBL-producing organisms among HIV-positive individuals is also strongly recommended.
Introduction
Antimicrobial resistance (AMR) is one of the top ten threats to global public health, and it is a major and serious issue in the 21st century [1]. Recent studies have provided updated insights into the global impact of antimicrobial resistance (AMR). In 2021, approximately 1.14 million deaths were directly attributable to bacterial AMR, with a total of 4.71 million deaths associated with AMR worldwide [2]. Projections indicate that, without significant intervention, AMR could be responsible for up to 10 million deaths annually by 2050 [3]. Regionally, the World Health Organization (WHO) African Region experiences a significant impact, with approximately 250,000 deaths attributable to bacterial AMR infections [4]. In the European region, about 133,000 deaths are attributed to bacterial AMR infections [5].
Although the overuse and misuse of antimicrobials in humans, animals and their environments are the main factors that increase AMR, limited AMR surveillance and poor infection prevention and control practices also contribute to the increase in AMR infections [6]. Furthermore, immunosuppressive conditions such as human immunodeficiency virus (HIV) infection create favorable conditions for the development of bacterial resistance. People living with HIV are more likely to have contact with healthcare facilities and be exposed to invasive medical procedures than the general population is. Moreover, these populations are vulnerable to bacterial AMR infections due to frequent hospital admissions, the consumption of many antimicrobial agents, and the weakened immune system to fight infections [7,8]. Additionally, antibiotic prophylaxis for the prevention of opportunistic infections among HIV patients is the major factor contributing to the development of resistance [9].
Human immunodeficiency virus remains a major global public health concern, with a significant burden in many low- and middle-income countries. As of 2023, an estimated 39.9 million people were living with HIV worldwide, and approximately 630,000 deaths were attributed to HIV-related causes. The cumulative number of individuals living with HIV has continued to rise, reaching an estimated 42.3 million to date. This persistent and widespread prevalence of HIV not only presents challenges for disease management and health systems but also has important implications for other infectious diseases, particularly in relation to antimicrobial resistance [10].
Acquired immunodeficiency syndrome (AIDS) occurs at the most advanced stage of HIV infection, and in this stage, people can also develop not only opportunistic infections such as tuberculosis, pneumocystis pneumonia and cryptococcal meningitis but also severe bacterial infections [11,12]. These severe bacterial infections are caused by common bacterial pathogens that are grouped under gram-positive cocci and gram-negative bacilli [12]. Among bacterial pathogens, Enterobacterales are multidrug resistant owing to their ability to produce various resistance mechanisms, including efflux pumps, porin modification, overexpression, and enzyme production [13]. Enzymatic inactivation due to extended-spectrum β-lactamase (ESBL) enzyme production is the predominant cause of resistance to β-lactam antibiotics [13,14]. These enzymes can hydrolyze penicillins, cephalosporins, and aztreonam. Penicillins and cephalosporins are widely accessible and frequently used to treat various infections globally, especially in developing countries. [14].
Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are the predominant Enterobacterales that produce ESBL enzymes [14,15]. Infections with these pathogens increase the risk of treatment failure and have led to increased use of last-resort antibiotics such as carbapenems [16] and combination therapies with more toxic antibiotics such as polymyxin, which cause nephrotoxicity, ototoxicity, and neuromuscular blockade [17].
To improve the management of HIV-positive patients and reduce mortality due to complications with bacterial infections, comprehensive data concerning resistant bacteria due to ESBL enzyme inactivation of antibiotics are paramount. Nevertheless, on the basis of previous studies, the proportion of ESBL-producing Enterobacterales among HIV-positive individuals varies from 2.3% to 57.4% [18,19]. In addition, there is a great discrepancy in the prevalence of ESBL-producing Enterobacterales among HIV-positive individuals in previously published studies [19–31].
Moreover, most existing systematic reviews and meta-analyses on HIV-related infections have focused on multidrug-resistant Mycobacterium tuberculosis [32] and methicillin-resistant Staphylococcus aureus [33]. However, pooled data on ESBL-producing Enterobacterales among HIV-positive individuals are still lacking. Therefore, this systematic review and meta-analysis aimed to provide comprehensive data concerning ESBL-producing Enterobacterales among HIV-positive individuals across the globe.
Methods
Study design and protocol registration
This systematic review and meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (SF 1 Table). The protocol is available on the International Prospective Register of Systematic Reviews (PROSPERO ID: CRD42024557981).
Search strategy
Two authors (MT, KT) searched the Medline/PubMed, Hinari, Google Scholar, and Scopus electronic databases for studies published from 1 January 2010–12 May 2024. We used keywords alone and in combination with Boolean operators such as “OR” or “AND”. For example, articles were identified via MeSH terms from keywords of the title on Medline/PubMed, as follows (((((((antimicrobial resistance) OR (antibiotic resistance)) OR (multidrug resistance)) OR (extended-spectrum β-lactamase)) OR (ESBL)) AND (((((((gram-negative) OR (bacteria)) OR (bacilli)) OR (enterobacteriaceae)) OR (enterobacterales)) OR (Escherichia coli)) OR (Klebsiella pneumoniae))) AND (((((immunocompromised host) OR (human immunodeficiency virus)) OR (acquired immunodeficiency syndrome)) OR (HIV)) OR (HIV/AIDS))) AND (“2010/01/01” to “2024/05/12”).
Outcome of interest
The main outcome was the prevalence of ESBL-producing Enterobacterales among people living with HIV across the globe. We estimated the prevalence by dividing the number of ESBL-producing Enterobacterales cases by the total sample size. For case-control studies, the prevalence was calculated only among the cases. The prevalence of ESBL-producing bacteria for individual species was calculated by dividing the number of ESBL-producing isolates by the total number of isolates for that specific species.
Eligibility criteria
We used the CoCoPop (Condition, Context, and Population) approach, in which the prevalence of ESBL-producing Enterobacterales was considered the condition (CO), people living with HIV were considered the population (POP), and the world served as the context (CO). To identify eligible articles, we declared the predetermined inclusion and exclusion criteria; all cross-sectional, case‒control, and cohort studies reported ESBL-producing Enterobacterales among people living with HIV worldwide. The review included studies with mixed populations which reported on ESBL-producing Enterobacterales prevalence by HIV status. Studies published between January 1, 2010, and May 12, 2024, which were written exclusively in the English language, and studies that were peer reviewed were included in this systematic review and meta-analysis. However, systematic reviews and meta-analyses, case reports, case series, conference papers and pilot studies were excluded. Preprint studies were also excluded.
Quality assessment
Three reviewers (MT, KT, and AB) independently and in duplicate screened the titles and abstracts of the studies and subsequently assessed the potential eligibility of the relevant full texts on the basis of the predefined inclusion criteria. The quality of studies was assessed via standard critical appraisal tools prepared by the Joanna Briggs Institute (JBI) for prevalence and case‒control studies [34]. The JBI appraisal checklist contains 9 and 10 questions for cross-sectional and case‒control studies, respectively. These critical appraisal tools have yes, ‘no’, ‘unclear’, and ‘not applicable’ options. For each question, a score of 0 was assigned for ‘no’, ‘unclear’, and ‘not applicable’ and a score of 1 was assigned for ‘yes’. The discrepancies were solved by taking the average score. The total score is calculated by counting the number of “yes” in each row. On the basis of the score of the quality assessment tool, the highest score had the minimum risk of bias. Overall scores ranging from 0–4, 5–6, and 7–9 for prevalence studies and from 0–4, 5–7, and 8–10 for case‒control studies are declared high, moderate, and low risk of bias, respectively [35]. Finally, studies with a score of five and above for “yes” (have moderate and low risk of bias) were included in the systematic review and meta-analysis (SF 2 Table).
Data extraction
All of the studies obtained from different electronic databases were combined and properly exported to EndNote version 9.2 (Clarivate Analytics, Philadelphia, PA, USA). Then, the articles were merged into one folder for identification, duplicate articles were removed, and the quality of the studies was checked. We subsequently assessed the eligibility of the studies imported into Microsoft Excel 2019 (Microsoft Corp., Redmond, WA, USA).
All important parameters were extracted from each study by three authors (MT, KT, & AB.) independently. Discrepancies between them were resolved by consensus. The data extraction format was prepared according to the Preferred Reporting Items for Systematic Reviews and Meta Analyses (PRISMA) guidelines. For each study, the primary author, year of publication, sample size, study design, study year, study country, age group, investigation type (phenotype or genotype), status (infection or colonization) and methods of detection for ESBL were extracted. Furthermore, data on the total number of ESBL-producing isolates, individual ESBL-producing species, and their corresponding total number of isolates were extracted.
Statistical analysis
The extracted data were exported to STATA software version 17 for analysis. We conducted a meta-analysis via the random effects DerSimonian and Laird model to estimate the pooled prevalence and 95% confidence intervals (CIs) [36]. The presence of between-study heterogeneity was checked by using the Cochrane Q statistic. The magnitude of heterogeneity between the included studies was quantitatively measured by the inverse variance (I2 statistic). I2 = 0, I2 = 0–25%, I2 = 50–75%, and I2 > 75% indicate no, low, moderate, and high heterogeneity, respectively. The significance of heterogeneity was determined by the p value of the Cochrane Q statistic, and a p value of less than 0.05 was evidence of heterogeneity [37]. The possible sources of heterogeneity were further investigated by performing a subgroup analysis in reference to the continents, publication year, age categories, and methods of confirmation for the ESBL. Additionally, a sensitivity analysis was conducted to determine the influence of single studies on the pooled estimates. Publication bias was checked by using a funnel plot test graphically and more objectively through Egger’s regression tests. A statistically significant Egger’s test (P value < 0.05) indicates the presence of a small study effect [38].
Results
Study selection and identification
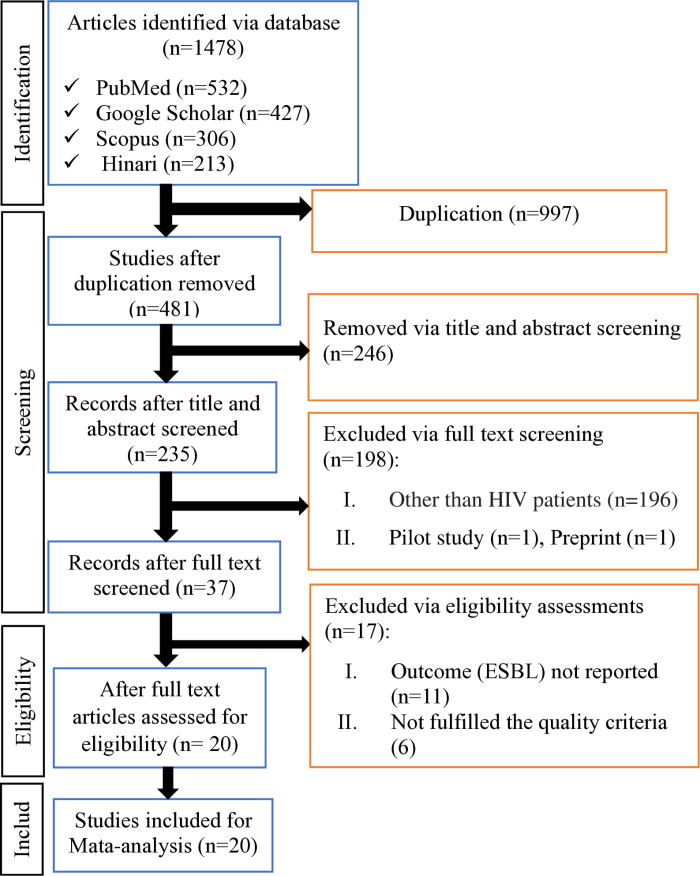
We identified a total of 1478 articles from available scientific databases such as PubMed, Hinari, Google Scholar, and Scopus. Of these, 997 studies were removed because they were duplications. Four hundred eighty-one studies were screened by reading their titles and abstracts. Two hundred forty-six articles were removed due to unrelated topics, incorrect publication periods, mixed or non-human populations, or inclusion of non-Enterobacterales or other pathogens. After that, 235 articles were screened by reading their full texts, and 198 studies were excluded because they were pilot studies, preprints, or involved study participants other than HIV patients. Thirty-seven studies passed the eligibility assessment, but 17 articles were removed because the outcomes were not reported or did not fulfill the quality criteria. Finally, 20 studies were eligible and included in the final meta-analysis [18–21,23–31,39–45], as presented in the PRISMA flow diagram (Fig 1).
Fig 1. A flow diagram of study selection for systematic review and meta-analysis of ESBL-producing Enterobacterales among HIV-positive individuals.
Summary of the risk of bias results
Out of the thirty-seven studies that passed the eligibility assessment, 11 were excluded because they did not meet the quality criteria. Among the studies that met the quality criteria, 13 out of 20 (65.0%) had a low risk of bias, while 7 (35.0%) had a moderate risk of bias. Six studies were excluded due to a high risk of bias (see SF 2 Table).
Characteristics of the studies included in the systematic review and meta-analysis
Among the included studies, 15 (75.0%) studies were published in and after 2020 [18–21,23–28,39,40,43–45]. Eighteen (90.0%) studies were cross-sectional [18–21,23–31,39,40,43–45], whereas the remaining two studies were case‒control studies [30,41]. Eleven (55.0%) studies focused on colonization [20,21,23,24,28–30,39,42,43,45], whereas the remaining 9 studies focused on infection [18,19,25–27,31,40,41,44]. All of the studies included in this review were from three continents: Africa [18,20–23,25,27,29,31,39,40,43–45], Asia [19,26,28,30,41], and Europe [24,42] (Table 1).
Table 1. Characteristics of individual studies included in the meta-analysis of ESBL-producing Enterobacterales among HIV-positive individuals across the globe, 2024.
| Study (Author, Year) | Study Year | Country | Design | Sample size | Specimen type | Age group | Status | Inv. M* | Meth** | ESBL case N (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Bayleyegn et al, 2021 [39] | 2020 | Ethiopia | CS | 161 | Stool | Children | Colonization | P | CDT | 32 (19.9) |
| Dimani et al, 2023 [20] | 2021 | Cameroon | CS | 185 | Stool and rectal swabs | All age | Colonization | P&G | DDST | 61 (32.9) |
| Endalamaw et al, 2020 [18] | 2017 | Ethiopia | CS | 387 | Urine | All age | Infection | P | DDST | 9 (2.3) |
| Falodun et al, 2021 [21] | 2017 | Nigeria | CS | 100 | Stool | Adult | Colonization | P | DDST | 56 (56.0) |
| Jerry et al, 2021 [40] | 2017 | Nigeria | CS | 205 | Urine | All age | Infection | P | DDST | 23 (11.2) |
| John-Onwe et al, 2022 [25] | 2019 | Nigeria | CS | 200 | Urine | All age | Infection | P | DDST | 58 (29.0) |
| M.R. Rameshkumar et al, 2021 [19] | NR | India | CS | 183 | Urine, pus, sputum, blood, and vaginal swabs | All age | Infection | P&G | CDT | 105 (57.4) |
| Maharjan et al, 2022 [26] | 2019 | Nepal | CS | 263 | Sputum | All age | Infection | P&G | CDT | 19 (7.2) |
| Manyahi et al, 2020 [23] | 2017-2018 | Tanzania | CS | 595 | Rectal swabs | Adult | Colonization | P&G | DDST | 244 (41.0) |
| Nwokolo et al, 2022 [27] | NR | Nigeria | CS | 363 | Urine | All age | Infection | P&G | CDT | 44 (12.1) |
| Osazuwa et al, 2011 [31] | 2009-2010 | Nigeria | CS | 948 | Urine & diarrheal stools | All age | Infection | P | DDST | 38 (4.0) |
| Padmavathy et al, 2011 [41] | NR | India | CC | 50 | Urine | All age | Infection | P | DDST | 13 (26.0) |
| Reinheimer et al, 2017 [42] | 2014-2016 | Germany | CC | 109 | Rectal swabs | Adult | Colonization | P | Vitek-2 | 7 (6.4) |
| Said et al, 2022 [43] | 2021 | Tanzania | CS | 236 | Stool and rectal swabs | Children | Colonization | P&G | CDT | 56 (23.7) |
| Simeneh et al, 2022 [44] | 2021 | Ethiopia | CS | 251 | Urine | Adult | Infection | P | DDST | 9 (3.6) |
| Singh et al, 2020 [28] | 2017-2018 | India | CS | 100 | Oral swabs | Adult | Colonization | P&G | CDT | 14 (14.0) |
| Subramanya et al, 2019 [30] | 2016-2017 | Nepal | CS | 119 | Rectal swabs | All age | Colonization | P&G | DDST | 46 (38.7) |
| Surgers et al, 2022 [24] | 2018-2019 | France | CS | 500 | Rectal swabs | Adult | Colonization | P&G | CDT | 61 (12.2) |
| Wilmore et al, 2017 [29] | 2014-2015 | Zimbabwe | CS | 175 | Stool | Children | Colonization | P | CDT | 24 (13.7) |
| 12 et al, 2022 [45] | 2021 | Cameroon | CS | 175 | Vaginal swabs | Adult | Colonization | P&G | DDST | 12 (6.9) |
Note: Inv. M
= Investigation methods, Meth
= Method used for phenotypic ESBL confirmation, CS = Cross-sectional, CC = Case‒control, CDT = Combination disk test, DDST = Double‒disc synergy test, NR = Not reported, P = Phenotypic, G = Genotypic, ESBL = Extended-spectrum β-lactamase.
A total of 5305 HIV-positive individuals from 20 studies were included in the meta-analysis of ESBL-producing Enterobacterales. Among 20 studies, only one detected ESBL by using Vitek-2 [42], but others used the combination disk test (CDT) or double disk synergy test (DDST). The minimum and maximum sample sizes were 50 [41] and 948 [31], respectively. A minimum (2.33%) and maximum (57.4%) prevalence of ESBL-producing Enterobacterales were reported by Endalamaw et al. [18] and M.R. Rameshkumar et al. [19], respectively (Table 1).
Pooled prevalence of ESBL-producing Enterobacterales among HIV positive individuals
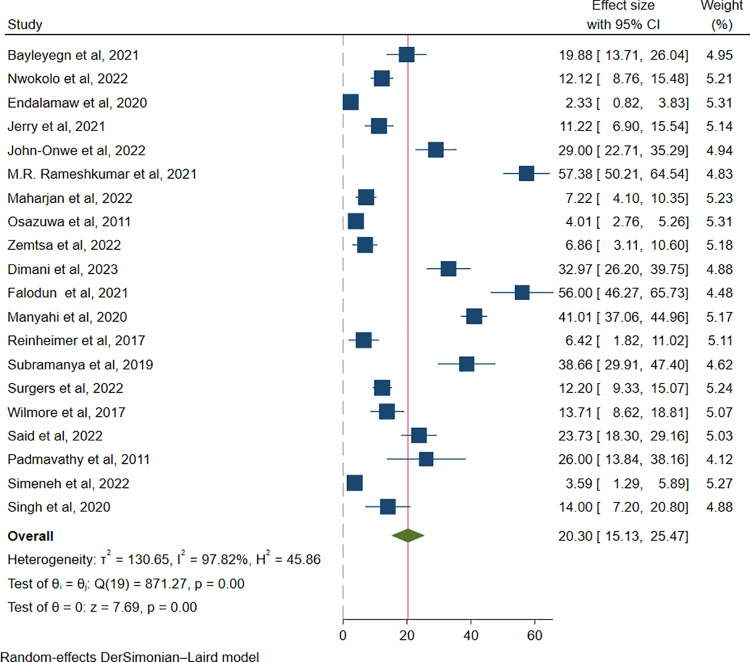
The overall pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals was 20.30% (931/5305; 95% CI: 15.13–25.47%, P < 0.001), with a high level of heterogeneity (I2 = 97.82%, P < 0.001), as presented in (Fig 2).
Fig 2. Forest plot showing overall pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals.
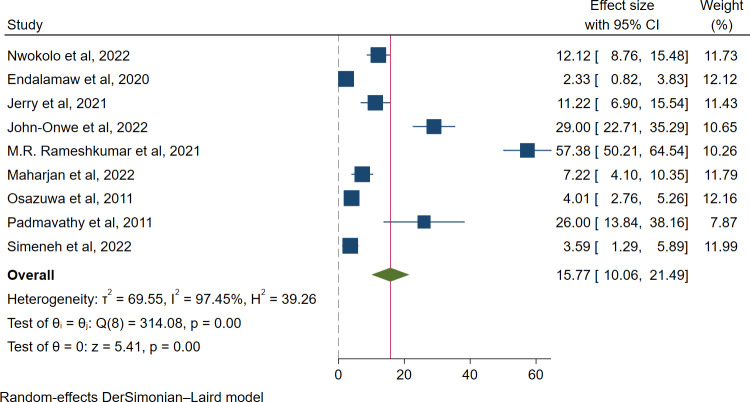
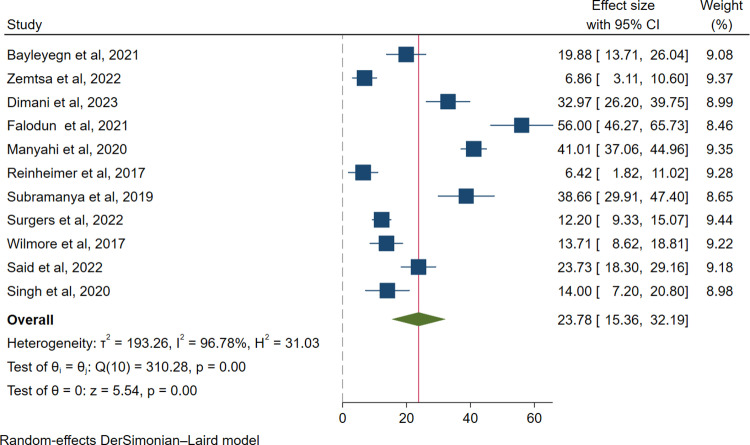
Moreover, the pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals with infection was 15.77% (318/2850; 95% CI: 10.06–21.49, I² = 97.45%, p < 0.001) (Fig 3), whereas among asymptomatic HIV-positive individuals, it was 23.78% (613/2455; 95% CI: 15.36–32.19, I² = 96.78%, p < 0.001) (Fig 4).
Fig 3. Forest plot for the pooled prevalence of ESBL-producing Enterobacterales among infection case.
Fig 4. Forest plot for the pooled prevalence of ESBL-producing Enterobacterales among colonization case.
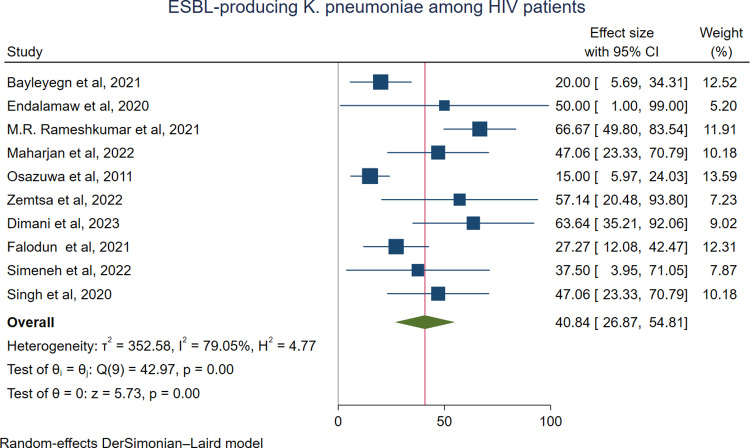
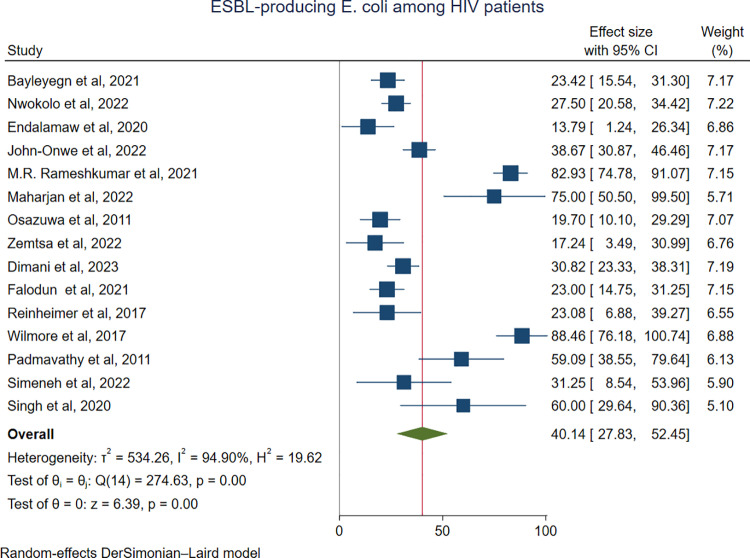
The predominant ESBL-producing pathogens among HIV-positive individuals were K. pneumoniae, with a pooled prevalence of 40.84% (76/217; 95% CI: 26.87–54.81%) (Fig 5), followed closely by E. coli at 40.14% (348/985; 95% CI: 27.83–52.45%) (Fig 6), and Proteus species at 37.2% (24/74; 95% CI: 8.03–66.36%) (SF 4 Fig). In contrast, Citrobacter species were the least commonly identified ESBL producers, with a prevalence of 9.68% (5/55; 95% CI: 1.26–18.10%) (SF 4 Fig). Three studies reported ESBL production in Salmonella spp. The pooled prevalence of ESBL-producing Salmonella spp. was 24.52% (10/42; 95% CI: 7.04–42.00; I² = 94.71%, p ≤ 0.019) (SF 4 Fig).
Fig 5. Forest plot showing the pooled prevalence of ESBL-producing K. pneumoniae among HIV-positive individuals.
Fig 6. Forest plot showing the pooled prevalence of ESBL-producing E. coli among HIV-positive individuals.
Regarding infection and colonization with ESBL-producing E. coli and K. pneumoniae among HIV-positive individuals, comparable pooled prevalence rates were observed in both infection and colonization cases. For infection cases, the prevalence of ESBL-producing E. coli was 37.0% (237/537; 95% CI: 20.08–53.94%), while that of K. pneumoniae was 38.76% (42/119; 95% CI: 22.69–54.83%) (SF 4 Fig). Similarly, among colonization cases, the prevalence of ESBL-producing E. coli was 42.93% (343/448; 95% CI: 24.16–61.69%), and that of K. pneumoniae was 42.36% (34/98; 95% CI: 15.97–68.74%) (SF 4 Fig).
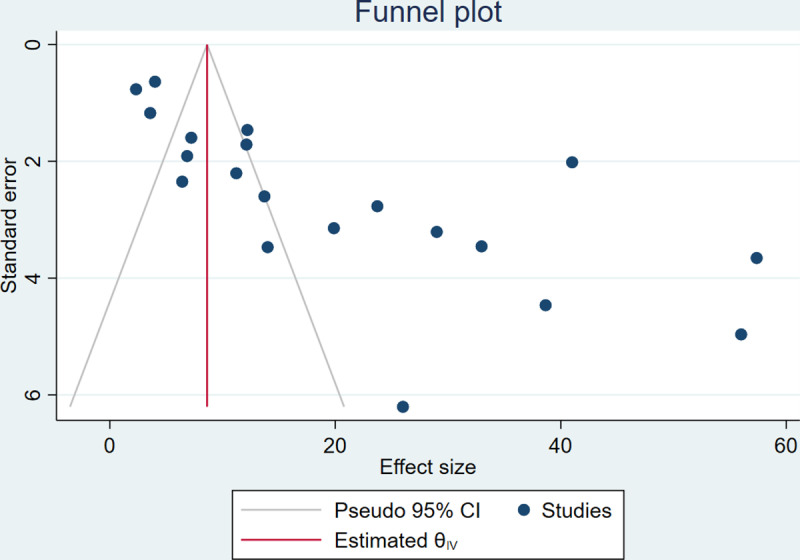
Publication bias
Publication bias was assessed to determine bias related to published and unpublished studies. The analysis produced an asymmetric funnel plot, which indicated the presence of publication bias (Fig 7). Moreover, Egger’s test revealed a significant publication bias (P < 0.001)(SF 3 Table).
Fig 7. Funnel plot showing publication bias for ESBL-producing Enterobacterales among HIV-positive individuals.
Nonparametric trim-and-fill analysis of the pooled prevalence of ESBL-producing Enterobacterales
A nonparametric trim-and-fill analysis was conducted to address the observed publication bias. After imputing data on the left, the pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals across the globe remained stable at 20.30% (95% CI: 15.13–25.47%) (SF 3 Table). Conversely, when analyzing 21 studies with one data points imputed on the right, the pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals was slightly higher at 21.58% (95% CI: 14.34–28.82%) (SF 3 Table).
Sensitivity analysis
A sensitivity analysis was performed via a random effects model to assess the impact of individual studies on the combined estimate. The pooled prevalence of ESBL-producing pathogens that was obtained after individual studies were excluded was within the 95% CI of the total combined estimate. This shows that no single study had an effect on the overall pooled prevalence (SF 3 Fig and Table).
Subgroup analysis
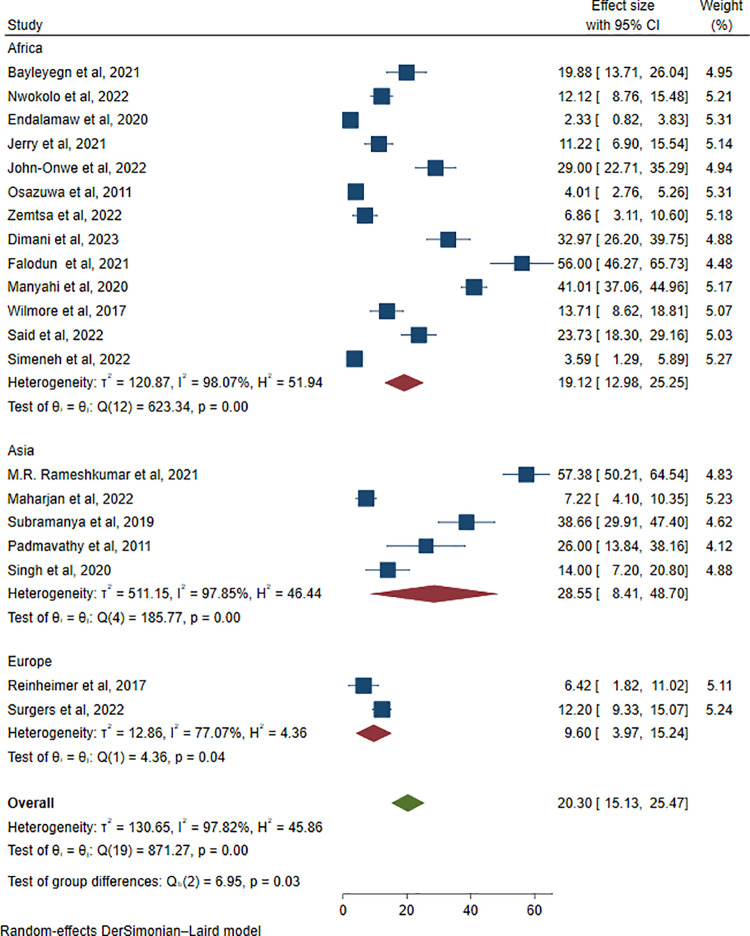
To address heterogeneity, subgroup analysis was carried out by geographical location, publication year, age group, and phenotypic ESBL confirmation methods. When subgroup analysis was performed by continent, the highest pooled prevalence of ESBL-producing pathogens among HIV patients was observed in Asia, at 28.55% (195/715; 95% CI: 8.41–48.70, I2 = 97.85%, P < 0.001), followed by Africa, at 19.12% (666/3981; 95% CI: 12.98–25.25, I2 = 98.07%, P < 0.001). On the other hand, the lowest pooled prevalence was found in Europe, at 9.60% (68/609; 95% CI: 3.97–15.24, I2 = 77.07%, P = 0.04) (Fig 8).
Fig 8. Subgroup analysis of ESBL-producing Enterobacterales among HIV-positive individuals by continent.
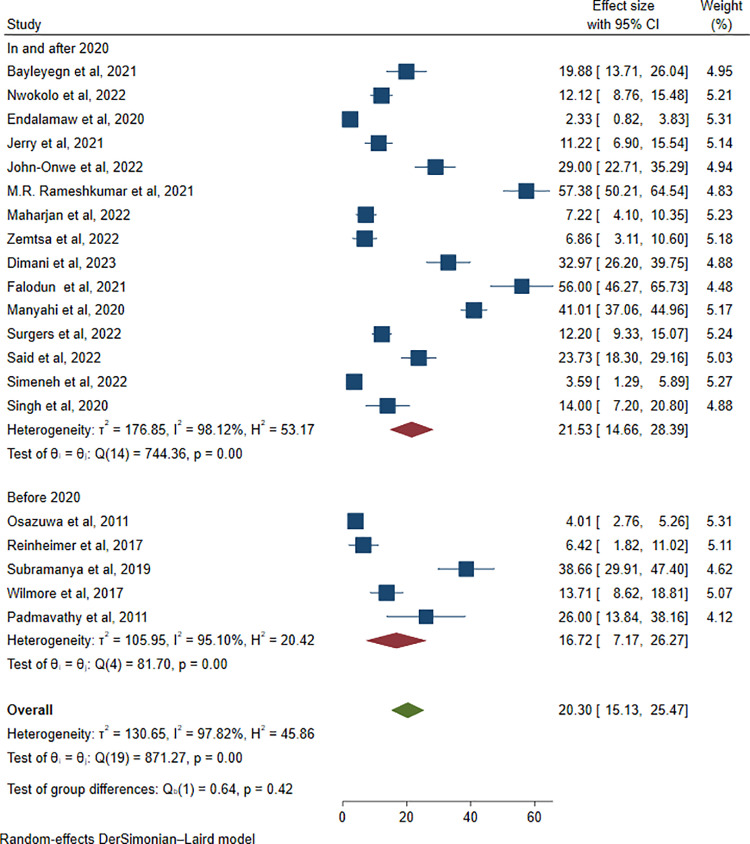
Similarly, subgroup analysis on the basis of the publication year of studies revealed that the highest pooled estimate was 21.53% (803/3904; 95% CI: 14.66–28.39, I2 = 98.12%, P < 0.001) in and after 2020, whereas for studies published before 2020, the prevalence was 16.72% (128/1401; 95% CI: 7.17–26.27, I2 = 95.10%, P = 0.001) (Fig 9).
Fig 9. Subgroup analysis of ESBL-producing Enterobacterales by year of publication.
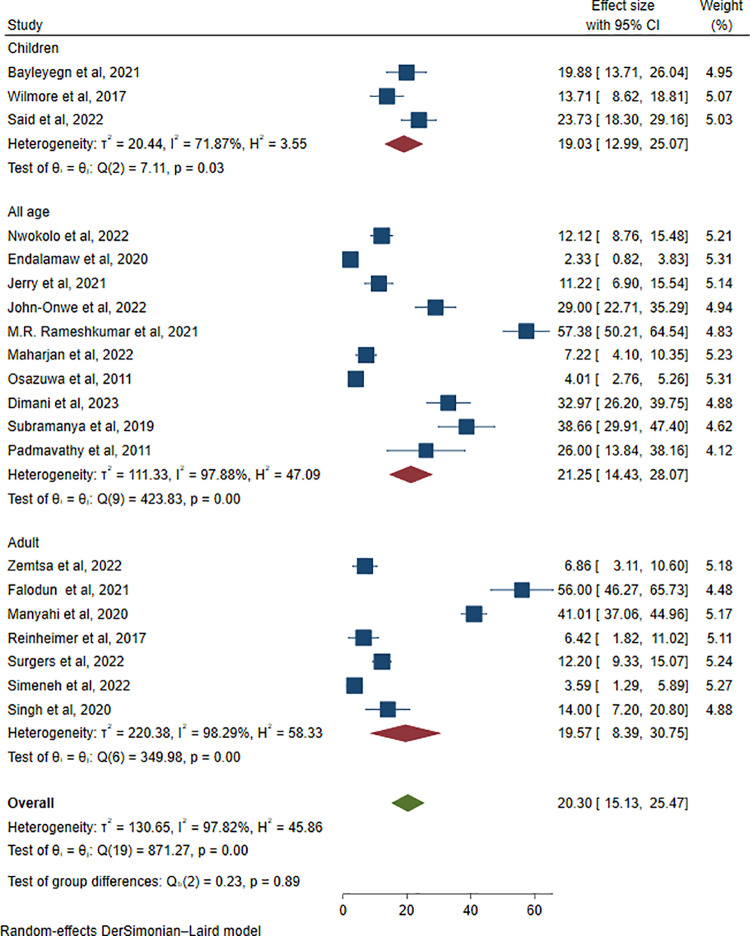
In addition, a subgroup analysis was conducted based on patient age groups. The pooled prevalence of ESBL-producing Enterobacterales did not show a substantial difference across age categories. The prevalence was reported as 19.03% (416/2903; 95% CI: 12.99–25.07, I² = 71.87%, P = 0.03) in children, 19.57% (112/572; 95% CI: 8.39–30.75, I² = 98.29%, P < 0.001) in adults, and 21.25% (403/1830; 95% CI: 14.43–28.07, I² = 97.88%, P < 0.001) in studies involving all age groups (Fig 10).
Fig 10. Forest plot showing subgroup analysis of ESBL-producing Enterobacterales via age categories.
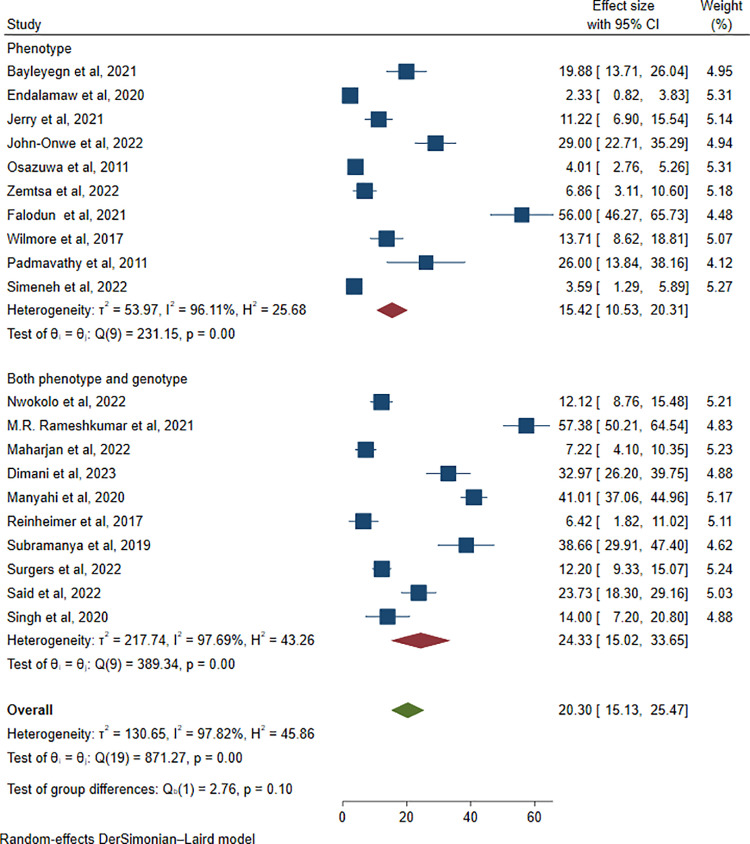
Furthermore, a subgroup analysis was conducted based on the type of investigation method. The highest pooled prevalence of ESBL-producing pathogens among HIV-positive patients was reported in studies employing genotypic methods (24.33%, 657/2653; 95% CI: 15.02–33.65; I² = 97.69%; P < 0.001), followed by studies using phenotypic methods (15.42%, 274/2652; 95% CI: 10.53–20.31; I² = 96.11%; P < 0.001) (Fig 11).
Fig 11. Forest plot showing subgroup analysis of ESBL-producing Enterobacterales via investigation methods.
We also performed subgroup analysis in terms of phenotypic ESBL detection methods. The highest pooled prevalence was observed using the double-disk synergy test (DDST), at 22.09% (569/3215; 95% CI: 14.80–29.37, I² = 98.38%, P < 0.001), followed by the combined disk test (CDT), at 19.70% (355/1981; 95% CI: 11.91–27.50, I² = 96.04%, P < 0.001). The lowest pooled prevalence was recorded using the Vitek-2 method, at 6.42% (7/109; 95% CI: 1.82–11.02, I² = 0.0%, P < 0.001) (SF 5 Fig).
Risk factors for the prevalence of ESBL producers among HIV-positive individuals
A meta-analysis could not be performed because of the small number of studies reporting risk factors. Among the 20 included studies, only 4 reported risk factors; among these, variability in variables such as a history of antibiotic use, hospital admission, and low cluster of differentiation 4 (CD4) T lymphocyte counts was a risk factor for a higher prevalence of ESBL-producing Enterobacterales among HIV-positive individuals (Table 2).
Table 2. Significant risk factors in some of the included studies.
| Study | Risk factors assessed | Analysis | Significant risk factors | Odds ratio (95% CI) |
|---|---|---|---|---|
| Subramanya et al, 2019 [30] | Age, sex, occupation, CD4 count, ART, ART duration, admitted to hospital in at 6 m, history of antibiotic use in at 6 m, animal contact (domestic and livestock), using medication over-the-counter, international travel, food habit | Multivariate | With ART | 1.537 (0.349,6.762) |
| Admitted to hospital in the previous 6 m | 3.638 (0.793,16.68) | |||
| Close contact with livestock | 1.153 (0.515,2.581) | |||
| Using medication over-the-counter | 1.988 (0.898,4.401) | |||
| Bayleyegn et al, 2021 [39] | Age, sex, residence, educational status, opportunistic infections, presence of fever, WHO stage of HIV, ART type, ART duration, history of antibiotic use, viral load, presence of diarrhea, eating uncooked products, eating row vegetable | Multivariate | History of antibiotic use | 3.2 (1.05–9.9) |
| Manyahi et al, 2020 [23] | Age, sex, residence, educational status, WHO stage of HIV, CD4 count, admitted to hospital in at 12 m, history of antibiotic use in at 1 m | Multivariate | CD4 count <350 | 1.78 1.03–3.09 |
| History of antibiotic use in the previous 1 m | 1.55 1.08–2.22 | |||
| Wilmore et al, 2017 [29] | Age, gender, CD4, viral load, ART duration, admitted to hospital with pneumonia in last 12 m, admitted to hospital in at 12 m | Multivariate | With ART ≤ 1year | 8.47 (2.22–2.27) |
| Admitted to hospital with pneumonia in last 12 m | 8.47 (1.12–64.07) |
Note: ART = antiretroviral therapy, HIV = human immunodeficiency virus, WHO = World Health Organization, CL = confidence interval, M = month, CD4 = cluster of differentiation 4.
Types of ESBL enzyme-encoding genes via Enterobacterales among HIV-positive individuals
Although the included studies employed heterogeneous genotyping techniques, blaCTX-M (73/150, 48.7%) was the most prevalent genotype, followed by blaTEM (49/150, 32.7%) among the commonly reported ESBL enzyme-encoding genes, as documented in five studies (Table 3).
Table 3. Types of genes in some studies that carry out genotypic analysis.
| Study (Author, Year) | No of isolates subjected to genotype test | Types of isolates | Types of genes | ||||
|---|---|---|---|---|---|---|---|
| Total ESBL gene | bla SHV | bla TEM | bla CTX-M | bla CTX-M + bla TEM | |||
| Nwokolo et al, 2022 [27] | 44 | E. coli | 26 | 7 (26.9) | 8 (30.8) | 11 (42.3) | |
| Maharjan et al, 2022 [26] | 12 | E. coli | 9 | – | 2 (22.2) | 3 (33.3) | 4 (44.5) |
| 17 | K.pneumoniae | 7 | – | 0 | 3 (42.9) | 4 (57.1) | |
| 12 et al, 2022 [45] | 14 | E. coli | 12 | – | 6 (50.0) | 1 (8.3) | 5 (41.7) |
| 5 | K. pneumoniae | 4 | – | – | 4 (100) | – | |
| Dimani et al, 2023 [20] | 45 | E. coli | 63 | 3 (4.7) | 27 (42.9) | 33 (52.4) | – |
| 11 | K. pneumoniae | 12 | 4 (33.3) | 3 (25.0) | 5 (41.7) | – | |
| Singh et al, 2020 [28] | 10 | E. coli | 9 | 0 | 2 (22.2) | 7 (77.8) | – |
| 17 | K. pneumoniae | 8 | 1 (12.5) | 1 (12.5) | 6 (75.0) | – | |
| Total | 175 | – | 150 | 15 (10.0) | 49 (32.7) | 73 (48.7) | 13 (8.6) |
Note: ESBL = extended-spectrum β-lactamase, blaTEM = Temoneira β-lactamase, blaSHV = sulfhydryl reagent variable β-lactamase, blaCTX-M = cefotaxim-hydrolizing β-lactamase..
Discussion
The management of antibiotic-resistant bacterial infections especially those caused by Enterobacterales poses a significant challenge in the context of infectious diseases like HIV [46]. Although not all ESBL-producing strains meet the strict definition of multidrug resistance (MDR), the production of ESBLs often confers resistance to multiple β-lactam antibiotics and is frequently associated with co-resistance to other antimicrobial classes. These pathogens are commonly linked to nosocomial infections [14]. In this context, our systematic review and meta-analysis aimed to estimate the global burden of ESBL-producing Enterobacterales among individuals living with HIV.
In our systematic review and meta-analysis, the overall pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals was 20.30% (95% CI: 15.13–25.47%, P < 0.001), with a high level of heterogeneity (I2 = 97.82%, p < 0.001). This significant prevalence of ESBLs among these populations may be due to the increased number of antibiotics consumed and increased risk of acquisition of resistant pathogens during healthcare visits and hospitalizations [47]. These populations are immunocompromised and need more frequent healthcare appointments and are vulnerable to comorbid conditions, which require hospital admissions, than are the general population [48].
The highest pooled prevalence of ESBL-producing pathogens among HIV-positive individuals was reported to be colonization 23.78% (95% CI: 15.36–32.19, I² = 96.78%, p < 0.001), followed by infection 15.77% (95% CI: 10.06–21.49, I² = 97.45%, p < 0.001). The higher prevalence may be explained by the methods used to report on colonization, sample size, method of detection, culture media. This may be because the gut microbiota is mostly composed of Enterobacterales, and HIV infection alters the composition of the microbiome and decreases the number of CD4 + T cells in the gut-associated lymphoid tissue, which is associated with microbiota dysbiosis that favors colonization and subsequent infections with resistant strains [49,50].
In the present meta-analysis, the predominant ESBL-producing pathogens among HIV-infected patients were K. pneumoniae, with a pooled prevalence of 40.84% (95% CI: 26.87–54.81%), followed closely by E. coli at 40.14% (95% CI: 27.83–52.45%). These organisms are known to resist beta-lactam antibiotics through the production of ESBL enzymes [51,52]. K. pneumoniae is most commonly associated with healthcare-associated infections, whereas E. coli is typically linked to community-acquired infections. As a result, infections caused by these pathogens may contribute to increased complications in HIV-infected patients [53].
In this meta-analysis, a high level of heterogeneity (I2 = 97.82%) was observed. This is not surprising given the differences in study settings, patient statuses, age groups, sample types, publication years and methods used for the detection of ESBLs. To determine the possible sources of heterogeneity, subgroup analysis was performed by continent, and the highest pooled prevalence of ESBL-producing pathogens among HIV-positive individuals was reported in Asia: 28.55% (95% CI: 8.41–48.70, I2 = 97.85%, P < 0.001), followed by the African continent 19.12% (95% CI: 12.98–25.25, I2 = 98.07%, P < 0.001). However, the lowest pooled prevalence was found in Europe, at 9.60% (95% CI: 3.97–15.24, I2 = 77.07%, P = 0.04). This is because the use of trimethoprim‒sulfamethoxazole prophylaxis in developed nations is decreasing because of the early diagnosis of HIV and well-controlled antiretroviral therapy (ART), which reduces immunosuppression and bacterial infections [54].
However, the high burden of HIV infection and weak health systems to diagnose HIV early, inadequate adherence and poorly controlled ART make the immune system weakened and susceptible to bacterial infections in low-income countries. Additionally, as opportunistic infections are common among HIV-positive individuals in these countries, trimethoprim‒sulfamethoxazole prophylaxis is widely used [55]. For this reason, this antibiotic can be used to coselect resistant strains among these populations [9]. Several studies have reported that trimethoprim-sulfamethoxazole prophylaxis is mostly associated with increased non-susceptibility of Enterobacterales to beta-lactam antibiotics and an increased risk of ESBL-producing Enterobacterales [9,56,57].
Another suggested reason for the highest prevalence of ESBL-producing pathogens among HIV-positive individuals in Asia and Africa is the poor antibiotic stewardship program and low control mechanism for antibiotic usage, which causes overuse and misuse of antibiotics in healthcare facilities that facilitate resistant infections [58]. Additionally, poor infection prevention mechanisms, such as poor hygiene and sanitation practices, environmental contamination and inadequate decontamination of medical devices, aggravate colonization and infection with resistant pathogens among HIV-positive individuals in developing countries [58,59].
According to the subgroup analysis by publication year, the highest pooled estimate of 21.53% (95% CI: 14.66–28.39, I2 = 98.12%, P < 0.001) was observed in studies published in and after 2020, whereas before 2020, the prevalence was 16.72% (95% CI: 7.17–26.27, I2 = 95.10%, P = 0.001). This gap may be due to the emergence of multidrug-resistant bacteria because the prevalence of ESBL-producing bacteria has increased over time, and recently studied papers may include a high number of ESBL-producing pathogens [60].
Furthermore, the subgroup analysis was carried out based on the investigation methods. Accordingly, the highest pooled prevalence of ESBL-producing pathogens among HIV patients was reported in studies that included genotypic investigations (24.33%, 95% CI: 15.02–33.65, I² = 97.69%, P < 0.001), followed by phenotypic investigations (15.42%, 95% CI: 10.53–20.31, I² = 96.11%, P < 0.001). The higher pooled prevalence of ESBL-producing pathogens reported in studies using genotypic methods may be attributed to the greater sensitivity and specificity of these techniques in detecting resistance genes, including those that are silent or weakly expressed. In contrast, phenotypic methods may underestimate prevalence due to their reliance on observable resistance, which can be affected by gene expression levels or overlapping resistance mechanisms. Additionally, genotypic studies may have been conducted in settings with higher antibiotic pressure or more advanced diagnostic capacity, contributing to the observed differences [61].
In our systematic review, among the 20 included studies, only 2 reported that a history of antibiotic use in the previous month was a risk factor for infection and/or colonization with ESBL-producing pathogens among HIV-positive individuals. Additionally, the highest prevalence of ESBL-producing pathogens was reported in HIV-positive individuals who were admitted to the hospital for the last 6 and 12 months. Several previous studies identified prior hospitalization and antibiotic exposure as risk factors for antibiotic-resistant infections [12,47,62]. Patients may acquire resistant pathogens from hospitals, which may be due to selective pressure; when patients use antibiotics, sensitive bacterial strains are eliminated, leaving behind or selecting those variants that can resist them [47]. As cephalosporins are the most commonly used antibiotics for the treatment of various infections, they favor ESBL-producing strains [63].
Additionally, some studies have shown that taking ART for less than one year is a risk factor for ESBL incidence, and the risk of infection with ESBL-producing pathogens among HIV-positive individuals varies according to the CD4 count and is greatest in HIV-positive individuals with CD4 counts <350 cells/mm3. Low immunity with low CD4 + T cells is associated with an increased risk of opportunistic infections, increasing the likelihood of antimicrobial use and hospitalization. Consequently, this phenomenon predisposes resistant pathogens to the emergence of ESBL-producing strains [64].Among the various ESBL enzyme-encoding genes, blaCTX-M is the most prevalent (48.7%), followed by blaTEM (32.7%). The dominance of blaCTX-M genes is largely attributed to their efficient dissemination through mobile genetic elements such as plasmids, transposons, insertion sequences, and integrons, which facilitate the horizontal transfer of resistance genes among Enterobacterales and other gram-negative bacilli in clinical settings worldwide [65]. In developing countries, where infection prevention and control measures are often limited, this gene transfer is even more widespread occurring not only in humans but also in animals and the environment [66]. Because these mobile genetic elements frequently carry multiple resistance determinants, pathogens harboring ESBL genes are also more likely to acquire additional MDR traits [65,66]. As a result, infections caused by these organisms are harder to treat. Therefore, infection with these pathogens complicates treatment options and intensifies the level of care required for immunocompromised patients, such as those living with HIV.
Limitations
Study restriction by language and year of publication are the main limitations of this study. Furthermore, few countries are overrepresented, as the majority of studies are from Africa, specifically, sub-Saharan Africa; however, owing to the high burden of HIV in this region, accurate generalization is difficult. Since a few studies reported the risk factors and the heterogeneous variables used, a meta-analysis was not performed on the associated factors.
Conclusion and recommendations
This study revealed a significant prevalence of ESBL-producing Enterobacterales among HIV-positive individuals, with K. pneumonia and E. coli being the dominant ESBL producers. The highest magnitude of ESBL-producing pathogens was observed in Asia and Africa. With respect to the types of ESBL enzyme-encoding genes, the most prevalent was blaCTX-M followed by blaTEM. Equitable allocation of resources across all regions is needed to reduce mortality due to complications in HIV patients with severe bacterial infections; the regions with the highest prevalence rates should pay attention to early diagnosis of HIV, adequate adherence and well-controlled ART. Consequently, this helps to reduce immunosuppression and bacterial infections. Early identification of infections and colonization with antibiotic-resistant pathogens among HIV patients is also needed.
Supporting information files
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
List of abbreviations
- AMR
Antimicrobial resistance
- ART
Antiretroviral therapy
- CDT
Combination disk test
- DDST
Double-disk synergy test
- ESBL
Extended-spectrum β-lactamase
- HIV
Human immunodeficiency virus
- MDR
Multidrug resistance
- WHO
World Health Organization.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.World Health Organization. Ten threats to global health 2019. [Available from: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019. [Google Scholar]
- 2.Naghavi M, Vollset SE, Ikuta KS, Swetschinski LR, Gray AP, Wool EE, et al. Global burden of bacterial antimicrobial resistance 1990-2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404(10459):1199–226. doi: 10.1016/S0140-6736(24)01867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh TR, Gales AC, Laxminarayan R, Dodd PC. Antimicrobial resistance: addressing a global threat to humanity. PLoS Med. 2023;20(7):e1004264. doi: 10.1371/journal.pmed.1004264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sartorius B GA, Weaver ND, Aguilar GR, Swetschinski LR, Ikuta KS et al. The burden of bacterial antimicrobial resistance in the WHO African region in 2019: a cross-country systematic analysis. Lancet Glob Health. 2024;12(2):e201–16. doi: 10.1016/S2214-109X(23)00539-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mestrovic T AG, Swetschinski LR, Ikuta KS, Gray AP, Weaver ND et al. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. Lancet Public Health. 2022;7(11):e897–913. doi: 10.1016/S2468-2667(22)00225-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulis G, Sayood S, Gandra S. Antimicrobial resistance in low- and middle-income countries: current status and future directions. Expert Rev Anti Infect Ther. 2022;20(2):147–60. doi: 10.1080/14787210.2021.1951705 [DOI] [PubMed] [Google Scholar]
- 7.Marbou WJ, Kuete V. Bacterial resistance and immunological profiles in HIV-infected and non-infected patients at Mbouda AD LUCEM Hospital in Cameroon. J Infect Public Health. 2017;10(3):269–76. doi: 10.1016/j.jiph.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 8.Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ. High prevalence of faecal carriage of ESBL-producing enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS One. 2016;11(12):e0168024. doi: 10.1371/journal.pone.0168024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egwuatu CC, Ogunsola FT, Iwuafor AA, Akujobi CN, Egwuatu TO, Nnachi AU, et al. Effect of trimethoprim-sulfamethoxazole prophylaxis on faecal carriage rates of resistant isolates of Escherichia coli in HIV-infected adult patients in Lagos. Afr J Infect Dis. 2016;11(1):18–25. doi: 10.4314/ajid.v11i1.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. HIV/AIDS. 2024. [Available from: https://www.who.int/news-room/fact-sheets/detail/hiv-aids. [Google Scholar]
- 11.Rameshkumar MR, Arunagirinathan N. Drug-resistant bacterial infections in HIV patients. AICA. 2018;83. [Google Scholar]
- 12.Olaru ID, Tacconelli E, Yeung S, Ferrand RA, Stabler RA, Hopkins H, et al. The association between antimicrobial resistance and HIV infection: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27(6):846–53. doi: 10.1016/j.cmi.2021.03.026 [DOI] [PubMed] [Google Scholar]
- 13.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- 14.Peirano G, Pitout JDD. Extended-spectrum β-Lactamase-producing enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs. 2019;79(14):1529–41. doi: 10.1007/s40265-019-01180-3 [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing enterobacteriaceae. Clin Microbiol Rev. 2018;31(2):e00079-17. doi: 10.1128/CMR.00079-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu D, Mao W. Efficacy and safety of intravenous combined with aerosolised polymyxin versus intravenous polymyxin alone in the treatment of multidrug-resistant gram-negative bacterial pneumonia: a systematic review and meta-analysis. Heliyon. 2023;9(5):e15774. doi: 10.1016/j.heliyon.2023.e15774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esposito S, Canevini MP, Principi N. Complications associated with antibiotic administration: neurological adverse events and interference with antiepileptic drugs. Int J Antimicrob Agents. 2017;50(1):1–8. doi: 10.1016/j.ijantimicag.2017.01.027 [DOI] [PubMed] [Google Scholar]
- 18.Endalamaw D, Amsalu A, Biset S, Mekonnen F, Balew M, Eshetie S. Extended-spectrum β-lactamases-producing enterobacteria and antimicrobial resistance pattern among HIV/AIDS patients in the university of gondar specialized hospital, Ethiopia. Recent Adv Biol Med. 2020;6(1):11909. [Google Scholar]
- 19.Rameshkumar MR, Arunagirinathan N, Senthamilselvan B, Swathirajan CR, Solomon SS, Vignesh R, et al. Occurrence of extended-spectrum β-lactamase, AmpC, and carbapenemase-producing genes in gram-negative bacterial isolates from human immunodeficiency virus infected patients. J Infect Public Health. 2021;14(12):1881–6. doi: 10.1016/j.jiph.2021.11.008 [DOI] [PubMed] [Google Scholar]
- 20.Dimani BD, Founou RC, Zemtsa JR, Mbossi A, Koudoum PL, Founou LL, et al. Faecal carriage of multidrug-resistant and extended-spectrum β-lactamase-producing Enterobacterales in people living with HIV in Yaoundé, Cameroon. J Glob Antimicrob Resist. 2023;35:26–34. doi: 10.1016/j.jgar.2023.07.021 [DOI] [PubMed] [Google Scholar]
- 21.Falodun OI, Ajayi O, Ademola AE, Bakarey SA. Antibiotic susceptibility patterns and extended spectrum beta-lactamase (ESBL) production in Enterobactericeae isolated from stool samples of HIV and AIDS patients in Ibadan, Nigeria. Janaki Med Coll J Med Sci. 2021;9(1):5–15. [Google Scholar]
- 22.Falodun OI, Musa IB, Oyelade AA. Prevalence of extended beta-lactamase-producing Pseudomonas aeruginosa isolated from human immunodeficiency virus patients in Ibadan, Nigeria. Gene Reports. 2021;23:101159. [Google Scholar]
- 23.Manyahi J, Moyo SJ, Tellevik MG, Langeland N, Blomberg B. High prevalence of fecal carriage of extended spectrum β-lactamase-producing enterobacteriaceae among newly HIV-Diagnosed adults in a community setting in Tanzania. Microb Drug Resist. 2020;26(12):1540–5. doi: 10.1089/mdr.2020.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surgers L, Chiarabini T, Royer G, Rougier H, Mercier-Darty M, Decré D, et al. Evidence of sexual transmission of extended-spectrum β-lactamase-producing enterobacterales: a cross-sectional and prospective study. Clin Infect Dis. 2022;75(9):1556–64. doi: 10.1093/cid/ciac218 [DOI] [PubMed] [Google Scholar]
- 25.John-Onwe BN, Iroha IR, Moses IB, Onuora AL, Nwigwe JO, Adimora EE, et al. Prevalence and multidrug-resistant ESBL-producing E. coli in urinary tract infection cases of HIV patients attending Federal Teaching Hospital, Abakaliki, Nigeria. Afr J Microbiol Res. 2022;16(5):196–201. doi: 10.5897/ajmr2022.9624 [DOI] [Google Scholar]
- 26.Maharjan R, Bastola A, Adhikari N, Rijal KR, Banjara MR, Ghimire P, et al. Multidrug-resistant bacteria with ESBL genes: a growing threat among people living with HIV/AIDS in Nepal. BMC Infect Dis. 2022;22(1):526. doi: 10.1186/s12879-022-07503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nwokolo CJ, Ugwu MC, Ejikeugwu CP, Iroha IR, Esimone CO. Incidence and antibiotic susceptibility profile of uropathogenic Escherichia coli positive for extended spectrum β-lactamase among HIV/AIDS patients in Awka metropolis, Nigeria. Iran J Microbiol. 2022;14(3):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh M, Chakraborty A. Prevalence of extended spectrum β-lactamase genes among the oral gram negative rods isolated from HIV infected patients. Indian J Public Health. 2020;11(6):505. [Google Scholar]
- 29.Wilmore SMS, Kranzer K, Williams A, Makamure B, Nhidza AF, Mayini J, et al. Carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in HIV-infected children in Zimbabwe. J Med Microbiol. 2017;66(5):609–15. doi: 10.1099/jmm.0.000474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosuru Subramanya S, Bairy I, Nayak N, Padukone S, Sathian B, Gokhale S. Low rate of gut colonization by extended-spectrum β-lactamase producing Enterobacteriaceae in HIV infected persons as compared to healthy individuals in Nepal. PLoS One. 2019;14(2):e0212042. doi: 10.1371/journal.pone.0212042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osazuwa F, Osazuwa E, Imade P, Dirisu J, Omoregie R, Okuonghae P. Occurrence of extended spectrum producing gram negative bacteria in HIV/AIDS infected patients with urinary and gastrointestinal tract infections in Benin metropolis. Res J Pharm Biol Chem Sci. 2011;2(2):230–4. [Google Scholar]
- 32.Sultana ZZ, Hoque FU, Beyene J, Akhlak-Ul-Islam M, Khan MHR, Ahmed S, et al. HIV infection and multidrug resistant tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2021;21(1):51. doi: 10.1186/s12879-020-05749-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabbagh P, Riahi SM, Gamble HR, Rostami A. The global and regional prevalence, burden, and risk factors for methicillin-resistant Staphylococcus aureus colonization in HIV-infected people: a systematic review and meta-analysis. Am J Infect Control. 2019;47(3):323–33. doi: 10.1016/j.ajic.2018.06.023 [DOI] [PubMed] [Google Scholar]
- 34.Joanna Briggs Institute. JBI’s critical appraisal tools assist in assessing the trustworthiness, relevance and results of published papers. 2024. [cited 2024, June,12]. Available from: https://jbi.global/critical-appraisal-tools. [Google Scholar]
- 35.Porritt K, Gomersall J, Lockwood C. JBI’s systematic reviews: study selection and critical appraisal. AJN Am J Nurs. 2014;114(6):47–52. [DOI] [PubMed] [Google Scholar]
- 36.DerSimonian R LN. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. [DOI] [PubMed] [Google Scholar]
- 37.Borenstein M, Cooper H, Hedges L, Valentine J. Heterogeneity in meta-analysis. The handbook of research synthesis meta-analysis. 2019;3(2):453–70 [Google Scholar]
- 38.Page MJ, Sterne JAC, Higgins JPT, Egger M. Investigating and dealing with publication bias and other reporting biases in meta-analyses of health research: a review. Res Synth Methods. 2021;12(2):248–59. doi: 10.1002/jrsm.1468 [DOI] [PubMed] [Google Scholar]
- 39.Bayleyegn B, Fisaha R, Kasew D. Fecal carriage of extended spectrum beta-lactamase producing Enterobacteriaceae among HIV infected children at the University of Gondar Comprehensive Specialized Hospital Gondar, Ethiopia. AIDS Res Ther. 2021;18(1):19. doi: 10.1186/s12981-021-00347-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jerry A, Ebele U, Tersagh S, Mwuese G. Prevalence of ESBL-producing and multi-drug resistant Escherichia coli isolated from urine of HIV patients in General Hospital Gboko, Benue State. WJBPHS. 2021;8(1):029–36. [Google Scholar]
- 41.Padmavathy K KPaSR. Detection of extended-spectrum β lactamases and AmpC β-lactamase production in Escherichia coli causing urinary tract infection among HIV and non-HIV patients. Am Med J. 2011;2(1):54–8. [Google Scholar]
- 42.Reinheimer C, Keppler OT, Stephan C, Wichelhaus TA, Friedrichs I, Kempf VAJ. Elevated prevalence of multidrug-resistant gram-negative organisms in HIV positive men. BMC Infect Dis. 2017;17(1):206. doi: 10.1186/s12879-017-2286-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Said MM, Msanga DR, Mtemisika CI, Silago V, Mirambo MM, Mshana SE. Extended spectrum β-Lactamase producing lactose fermenting bacteria colonizing children with human immunodeficiency virus, sickle cell disease and diabetes mellitus in Mwanza City, Tanzania: a cross-sectional study. Trop Med Infect Dis. 2022;7(8):144. doi: 10.3390/tropicalmed7080144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simeneh E, Gezimu T, Woldemariam M, Alelign D. Magnitude of multidrug-resistant bacterial uropathogens and associated factors in urinary tract infection suspected adult HIV-positive patients in Southern Ethiopia. TOMICROJ. 2022;16(1). doi: 10.2174/18742858-v16-e2208180 [DOI] [Google Scholar]
- 45.Zemtsa RJ, Noubom M, Founou LL, Dimani BD, Koudoum PL, Mbossi AD, et al. Multidrug-RESISTANT AND EXTENDED-SPECTRUm β-Lactamase (ESBL) - producing enterobacterales isolated from carriage samples among HIV infected women in yaoundé, Cameroon. Pathogens. 2022;11(5):504. doi: 10.3390/pathogens11050504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olaru ID, Tacconelli E, Yeung S, Ferrand RA, Stabler RA, Hopkins H, et al. The association between antimicrobial resistance and HIV infection: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27(6):846–53. [DOI] [PubMed] [Google Scholar]
- 47.Bell BG, Schellevis F, Stobberingh E, Goossens H, Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect Dis. 2014;14:13. doi: 10.1186/1471-2334-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godongwana M, De Wet-Billings N, Milovanovic M. The comorbidity of HIV, hypertension and diabetes: a qualitative study exploring the challenges faced by healthcare providers and patients in selected urban and rural health facilities where the ICDM model is implemented in South Africa. BMC Health Serv Res. 2021;21(1):647. doi: 10.1186/s12913-021-06670-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ako-Nai KAF, Adejuyigbe E, Ebhodaghe B, Osho P, Kassim O. The dynamics of bacteria population on the skin, throat, and gastrointestinal tract of HIV-seropositive patients. Ann Trop Med Public Health. 2015;8(5):164. [Google Scholar]
- 50.Baltazar-Díaz TA, Amador-Lara F, Andrade-Villanueva JF, González-Hernández LA, Cabrera-Silva RI, Sánchez-Reyes K, et al. Gut bacterial communities in HIV-infected individuals with metabolic syndrome: effects of the therapy with integrase strand transfer inhibitor-based and protease inhibitor-based regimens. Microorganisms. 2023;11(4):951. doi: 10.3390/microorganisms11040951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tigabie M, Ayalew G, Demoze L, Tadesse K, Gashaw Y, Assefa M. Extended-spectrum beta-lactamase and carbapenemase-producing Gram-negative bacteria in urinary tract infections in Ethiopia: a systematic review and meta-analysis. BMC Urol. 2025;25(1):11. doi: 10.1186/s12894-025-01695-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tigabie M, Girmay G, Gashaw Y, Bitew G, Birhanu A, Getaneh E, et al. Colonization with extended-spectrum β-lactamase and carbapenemase-producing Enterobacterales in Ethiopia: a systematic review and meta-analysis. PLoS One. 2025;20(4):e0316492. doi: 10.1371/journal.pone.0316492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bammigatti C, Doradla S, Belgode HN, Kumar H, Swaminathan RP. Healthcare associated infections in a resource limited setting. J Clin Diagn Res. 2017;11(1):OC01–4. doi: 10.7860/JCDR/2017/23076.9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daniels B, Kuhn L, Spooner E, Mulol H, Goga A, Feucht U, et al. Cotrimoxazole guidelines for infants who are HIV-exposed but uninfected: a call for a public health and ethics approach to the evidence. Lancet Glob Health. 2022;10(8):e1198–203. doi: 10.1016/S2214-109X(22)00120-6 [DOI] [PubMed] [Google Scholar]
- 55.Church JA, Fitzgerald F, Walker AS, Gibb DM, Prendergast AJ. The expanding role of co-trimoxazole in developing countries. Lancet Infect Dis. 2015;15(3):327–39. doi: 10.1016/S1473-3099(14)71011-4 [DOI] [PubMed] [Google Scholar]
- 56.Cotton MF, Wasserman E, Smit J, Whitelaw A, Zar HJ. High incidence of antimicrobial resistant organisms including extended spectrum beta-lactamase producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus in nasopharyngeal and blood isolates of HIV-infected children from Cape Town, South Africa. BMC Infect Dis. 2008;8:40. doi: 10.1186/1471-2334-8-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanc P, Bonnet F, Leleux O, Perrier A, Bessede E, Pereyre S, et al. Severe bacterial Non-AIDS infections in persons with HIV: the epidemiology and evolution of antibiotic resistance over an 18-year period (2000-2017) in the ANRS CO3 AquiVih-Nouvelle-aquitaine cohort. Clin Infect Dis. 2023;76(10):1814–21. doi: 10.1093/cid/ciac978 [DOI] [PubMed] [Google Scholar]
- 58.Abbas S. The challenges of implementing infection prevention and antimicrobial stewardship programs in resource-constrained settings. Antimicrob Steward Healthc Epidemiol. 2024;4(1):e45. doi: 10.1017/ash.2024.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Josephs-Spaulding J, Singh OV. Medical device sterilization and reprocessing in the era of multidrug-resistant (MDR) bacteria: issues and regulatory concepts. Front Med Technol. 2021;2:587352. doi: 10.3389/fmedt.2020.587352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bharadwaj A, Rastogi A, Pandey S, Gupta S, Sohal JS. Multidrug-resistant bacteria: their mechanism of action and prophylaxis. Biomed Res Int. 2022;2022:5419874. doi: 10.1155/2022/5419874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muntean MM, Muntean A-A, Preda M, Manolescu LSC, Dragomirescu C, Popa M-I, et al. Phenotypic and genotypic detection methods for antimicrobial resistance in ESKAPE pathogens (Review). Exp Ther Med. 2022;24(2):508. doi: 10.3892/etm.2022.11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bryce AHA, Lane IF, Thornton HV, Wootton M, Costelloe C. Global prevalence of antibiotic resistance in paediatric urinary tract infections caused by Escherichia coli and association with routine use of antibiotics in primary care: systematic review and meta-analysis. BMJ. 2016;352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corcione SLT, De Rosa FG. Novel cephalosporins in septic subjects and severe infections: present findings and future perspective. Front Med. 2021;8:617378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weissberg D, Mubiru F, Kambugu A, Fehr J, Kiragga A, von Braun A, et al. Ten years of antiretroviral therapy: Incidences, patterns and risk factors of opportunistic infections in an urban Ugandan cohort. PLoS One. 2018;13(11):e0206796. doi: 10.1371/journal.pone.0206796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hussain HI, Aqib AI, Seleem MN, Shabbir MA, Hao H, Iqbal Z, et al. Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb Pathog. 2021;158:105040. doi: 10.1016/j.micpath.2021.105040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katale BZ, Mshana SE, Chiyangi H, Campino S, Clark TG, Good L, et al. Genetic diversity and risk factors for the transmission of antimicrobial resistance across human, animals and environmental compartments in East Africa: a review. Antimicro Resist Infect Control. 2020;8(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]