Abstract
Genomic instability, including the ability to undergo gene amplification, is a hallmark of neoplastic cells. Similar to normal cells, “nonpermissive” REF52 cells do not develop resistance to N-(phosphonacetyl)-l-aspartate (PALA), an inhibitor of the synthesis of pyrimidine nucleotides, through amplification of cad, the target gene, but instead undergo protective, long-term, p53-dependent cell cycle arrest. Expression of exogenous MYC prevents this arrest and allows REF52 cells to proceed to mitosis when pyrimidine nucleotides are limiting. This results in DNA breaks, leading to cell death and, rarely, to cad gene amplification and PALA resistance. Pretreatment of REF52 cells with a low concentration of PALA, which slows DNA replication but does not trigger cell cycle arrest, followed by exposure to a high, selective concentration of PALA, promotes the formation of PALA-resistant cells in which the physically linked cad and endogenous N-myc genes are coamplified. The activated expression of endogenous N-myc in these pretreated PALA-resistant cells allows them to bypass the p53-mediated arrest that is characteristic of untreated REF52 cells. Our data demonstrate that two distinct events are required to form PALA-resistant REF52 cells: amplification of cad, whose product overcomes the action of the drug, and increased expression of N-myc, whose product overcomes the PALA-induced cell cycle block. These paired events occur at a detectable frequency only when the genes are physically linked, as cad and N-myc are. In untreated REF52 cells overexpressing N-MYC, the level of p53 is significantly elevated but there is no induction of p21waf1 expression or growth arrest. However, after DNA is damaged, the activated p53 executes rapid apoptosis in these REF52/N-myc cells instead of the long-term protective arrest seen in REF52 cells. The predominantly cytoplasmic localization of stabilized p53 in REF52/N-myc cells suggests that cytoplasmic retention may help to inactivate the growth-suppressing function of p53.
Normal mammalian cells have complex growth controls which prevent them from progressing through the cell cycle when conditions are unfavorable or when their DNA has been damaged. In tumor cells, loss of these controls allows various chromosomal abnormalities, including gene amplification, to accumulate. The tumor suppressor protein p53 mediates cell cycle arrest or apoptosis in response to DNA damage (reviewed in references 9, 28, and 31). p53 also mediates the reversible, protective arrest of normal cells in response to starvation for DNA or RNA precursors (32). This G1 arrest occurs without replicative DNA synthesis or detectable DNA damage and thus contrasts with the response of normal cells to DNA damage (15, 32). p53 is also involved in G2/M arrest (1, 57), in ensuring that mitosis is complete before the next S phase begins (10), and in ensuring that DNA synthesis is complete before mitosis begins (59). The p53 protein, induced and activated by stress, stimulates transcription of a set of genes that regulate growth arrest and apoptosis (reviewed by Ko and Prives [28] and Cox and Lane [9]). p53 mediates growth arrest in large part by inducing the cyclin-dependent kinase inhibitor p21waf1 (16, 69) and also gadd45, which is thought to mediate arrest through its interactions with p21 and proliferating-cell nuclear antigen (27, 53). p53 can also induce the expression of bax (36) and fas (40) and reduce the expression of bcl-2 (36), thus promoting apoptosis. These activities help to account for the connection between the loss of p53 and the genesis of aneuploidy, chromosomal aberrations, and gene amplification in tumors and cell lines.
Inactivation of p53 through deletion, mutation, or the action of viral oncogenes is required to allow cells to tolerate chromosomal aberrations such as gene amplification (reviewed by Chernova et al. [8]). We now understand amplification mechanisms well enough to appreciate that breakage of chromosomes is an important initial step (44, 63). Normal cells are very sensitive to broken DNA, arresting when very few double-strand breaks or large gaps are present (24); this helps to explain why gene amplification has not been detected in normal cells (62, 68) and why the loss of p53 is required to make them permissive for amplification (33, 71). In contrast, amplification is a frequent mechanism for overexpressing oncogenes or genes mediating drug resistance in tumors or cell lines in which the p53 response has been lost (3, 56). Most immortal cell lines, especially those of rodent origin, develop resistance to N-(phosphonacetyl)-l-aspartate (PALA) or methotrexate (MTX) through amplification of the target genes for carbamyl-P synthetase, aspartate transcarbamylase, dihydro-orotase (cad), or dihydrofolate reductase (dhfr) (48, 56). Inactivation of the p53 gene by deletion (33, 71) or mutation (26) or inactivation of the response to p53 by oncoproteins (42, 66) is required to achieve PALA resistance and cad amplification in normal cells.
The REF52 cell line is unusual because no resistant colonies are formed upon selection with PALA or MTX (the frequency of resistance is less than 10−9 [42]). Therefore, REF52 cells have been useful for identifying genes involved in regulating permissivity for gene amplification. For example, the expression of activated ras plus adenovirus E1A or simian virus 40 (SV40) T-antigen alone converts REF52 cells to a state permissive for cad amplification (42), as does a dominant negative mutant p53 protein (26). c-MYC is an important regulator of cellular proliferation, differentiation, and apoptosis (reviewed by Grandori and Eisenman [20], Packham and Cleveland [41], and Amati and Land [4]), and it is frequently overexpressed in tumors. Deregulated expression of c-myc induces cell cycle progression in quiescent cells and, in the absence of survival factors, p53-mediated apoptosis (17, 22). The mechanisms of these diverse activities of c-MYC are not yet well understood. C-MYC functions as a transcriptional activator when complexed with MAX (reviewed in reference 4), inducing genes important for cell cycle progression, including cad (reviewed in reference 20). C-MYC also plays an important role as a repressor of the expression of genes such as cyclinD1 (43), gadd45 (35), and c-myc itself (18). Fewer data are available regarding N-MYC, which is overexpressed in neuroblastomas (49), retinoblastomas (30), and rhabdomyosarcomas (14).
Since there is evidence that overexpression of c-MYC increases the frequencies of MTX resistance and dihydrofolate reductase amplification in established cell lines (12, 34), we decided to study whether deregulated expression of MYC might have a different function in overcoming the lack of permissivity of REF52 cells for drug resistance and gene amplification. We introduced the N-myc or c-myc genes into REF52 cells and selected the resulting cell lines with PALA or MTX. The data reveal that overexpression of exogenous MYC abrogates PALA-induced, p53-mediated cell cycle arrest and facilitates cad amplification. Using a selection protocol involving pretreatment with a low concentration of PALA, we have also shown that two distinct events are required to form PALA-resistant REF52 cells: amplification of the target gene cad and greatly increased expression of endogenous N-myc through coamplification with cad.
MATERIALS AND METHODS
Plasmids.
pSV40myc, containing exons 2 and 3 of the human N-myc gene under the control of an SV40 promoter (6), was kindly provided by William E. Fahl (University of Wisconsin, Madison, Wis.). To obtain a retroviral construct containing the c-myc gene, the HindIII fragment of mouse c-myc, from pMc-myc54 (55), was ligated into the HincII site of pBluescript KSII. EcoRI linkers were ligated to the HindIII site, and the 1.8-kb EcoRI fragment was transferred to the EcoRI site of pBabeHygro (39).
Cells and transfection.
Low-passage REF52 cells (19) were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (Gibco BRL) at 37°C in an atmosphere containing 10% CO2. REF52 cells were cotransfected with a mixture of pSV40myc and pSV2puro in a 10:1 ratio by using the modified calcium phosphate method of Chen and Okayama (7). Stably transfected clones were selected with puromycin (1 μg/ml). For retroviral transfer of the c-myc gene, subconfluent cells were treated with Polybrene (10 μg/ml; Sigma) and incubated as described by Perry et al. (42). Clones with a retroviral integration were selected with hygromycin (200 μg/ml). Expression of myc mRNAs was confirmed by Northern analysis. C11 cells were derived from p53-null MDAH041 cells by introduction of the wild-type p53 gene under its own promoter (2). These cells were grown, transfected, and selected with PALA similarly to REF52 cells.
Drug selections and frequencies of gene amplification.
PALA was obtained from the Drug Synthesis and Chemistry branch of the Division of Cancer Treatment, National Cancer Institute, and MTX was obtained from Sigma. Selections were performed as described by Perry et al. (42). Briefly, 5 × 104 cells, plated on 10-cm dishes, were grown in medium containing dialyzed fetal calf serum in the presence of PALA or MTX at concentrations three times the 50% inhibitory concentration (IC50). Drug-resistant colonies, detectable after 4 to 5 weeks, were cloned or, alternatively, fixed, stained, and counted. In pretreatment experiments, fewer cells (5 × 103) were plated on 10-cm dishes and exposed to a low, nonselective concentration of drug (10 or 15 μM PALA, 15 or 20 nM MTX). After 72 h, the amount of drug was increased to a selective concentration (30 μM PALA or 40 to 50 nM MTX), and the cells were kept in this selection for 5 to 6 weeks. Control cells were selected directly at the high concentrations. Individual drug-resistant colonies were expanded to 105 to 106 cells at the selective concentration of each drug and analyzed by fluorescent in situ hybridization (FISH). Plating efficiencies and IC50s were determined as described by Perry et al. (42).
Analysis of metaphase cells by FISH.
PALA-resistant cells were analyzed for cad or N-myc amplification essentially as described by Smith et al. (51). As probes, we used genomic clones of the rat cad or N-myc genes, isolated from a rat cosmid library, with pCAD142 (50) and the mouse N-myc gene as probes. The probes were labeled by nick translation with digoxigenin-11-dUTP (Boehringer Mannheim) or with biotin-11-dUTP (BioNick labeling system; Gibco BRL), and repetitive sequences were competed out with sonicated rat genomic DNA. Hybridization was detected as described previously (51). The hybridization mixture (15 μl/slide), containing one or two labeled cosmid probes (150 to 200 ng of each) and 20 μg of sonicated rat genomic DNA in 50% formamide–2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10% dextran sulfate was heated, preannealed, placed on the slides, covered, and incubated overnight at 37°C. After the slides were washed, biotin-labeled probes were detected by incubation with fluorescein isothiocyanate-avidin (5 μg/ml; Vector Laboratories). Digoxigenin-labeled probes were detected with fluorescein isothiocyanate-conjugated sheep anti-digoxigenin serum (10 μg/ml; Boehringer Mannheim). DNA was counterstained with 0.2 μg of propidium iodide or 4′,6-diamidino-2-phenylindole (DAPI) per ml. The images were obtained with a Nikon Optiphot epifluorescence microscope coupled to a cooled computer-controlled charge-coupled device camera (Oncor Imaging System, Gaithersburg, Md.).
Flow cytometric analysis.
After treatment with PALA or MTX, adherent and detached cells were combined, fixed with methanol, stained with propidium iodide by using a Cycletest kit (Becton Dickinson), and analyzed for DNA content by using a FACScan instrument (Becton Dickinson). The cell cycle distribution was determined with the SOBR model of the CellFIT program.
RNA analysis.
Total RNA was extracted with the Trizol reagent (Gibco BRL) as specified by the manufacturer. Northern and RNase protection assays were performed as described by Sambrook et al. (47). To obtain an N-myc probe for RNase protections, the 580-bp fragment of exon 2 was amplified by PCR from primers containing XbaI and KpnI adapters, with a cosmid containing the rat N-myc gene as a template. This fragment was recloned into pSP72 (Promega). The inserts from pSV2dhfr (47) and pCAD142 (50), which contain the mouse dhfr and hamster cad cDNAs, respectively, were used as probes. Radioactive bands were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Protein preparation and Western analyses.
Soluble proteins were extracted essentially as described by Harlow and Lane (21). Briefly, cells were lysed in ice-cold NET buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 0.5 mM phenylmethanesulfonyl fluoride, 2 mM benzamidine). After incubation for 10 min on ice, the cells were resuspended by vortexing and the soluble proteins were separated by centrifugation at 16,000 × g for 15 min at 4°C. The extracts were stored at −80°C. Nuclear and cytoplasmic extracts were prepared essentially as described by Tishler et al. (61). Portions of lysates containing 20 μg of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8 or 12% polyacrylamide) and transferred to a polyvinylidene difluoride membrane (Stratagene). After the transfer, the gels were stained with Coomassie blue and the membranes were stained reversibly with Ponceau S to verify equal loading. The membranes were probed with the monoclonal antibody PAb421, directed against p53 (a kind gift of Arnold Levine, Princeton University, Princeton, N.J.), or polyclonal rabbit antibodies C-19 and L-17, directed against p21waf1 (Santa Cruz Biotechnology). Secondary horseradish peroxidase-conjugated goat anti-mouse antibodies (Bio-Rad) or goat anti-rabbit antibodies (Pierce) were visualized by enhanced chemiluminescence as specified by the manufacturer (Renaissance reagents; DuPont NEN).
RESULTS
Overexpression of N-MYC or c-MYC permits cad but not dhfr amplification in REF52 cells.
As shown by Perry et al. (42) and confirmed here, REF52 cells are not permissive for amplification of cad or dhfr when selected with PALA (30 μM) or MTX (50 nM) at three times the IC50 (frequencies less than 10−9). Upon incubation with selective concentrations of these agents, the cells arrest, remaining attached to the plates and nearly constant in number for several weeks. We studied the ability of exogenous N-MYC to abrogate this arrest and thus to facilitate gene amplification in REF52 cells. The cells were cotransfected with human N-myc and pSV2puro. Several puromycin-resistant clones were isolated, and the levels of N-MYC expression were analyzed. We were not able to detect endogenous N-myc mRNA in these cells. Four REF52/N-myc clones (numbered 8, 11, 12, and 16) with different expression levels were used for PALA and MTX selections. As a first step, we determined the relative sensitivities of the cells to the selective drugs. Compared to REF52 and control REF52/puro cells, the REF52/N-myc clones had higher IC50s of PALA and lower IC50s of MTX, in proportion to their levels of N-MYC expression (Fig. 1). The highest level of expression of N-MYC (in clone 11) was accompanied by a 3.5-fold increase in the IC50 of PALA and a 2.6-fold decrease in the IC50 of MTX.
FIG. 1.
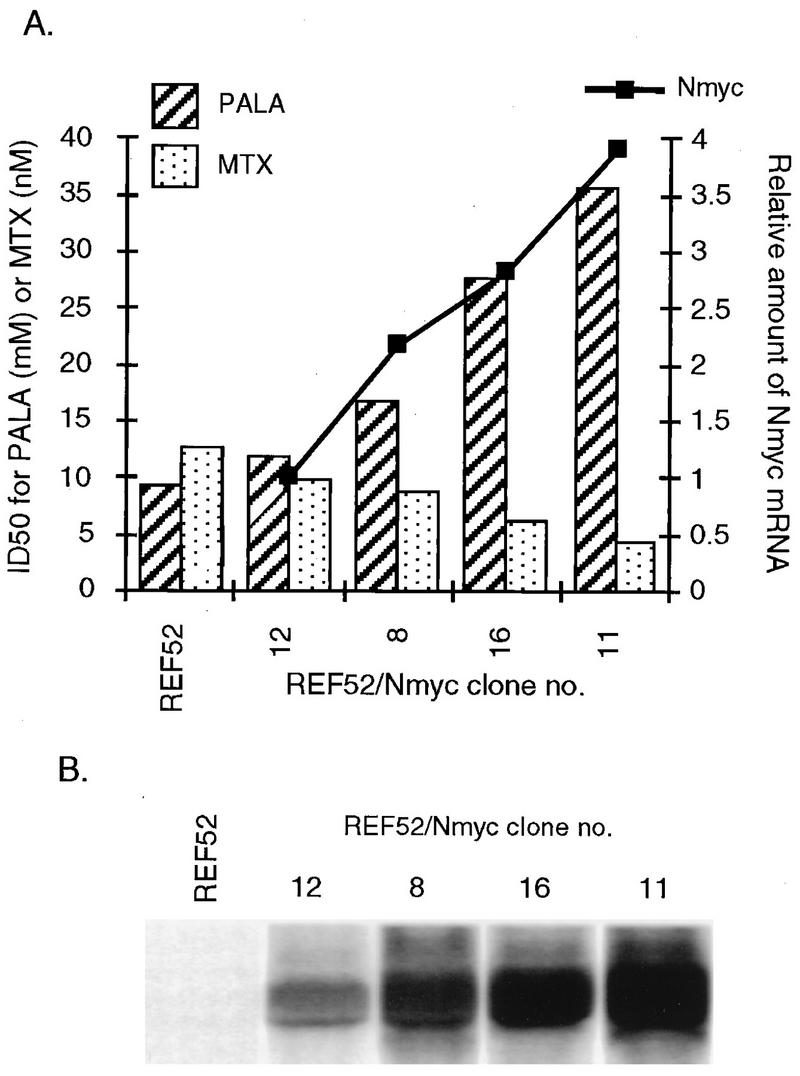
Levels of exogenous N-myc mRNA and resistance to PALA or MTX in REF52/N-myc clones. (A) Bars represent the IC50s of PALA or MTX. The levels of N-myc mRNA, obtained by using a PhosphorImager, were normalized to the level in clone 12. (B) Northern transfers (15 μg of total RNA) were hybridized with a human N-myc probe.
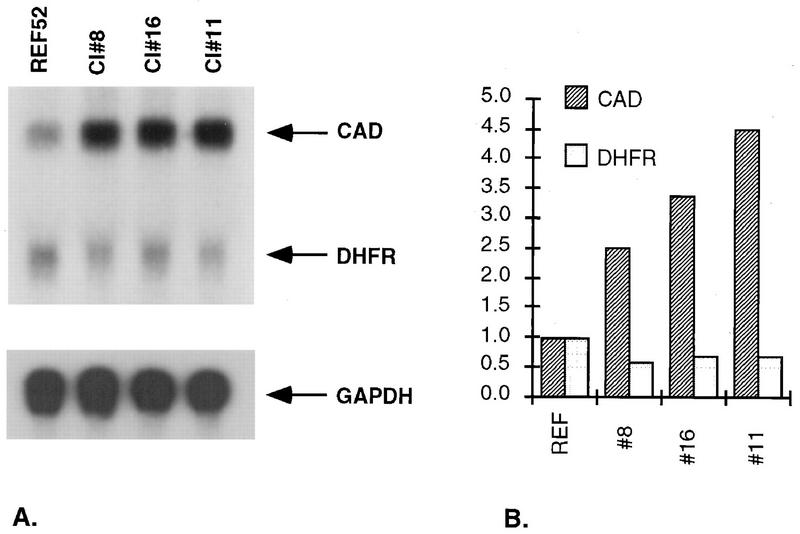
To analyze the basis for the differences in sensitivity, we measured the levels of cad and dhfr mRNAs. In untreated cells, increased levels of N-MYC (Fig. 2) or c-MYC (data not shown) enhanced the expression of cad mRNA in proportion to the levels of myc mRNA expression. In all of these clones, the levels of dhfr mRNA were decreased by about one-half (Fig. 2), which correlates with the increased sensitivity to MTX (Fig. 1).
FIG. 2.
Levels of cad and dhfr mRNAs in untreated REF52 and REF52/N-myc cells. (A) Northern transfers (15 μg of total RNA) were hybridized with cad or dhfr probes. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was analyzed as a loading control. (B) Quantitative data from a PhosphorImager were normalized to the levels in REF52 cells.
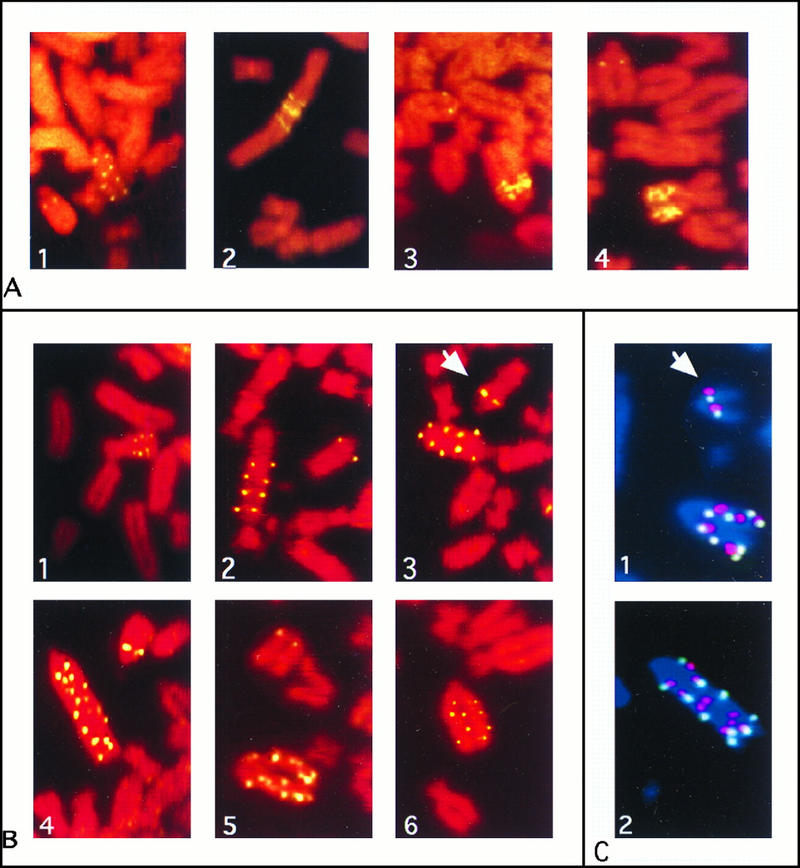
Taking these differences into account, we selected each clone in a drug concentration corresponding to three times the IC50. All four REF52/N-myc clones produced PALA-resistant colonies at similar frequencies of ∼10−4, more than 5 orders of magnitude higher than for REF52 cells. To determine the nature of the event responsible for resistance, four or five PALA-resistant colonies from each REF52/N-myc clone were analyzed by FISH. They all showed chromosomal cad amplification in which the extra copies (range, 3 to 10 per cell) were present as condensed repeating units on rearranged marker chromosomes (examples are shown in Fig. 3A). The extent of cad gene amplification was confirmed by Southern analysis of genomic DNA from the PALA-resistant colonies (data not shown). Similar results were obtained with REF52 cells transfected with the mouse c-myc gene, where PALA selection gave resistant colonies with amplified cad at frequencies of ∼10−5.
FIG. 3.
FISH of metaphase chromosomes from PALA-resistant cells. (A) Partial metaphase spreads representing cad amplification in PALA-resistant cells selected from REF52/N-myc clones 12, 8, 16, and 11 (panels 1 to 4, respectively). An unrearranged chromosome shows two green signals for cad. (B) cad amplification in PALA-resistant REF52 cells selected after preexposure to PALA. Panels 1 to 6 represent independent clones 2, 4, 7, 11, 12, and 24, respectively. (C) Coamplification of cad and N-myc in PALA-resistant REF52 cells selected after preexposure to PALA. Panels 1 and 2 represent clones 7 and 21, respectively. Chromosomes were hybridized simultaneously with a biotin-labeled cad probe (green) and a digoxigenin-labeled N-myc probe (red). Chromosomes were counterstained with DAPI (blue). An unrearranged chromosome 6, carrying single copies of cad and N-myc, is indicated by an arrow.
To extend these observations to cells of another species, we developed a nonpermissive human cell line, C11, by reintroducing wild-type p53 into p53-null Li-Fraumeni MDAH041 cells (2). While MDAH041 cells form PALA-resistant colonies at three times the IC50 at a frequency of 5 × 10−4, the introduction of p53 converted these cells to a nonpermissive state, with a frequency of less than 10−8 when selected in 200 μM PALA (three times the IC50). To discover if deregulated expression of N-myc allows these nonpermissive human cells to develop PALA resistance, we performed experiments similar to those described above for REF52 cells. The human N-myc gene was introduced into C11 cells, and two individual clones that expressed it were subjected to PALA selection. As for REF52/N-myc cells, the IC50s of PALA (80 and 85 μM) were higher for both clones than for parental C11 cells (60 μM). Both clones, selected in 300 μM PALA for 4 weeks, formed PALA-resistant colonies at a frequency of 5 × 10−5.
To see if MYC acts more generally to regulate permissivity for gene amplification in REF52 cells, we selected the REF52/N-myc clones with MTX at concentrations equal to three times the IC50 for each clone. No resistant colonies were observed (frequency less than 3 × 10−7). Therefore, the expression of N-MYC or c-MYC in REF52 cells facilitates amplification of cad but not dhfr.
Pretreatment with PALA induces resistance in REF52 cells.
In trying different selection protocols, we found that pretreatment with a low concentration of PALA (1.0 or 1.5 times the IC50 [10 or 15 μM]) for 3 days before exposure to a selective concentration (3 times the IC50) allowed PALA-resistant REF52 colonies to arise at the high frequency of ∼3 × 10−4 (Table 1). Pretreatment with a low concentration of PALA (60 μM) for 3 days followed by selection in 200 μM PALA also overcame the lack of permissivity of C11 cells, where resistant colonies were formed at a frequency of 3 × 10−4. Thus, a short pretreatment with a low, nonselective, concentration of PALA significantly facilitates the appearance of PALA-resistant REF52 cells. Low concentrations of PALA inhibited cell growth only partially; many REF52 cells divided two or three times before arrest, forming microcolonies of four to eight cells (data not shown). Counting REF52 cells cultured for 3 days in PALA revealed that in 10, 15, or 30 μM PALA, the total number of cells increased by 2.6-, 1.7-, or 1.1-fold, respectively, while the number of untreated cells increased by 5.8-fold during the same period. In contrast to the results with PALA, a similar two-step selection of REF52 cells with MTX did not generate any MTX-resistant colonies (Table 1). Although a few such colonies were observed in the control selection (Table 1), there was no increase in dhfr copy number, as revealed by Southern and FISH analyses (data not shown; also observed previously by Perry et al. [42]). We also performed mixed two-step selections. REF52 cells were pretreated with PALA at 1.5 times the IC50 for 72 h and then subjected to selection in MTX at 3 times the IC50, and vice versa. Neither of these mixed selections gave rise to any resistant colonies (Table 1).
TABLE 1.
Gene amplification in REF52 cells pretreated and selected with PALA or MTXa
| Pretreatment | Selection | No. of colonies observedb | No. of cells analyzedb | Amplification frequency |
|---|---|---|---|---|
| None | PALA (30 μM) | 0 | 5 × 107 | <2 × 10−8 |
| MTX (50 nM) | 0 | 5 × 106 | <2 × 10−7 | |
| MTX (40 nM) | 3c | 5 × 106 | <2 × 10−7 | |
| PALA (15 μM) | PALA (30 μM) | 82 | 7 × 105 | 2.9 × 10−4 |
| PALA (10 μM) | PALA (30 μM) | 98 | 6 × 105 | 4.1 × 10−4 |
| PALA (15 μM) | MTX (40 nM) | 0 | 1 × 106 | <10−6 |
| MTX (50 nM) | 0 | 1 × 106 | <10−6 | |
| MTX, 15 nM | MTX (40 nM) | 0 | 1 × 106 | <10−6 |
| MTX, 20 nM | MTX (50 nM) | 0 | 1 × 106 | <10−6 |
| MTX, 15 nM | PALA, 30 μM | 0 | 1 × 106 | <10−6 |
| MTX, 20 nM | PALA | 0 | 1 × 106 | <10−6 |
REF52 cells were seeded at a low density (5 × 103 cells per 10-cm plate) in low, nonselective concentrations of PALA or MTX for 72 h and then exposed to selective concentrations. Resistant colonies appeared after about 4 weeks.
Totals from two or three independent selections.
dhfr was not amplified in these colonies.
Coamplification of cad and endogenous N-myc in PALA-resistant REF52 clones.
To understand the mechanisms of PALA resistance in REF52 cells selected after pretreatment, we examined both the cad copy number and the structure of amplified cad DNA in PALA-resistant cells. Twenty colonies from three independent two-step PALA selections of REF52 cells were analyzed at the 105- to 106-cell stage. The selection scheme used (see Materials and Methods) ensures that clones derived from different plates are independent. Analysis by FISH revealed that the cad gene was amplified in all 20 clones and also that the amplified cad genes were present as intrachromosomal ladder-like structures, most often on one chromosome and often occupying most of this chromosome (representative metaphase spreads are shown in Fig. 3B). Parental REF52 cells were nearly tetraploid, with four chromosomes carrying cad, each copy represented by a hybridization spot on each chromatid. The chromosomes carrying amplified cad, usually one per cell, had 1 to 10 additional copies, most often 4 to 8 (16 clones analyzed). Similar numbers were obtained by Southern analysis of genomic DNA from PALA-resistant colonies (data not shown).
The early steps of gene amplification often involve chromosome breaks, which are recognized by p53-dependent pathways. How might pretreatment of REF52 cells with PALA allow them to tolerate such breaks in the presence of p53? Replication of DNA under suboptimal conditions in a low concentration of PALA may lead to amplification and increased expression of genes such as myc, which are involved in cell cycle regulation, thus disrupting the normal inhibition of growth in response to DNA damage. The known colocalization of cad and N-myc on human chromosome 2p (54) prompted us to study the involvement of endogenous N-myc in cad amplification in rat REF52 cells. The rat N-myc gene has been mapped to chromosome 6 (25), but the chromosomal localization of the rat cad gene has not been reported. When we cohybridized rat genomic cad and N-myc probes to metaphase spreads of REF52 cells, we found that these two genes were linked in rat cells as well as in human cells (Fig. 3C). To test for coamplification, metaphase spreads from 16 PALA-resistant REF52 clones, selected after pretreatment with a low concentration of PALA, were hybridized with both probes. In all of these, both cad and N-myc were amplified on the same chromosome (Fig. 3C).
To determine if the amplification of N-myc induced its expression, we analyzed mRNA from REF52 cells and eight PALA-resistant clones. N-myc mRNA, undetected in parental REF52 cells, was observed in all the PALA-resistant clones tested (four examples are shown in Fig. 4), supporting the conclusion that activation of endogenous N-myc follows its amplification and the idea that N-MYC allows nonpermissive REF52 cells to escape growth arrest and to give rise to PALA-resistant colonies. Remarkably, only one of the four PALA-resistant REF52/N-myc colonies selected from clones 11 and 16 (highest expression of exogenous N-myc) exhibited coamplification of cad and endogenous N-myc (data not shown), consistent with the shorter amplicons found in PALA-resistant cells derived from these clones (compare Fig. 3A, panels 3 and 4, and B). Since there is no need to express endogenous N-myc when exogenously expressed N-myc is already present, the cad gene can be amplified independently of N-myc amplification in this situation.
FIG. 4.
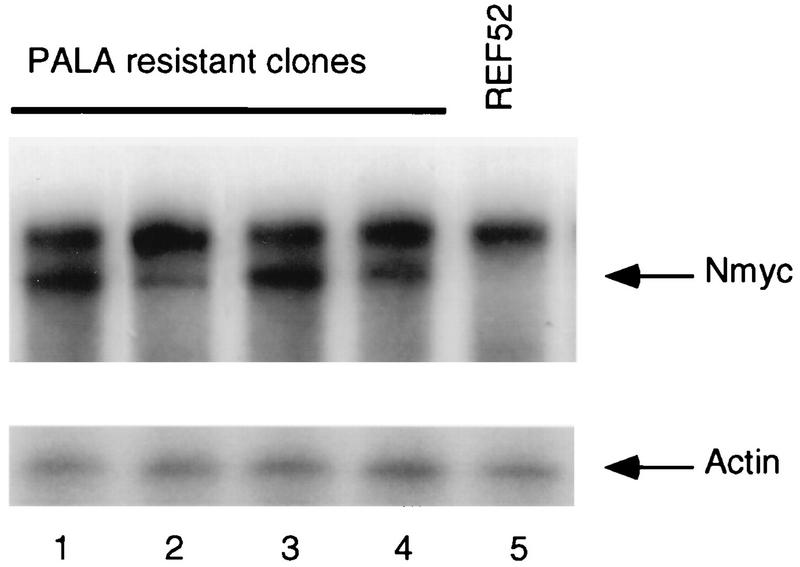
Expression of N-myc mRNA in PALA-resistant REF52 clones. Total RNA from PALA-resistant clones selected after preexposure to PALA (lanes 1 to 4) and parental REF52 cells (lane 5) was analyzed by RNAse protection, using as a probe a 585-bp fragment derived from the second exon of the rat N-myc gene. The amount of RNA was normalized with a β-actin probe.
Cell cycle regulation of REF52/N-myc cells in response to PALA or MTX.
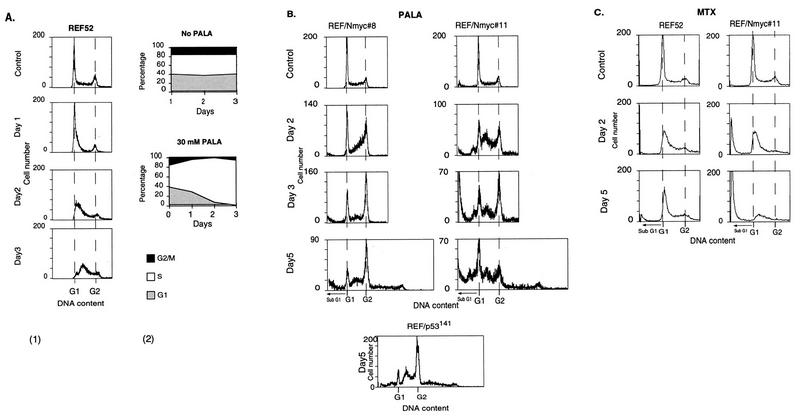
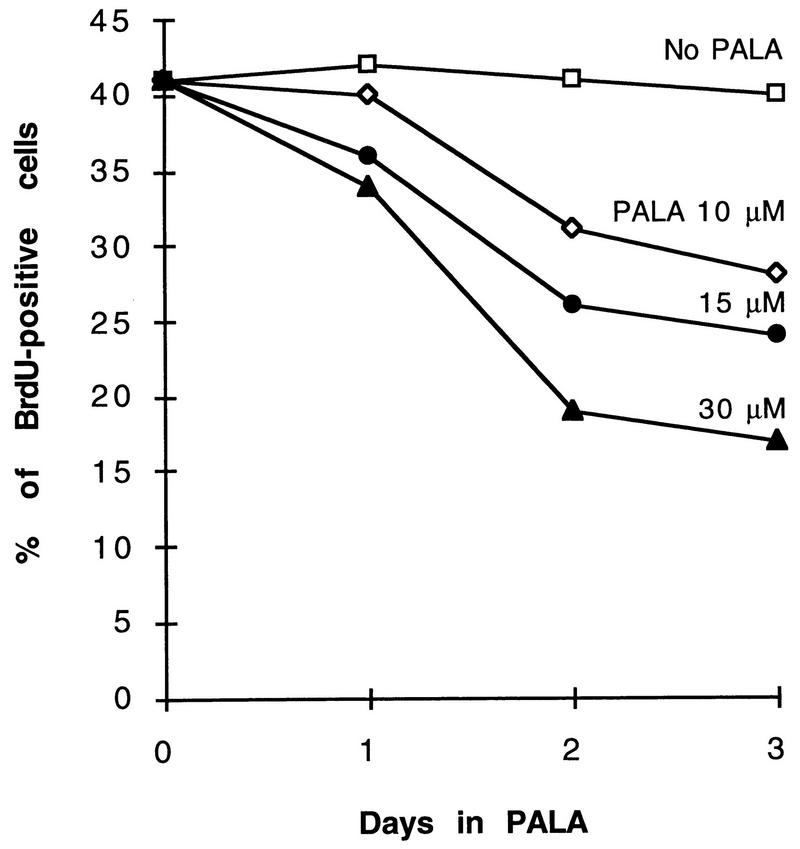
To understand how MYC affects cell cycle arrest and thus the potential for gene amplification, we performed a cell cycle analysis of PALA-treated REF52 and REF52/N-myc cells by flow cytometry. At 24 h after adding a selective concentration of PALA (30 μM) to an exponentially growing population of REF52 cells, some of the cells accumulated in early S-phase (Fig. 5A). The fraction of cells in S phase increased gradually with time, mainly at the expense of cells in G1, so that after 72 h about 95% of the population had a DNA content corresponding to early to mid-S (Fig. 5A). In the continuous presence of PALA for 5 or 7 days, the cell cycle distribution was very similar to that observed after 3 days (data not shown). In parallel experiments, we measured the percentage of PALA-treated REF52 cells that were able to incorporate bromodeoxyuridine (BrdU) into their DNA during a 2-h pulse. As seen in Fig. 6, the number of BrdU-positive cells declined with increasing duration of treatment or drug concentration. The staining was much weaker than in untreated control cells, reflecting inhibited DNA synthesis (note that PALA-treated cells have very little dTTP, allowing significant, albeit low-level incorporation of BrdU in place of dTTP). Although up to 17% of the REF52 cells were BrdU positive after 3 days in 30 μM PALA, they all had the morphology of arrested cells. Arrest was confirmed by counting the total number of cells after exposure to 30 μM PALA for 3 days (1.1-fold increase) or by counting the numbers of cells in marked areas of the plates at 24-h intervals for 5 days (1.1-fold increase). No increase in the number of dead cells was detected by cell cycle analysis (Fig. 5A) or visual observation. To test the reversibility of arrest, after 1 week PALA was removed and uridine was added (to allow renewed DNA and RNA synthesis). We found that 15% of the cells formed colonies, corrected for the plating efficiency of untreated control cells.
FIG. 5.
Flow-cytometric analyses of REF52 and REF52/N-myc cells treated with PALA or MTX. (A) In panel 1, REF52 cells were treated with 30 μM PALA for 24, 48, or 72 h; fixed; stained with propidium iodide; and analyzed for cell cycle distribution with a FACScan instrument. In panel 2, the percentage of cells in each phase of the cell cycle was calculated as a function of the number of days in PALA. (B) Analysis of REF52/N-myc clones 8 and 11 treated with PALA. Cells were exposed to 50 μM (clone 8) or 100 μM (clone 11) PALA for 2, 3, or 5 days and analyzed as in panel A. REF52/p53C141Y cells were treated with 30 μM PALA for 5 days. (C) Analysis of REF52 cells and REF52/N-myc clone 16 treated with MTX. Cells were exposed to 40 nM (REF52) or 30 nM (clone 16) MTX for 2 or 5 days and analyzed as for panel A. Apoptotic cells have less DNA than G1-phase cells.
FIG. 6.
Analysis of DNA synthesis in REF52 cells treated with PALA. The cells were exposed to 10, 15, or 30 μM PALA for 24, 48, or 72 h; labeled with BrdU for 2 h; fixed; and stained with a cell proliferation kit (Amersham). The percentage of nuclei labeled with BrdU is shown at each time point.
A similar cell cycle analysis was performed with the four REF52/N-myc clones, each treated with PALA at the appropriate concentration (three times the IC50 [see below]) for 2, 3, or 5 days. Since all four lines behaved similarly, data for only two are shown (Fig. 5B). PALA-treated REF52/N-myc cells proceeded slowly through S phase for the first 1 to 2 days, eventually entering G2 and mitosis. (Note that net DNA synthesis can occur in PALA-treated cells, probably through conversion of rRNA to deoxynucleoside triphosphates.) Some of the cells even managed to rereplicate their DNA, giving rise to a small peak with twice the G2 DNA content (Fig. 5B) (note that the REF52 cells are nearly tetraploid). There was a parallel increase in the number of apoptotic cells (Fig. 5B), represented by the sub-G1 fraction (11). The extent of apoptosis in REF52/N-myc clones correlated with the level of N-myc expression, so that clone 11 had more apoptotic cells than did clone 8. This result contrasts with that obtained for REF52 cells, which are tightly arrested by a selective concentration of PALA (Fig. 5A), demonstrating that overexpression of MYC enhances the ability of the cells to undergo apoptosis when starved for pyrimidine nucleotides.
To test whether inactivation of p53 in REF52 cells produces a similar effect on the cell cycle distribution in PALA, we analyzed REF52/p53C141Y cells, which express the dominant negative p53 mutant protein C141Y and are permissive for cad gene amplification (26). Similar to REF52/N-myc clones, REF52/p53C141Y cells did not arrest in response to PALA (Fig. 5B; only one time point is shown). Most of these cells reached mitosis, and, as in REF52 cells expressing N-MYC, abnormal mitoses gave rise to cells with a range of DNA contents. These data confirm that the PALA-induced arrest of REF52 cells is mediated by p53. The arrest prevents REF52 cells from entering mitosis when pyrimidine nucleotides are limiting, and the failure of REF52/N-myc cells to arrest in PALA leads to aberrant DNA replication, abnormal mitosis, DNA damage, and cell death.
Induction of p53 in PALA-treated, MTX-treated, or UV-irradiated REF52 and REF52/N-myc cells.
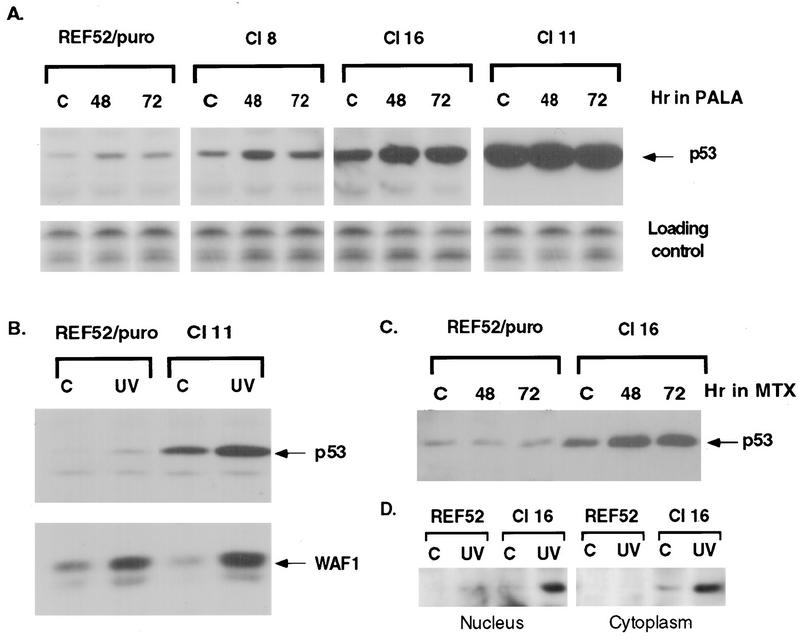
Inactivation of the p53-mediated cell growth arrest pathway in normal human or mouse fibroblasts (33, 71) or REF52 cells (26) is required for the cells to be permissive for PALA resistance and cad amplification. Since REF52/N-myc cells fail to arrest in PALA, thus giving rise to PALA-resistant colonies, we evaluated the induction and function of p53 by comparing the abilities of control REF52/puro cells and three REF52/N-myc clones to induce p53 and the p53-dependent gene p21waf1 in response to PALA or UV radiation. The level of p53 protein was much higher in untreated REF52/N-myc cells than in control REF52/puro cells, in proportion to the level of N-MYC expression (Fig. 7A). Cells exposed to PALA at three times the IC50 were analyzed after 24, 48 or 72 h, and cells irradiated with UV were analyzed after 8 h. Treatment with PALA or UV led to an increase in the amount of p53 protein in all the cells (Fig. 7A and B). In the PALA-treated cells, the increase in the amount of p53 was detected after 24 h (data not shown); the level of p53 reached a maximum after 48 h and stayed high for 72 h (Fig. 7A). The level of p21waf1 in untreated REF52/N-myc cells was not elevated, despite the high level of p53, but did increase in response to p53 induction after exposure to UV (Fig. 7B) or PALA (data not shown).
FIG. 7.
Induction of p53 and p21waf1 in cells treated with PALA, MTX, or UV. (A) Immunoblot analysis of p53 expression in treated or untreated control (lanes C) REF52/puro and REF52/N-myc cells. The cells were exposed to PALA for 48 or 72 h (30 μM for REF52/puro, 55 μM for REF52/N-myc clone 8, 85 μM for clone 16, and 110 μM for clone 11). The film for clone 11 was exposed three times less than were the films for the other clones shown. The untransferred part of the gel, stained with Coomassie blue, is presented as a loading control. (B) REF52/puro and REF/N-myc clone 11 cells were irradiated with UV (24 J/m2), and the proteins were extracted 8 h later. p53 was detected with antibody PAb421, and p21waf1 was detected with a mixture of antibodies C-19 and L-17. (C) REF52/puro and REF52/N-myc clone 16 cells were treated with 50 nM MTX for 48 or 72 h and analyzed as in panel A. (D) Western analysis of the relative amounts of p53 in nuclear and cytoplasmic extracts from REF52 and REF52/N-myc clone 16 cells. The amount of nuclear extract assayed corresponded to three times as many cells as the amount of cytoplasmic extract.
Both REF52/puro and REF52/N-myc cells arrested early in S phase when treated with MTX, but the REF52/N-myc cells progressed to apoptosis within 1 day (data not shown), with appreciable apoptosis after 2 days (Fig. 5C). There was no induction of p53 in REF52/puro cells after 3 days of treatment (Fig. 7C). Induction of p53 and p21waf1 by PALA but not by MTX has also been reported for primary human fibroblasts (32). In contrast, the p53 level in MTX-treated REF52/N-myc cells increased after 24 h and stayed high for 72 h (Fig. 7B). The high level of p53 in REF52/N-myc cells correlates well with their ability to undergo apoptosis in response to serum deprivation, nucleotide starvation, or DNA damage. However, untreated REF52/N-myc cells have the same low level of p21waf1 as do parental REF52 cells. Therefore, we examined whether the p53 in untreated REF52/N-myc cells might be prevented from functioning by retention in the cytoplasm (37, 58). We analyzed both nuclear and cytoplasmic extracts from untreated or UV-irradiated cells (Fig. 7D). The level of p53 was very low in untreated REF52 cells, and after UV irradiation, it accumulated in the nucleus. On the other hand, in REF52/N-myc cells, p53 is more abundant in the cytoplasm and UV irradiation of these cells resulted in the accumulation of p53 in both the cytoplasm and the nucleus. Our data agree well with the recent finding that the cytoplasmic retention of wild-type p53 impairs the G1 checkpoint after DNA damage in neuroblastomas (38). Altogether, the data suggest that, in spite of the significantly elevated level of p53 in untreated REF52/N-myc cells, its predominantly cytoplasmic localization prevents the induction of p21. After DNA damage, activated p53 accumulates in nuclei and induces the expression of p21, but the cell cycle arrest is still compromised. However, the activated p53 is able to induce apoptosis.
DISCUSSION
Overexpression of MYC overcomes cell cycle arrest and permits cad amplification in REF52 cells.
The first line of defense in response to the pyrimidine nucleotide depletion caused by exposure to PALA of normal cells, which are not permissive for gene amplification or other DNA rearrangements, is reversible p53-mediated cell cycle arrest, a process quite distinct from the irreversible p53-mediated response of the same cells to agents that damage DNA directly (32). Reversible arrest protects cells from the DNA damage that accompanies attenuated mitosis when DNA synthesis is not complete (59), while irreversible arrest protects the organism from abnormal cells that have suffered unrepaired damage. PALA-treated REF52 cells first accumulate at the beginning of S phase and then shift toward the middle of S phase. The arrest is tight: the cells do not reach G2 even after 5 days (Fig. 5A) or 9 days (data not shown) in PALA. Although the arrest is reversible for only 15% of the treated cells, it is protective for the cell populations, since it prevents the propagation of damaged cells. Abrogation of arrest in REF52 cells by the mutant p53 protein C141Y (Fig. 5B) or by SV40 large T antigen (26) confirms the dependence on p53. Deregulated expression of N-MYC or c-MYC abolishes arrest in REF52 cells, allowing the cells to enter mitosis under pyrimidine nucleotide-limiting conditions. Thus, the failure to arrest in PALA can lead to broken DNA, chromosomal aberrations, and death of the great majority of cells but can also promote the genesis of rare cells with amplified cad genes. In contrast, both REF52 and REF52/N-myc cells are arrested by MTX early in S phase, do not replicate their DNA, and fail to amplify dhfr. These results agree well with the recently reported MTX-induced arrest early in the S phase of both p53+ and p53− cells (32), confirming that this arrest is not mediated by p53. The differences in cell cycle regulation in response to the very different nucleotide deprivations caused by PALA or MTX may provide the basis for the different abilities of the cells to form colonies resistant to these drugs. Deregulated MYC expression leads to two changes in REF52 cells: failure to arrest in response to ribonucleotide starvation, resulting in the initiation of gene amplification, and failure to arrest in response to the DNA breaks that accompany amplification, thus allowing the propagation of cells carrying broken DNA. The importance of the second control has been demonstrated in REF52 cells transformed by tsA58, a temperature-sensitive mutant of SV40 T antigen (26). When selected with PALA at 33°C (a permissive temperature), these cells develop colonies with amplified cad. Inactivation of T antigen at 39.5°C (a nonpermissive temperature) soon after the genesis of PALA-resistant cells (less than 1,000 cells per colony) was followed by rapid p53-mediated cell growth arrest, which could be reversed by shifting the temperature back to 33°C. The effect of N-MYC in allowing the formation of PALA-resistant colonies is not unique to REF52 cells. We also analyzed human Li-Fraumeni fibroblasts with restored wild-type p53 (2). These C11 cells do not give rise to PALA-resistant colonies at a detectable frequency (less than 10−8), and, as for REF52 cells, introduction of exogenous N-myc does permit PALA-resistant colonies to form at a relatively high frequency. The data for both REF52 and C11 cells confirm the crucial role of p53 in preventing resistance to PALA and demonstrate the ability of deregulated N-myc to abrogate this defensive mechanism.
Effects of preexposing cells to PALA.
The success of gene amplification in REF52 cells requires that two events occur in the same cell: inactivation of checkpoints that respond to DNA damage or nucleotide starvation and amplification of the target gene. The probability of achieving this situation is very low except when the events are not independent. Pretreatment of REF52 cells with a low concentration of PALA for two to three cell divisions before selection induces the formation of PALA-resistant colonies with amplified cad at a frequency many orders of magnitude higher than that observed in a one-step selection. All of the PALA-resistant colonies so obtained have large amplicons which include the physically linked cad and N-myc genes. In these cells, the amplification of endogenous N-myc is accompanied by activation of its expression, thus permitting cells with amplified cad to overcome p53-mediated growth arrest.
We do not know the mechanism of activation of N-myc expression as a result of an increase in copy number, but we speculate that expression could be stimulated if a factor that negatively regulates N-myc is titrated out by an increase in gene copy number. The N-MYC protein is expressed at a high level in several neonatal mouse tissues (13), but only low levels of N-myc mRNA are detected in most adult tissues. The N-myc promoter is active in many cell types, even those with undetectable levels of mRNA (5). Down regulation of N-myc mRNA occurs through trans-acting proteins which negatively regulate transcriptional initiation (60, 67), transcriptional elongation (70), and the stability of mature transcripts (5).
When any cell is starved for deoxynucleoside triphosphates under conditions where DNA synthesis is not inhibited completely, the DNA is likely to be broken through misincorporation and unsuccessful attempts at repair (44). For a permissive cell line such as BHK, which does not arrest efficiently when DNA is broken, pretreatment with either PALA or MTX can stimulate amplification of either cad or dhfr, since amplification of the target gene alone is sufficient for resistance (44). When nonpermissive cells are used, the situation and results are different: pretreatment with PALA permits only cad amplification, whereas pretreatment with MTX does not cause the cells to become permissive at all. Our cell cycle data show that REF52 cells do not arrest in response to a low nonselective concentration of PALA, continuing to replicate their DNA for a few days. The DNA breaks likely to result from replication when DNA precursors are limiting can provide starting points for gene amplification, probably through bridge-breakage-fusion mechanisms, as indicated by the structure of the amplified cad genes, which are arranged in large amplicons on marker chromosomes (52). The activation of N-myc expression allows REF52 cells to escape from the arrest induced by broken DNA and facilitates further cad and N-myc coamplification. It is interesting that the amplified DNA is unstable, since culturing the cells without PALA for 2 to 3 weeks resulted in the loss of both amplified cad and N-myc genes (data not shown), indicating that PALA-resistant REF52 cells do not tolerate amplified DNA well, probably because of the associated DNA breaks that are a necessary part of continuing bridge-breakage-fusion cycles (44, 52). Failure to detect dhfr amplification upon pretreatment with a low nonselective concentration of MTX, followed by selection at a higher concentration, may be due to a very low frequency of the two required events if there is no gene near dhfr that can overcome the arrest.
Overexpression of MYC abrogates the ability of p53 to cause growth arrest.
We have shown previously that the inactivation of wild-type p53 in REF52 cells by expression of a mutant p53 protein or SV40 T antigen (26) permits the selection of PALA-resistant cells with amplified cad genes. The present data demonstrate that a similar effect can be achieved through overexpression of MYC, which overcomes the p53-mediated cell cycle arrest induced in response to PALA treatment. In contrast, MTX-induced early S-phase arrest does not depend on p53 (32); N-myc is not able to overcome this arrest, and, as a result, REF52/N-myc cells do not give rise to MTX-resistant colonies. Analysis of untreated REF52/N-myc cells reveals that the p53 level is significantly increased, probably through protein stabilization, in proportion to N-MYC expression. The accumulation of wild-type or mutant p53 has been found in cells with deregulated expression of c-MYC (23, 45), but the mechanism is unknown. Despite their high levels of p53, untreated REF52/N-myc cells have a low basal level of p21waf1, suggesting that the p53 present is unable to activate p21waf1 transcription. DNA damage or treatment with PALA does lead to p53-dependent activation of p21waf1 expression, which, however, does not lead to efficient cell growth arrest. Instead, the REF52/N-myc cells became very sensitive to apoptosis, and their ability to undergo apoptosis correlated with the levels of N-myc and p53. These data are consistent with the observations that MYC-mediated, p53-dependent apoptosis is independent of cell cycle arrest and the induction of p21waf1 (65). However, the involvement of p53-independent mechanisms of MYC-mediated apoptosis (46) is also possible.
The predominantly cytoplasmic localization of p53 in REF52/N-myc cells, even after UV irradiation, prompted us to suggest cytoplasmic retention as a p53-inactivating mechanism. Many tumors and cell lines have been identified recently which use cytoplasmic retention of wild-type p53 as a way to inactivate the ability of p53 to suppress growth (37, 38, 58). However, this mechanism has not been connected to the deregulated expression of N-MYC. Interestingly, cytoplasmic retention of p53 is also observed in neuroblastomas, tumors in which N-MYC is overexpressed frequently. The absence of p53 mutations in primary neuroblastomas (29, 64) supports the idea that these tumors may have developed other ways to inactivate the growth-suppressive function of p53. Recent data showing that c-MYC represses growth arrest by suppressing the transcription of gadd45 mRNA (35) represents another possible mechanism, which we are now testing.
In summary, we have demonstrated that activation of a myc proto-oncogene, which can be stimulated under relatively mild conditions, allows cells to overcome the p53-mediated cell cycle arrest that follows DNA damage, thus promoting gene amplification and genomic instability.
ACKNOWLEDGMENTS
We are grateful to William E. Fahl for pSV40myc, Kenneth B. Marcu for pMC-myc, Yoichi Taya for mouse N-myc, and Arnold Levine for PAb421. We thank Theresa Bendele for help with the flow cytometry and Gloria Umoh and Galina Ilyinskaya for technical assistance.
This research was funded by NIH grant R01 GM49345.
Olga B. Chernova and Michail V. Chernov contributed equally to this work.
REFERENCES
- 1.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal, M. L., O. B. Chernova, A. Agarwal, W. R. Taylor, Y. K. Sharma, and G. R. Stark. Unpublished data.
- 3.Alitalo K, Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. doi: 10.1016/s0065-230x(08)60201-8. [DOI] [PubMed] [Google Scholar]
- 4.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 5.Babiss L E, Friedman J M. Regulation of N-myc gene expression: use of an adenovirus vector to demonstrate posttranscriptional control. Mol Cell Biol. 1990;10:6700–6708. doi: 10.1128/mcb.10.12.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brondyk W H, Boeckman F A, Fahl W E. N-myc oncogene enhances mitogenic responsiveness of diploid human fibroblasts to growth factors but fails to immortalize. Oncogene. 1991;6:1269–1276. [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernova O B, Chernov M V, Agarwal M L, Taylor W R, Stark G R. The role of p53 in regulating genomic stability when DNA and RNA synthesis are inhibited. Trends Biochem Sci. 1995;20:431–434. doi: 10.1016/s0968-0004(00)89094-5. [DOI] [PubMed] [Google Scholar]
- 9.Cox L S, Lane D P. Tumour suppressors, kinases and clamps: how p53 regulates the cell cycle in response to DNA damage. Bioessays. 1995;17:501–508. doi: 10.1002/bies.950170606. [DOI] [PubMed] [Google Scholar]
- 10.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 11.Darzynkiewicz Z, Bruno S, Del-Bino G, Gorcyzyca W, Hotz M A, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 12.Denis N, Kitzis A, Kruh J, Dautry F, Corcos D. Stimulation of methotrexate resistance and dihydrofolate reductase gene amplification by c-myc. Oncogene. 1991;6:1453–1457. [PubMed] [Google Scholar]
- 13.DePinho R A, Legouy E, Feldman L B, Kohl N E, Yancopolous G D, Alt F W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci USA. 1986;83:1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias P, Kumar P, Marsden H B, Gattamaneni H R, Heighway J, Kumar S. N-myc gene is amplified in alveolar rhabdomyosarcomas (RMS) but not in embryonal RMS. Int J Cancer. 1990;45:593–596. doi: 10.1002/ijc.2910450403. [DOI] [PubMed] [Google Scholar]
- 15.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 18.Facchini L M, Chen S, Marhin W W, Lear J N, Penn L Z. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol Cell Biol. 1997;17:100–114. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franza B, Jr, Maruyama K, Garrels J I, Ruley H E. In vitro establishment is not a sufficient prerequisite for transformation by activated ras oncogenes. Cell. 1986;44:409–418. doi: 10.1016/0092-8674(86)90462-9. [DOI] [PubMed] [Google Scholar]
- 20.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 22.Harrington E A, Bennett M R, Fanidi A, Evan G I. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 24.Huang L-C, Clarkin K C, Wahl G M. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingvarsson S, Asker C, Wirschubsky Z, Szpirer J, Levan G, Klein G, Sumegi J. Mapping of Lmyc and Nmyc to rat chromosomes 5 and 6. Somatic Cell Mol Genet. 1987;13:335–339. doi: 10.1007/BF01534927. [DOI] [PubMed] [Google Scholar]
- 26.Ishizaka Y, Chernov C M, Burns C M, Stark G R. p53-dependent growth arrest of REF52 cells containing newly amplified DNA. Proc Natl Acad Sci USA. 1995;92:3224–3228. doi: 10.1073/pnas.92.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearsey J M, Coates P J, Prescott A R, Warbrick E, Hall P A. Gadd45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- 28.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 29.Komuro H, Hayashi Y, Kawamura M, Hayashi K, Kaneko Y, Kamoshita S, Hanada R, Yamamoto K, Hongo T, Yamada M, et al. Mutations of the p53 gene are involved in Ewing’s sarcomas but not in neuroblastomas. Cancer Res. 1993;53:5284–5288. [PubMed] [Google Scholar]
- 30.Lee W H, Murphree A L, Benedict W F. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–460. doi: 10.1038/309458a0. [DOI] [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Linke S P, Clarkin K C, DiLeonardo A, Tsou A, Wahl G M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 33.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 34.Mai S, Hanley-Hyde J, Fluri M. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene. 1996;12:277–288. [PubMed] [Google Scholar]
- 35.Marhin W W, Chen S, Facchini L M, Fornace A J, Jr, Penn L Z. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 36.Miyashita T, Krajewska M, Wang H G, Lin H K, Liebermann D A, Hoffman B, Reed J C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 37.Moll U M, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci USA. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moll U M, Ostermeyer A G, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern J P, Land H. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W-W, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Packham G, Cleveland J L. c-Myc and apoptosis. Biochim Biophys Acta. 1995;1242:11–28. doi: 10.1016/0304-419x(94)00015-t. [DOI] [PubMed] [Google Scholar]
- 42.Perry M E, Commane M, Stark G R. Simian virus 40 large tumor antigen alone or two cooperating oncogenes convert REF52 cells to a state permissive for gene amplification. Proc Natl Acad Sci USA. 1992;89:8112–8116. doi: 10.1073/pnas.89.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poupon M-F, Smith K A, Chernova O B, Gilbert C, Stark G R. Inefficient growth arrest in response to dNTP starvation stimulates gene amplification through bridge-breakage-fusion cycles. Mol Biol Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy B, Beamon J, Balint E, Reisman D. Transactivation of the human p53 tumor suppressor gene by c-Myc/Max contributes to elevated mutant p53 expression in some tumors. Mol Cell Biol. 1994;14:7805–7815. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamuro D, Eviner V, Elliott K J, Showe L, White E, Prendergast G C. c-Myc induces apoptosis in epithelial cells by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:2411–2418. [PubMed] [Google Scholar]
- 47.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Schimke R T. Gene amplification in cultured cells. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- 49.Schwab M, Ellison J, Busch M, Rosenau W, Varmus H E, Bishop J M. Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc Natl Acad Sci USA. 1984;81:4940–4944. doi: 10.1073/pnas.81.15.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigesada K, Stark G R, Maley J A, Niswander L A, Davidson J N. Construction of cDNA to the hamster CAD gene and its application toward defining the domain for aspartate transcarbamylase. Mol Cell Biol. 1985;5:1735–1742. doi: 10.1128/mcb.5.7.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith K A, Chernova O B, Groves R P, Stark M B, Martinez J L, Davidson J N, Trent J M, Patterson T E, Agarwal A, Duncan P, Agarwal M L, Stark G R. Multiple mechanisms of N-phosphonacetyl-l-aspartate resistance in human cell lines: carbamyl-P-synthetase/aspartate transcarbamylase/dihydro-orotase gene amplification is frequent only when chromosome 2 is rearranged. Proc Natl Acad Sci USA. 1997;94:1816–1821. doi: 10.1073/pnas.94.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith K A, Stark M B, Gorman P A, Stark G R. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith M L, Chen I-T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O’Connor P M, Fornace A J., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 54.Spurr N K, White R. Report of the committee on the genetic constitution of chromosome 2. Cytogenet Cell Genet. 1991;58:142–169. doi: 10.1159/000132998. [DOI] [PubMed] [Google Scholar]
- 55.Stanton L W, Watt R, Marcu K B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983;303:401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- 56.Stark G R. Regulation and mechanisms of mammalian gene amplification. Adv Cancer Res. 1993;61:87–113. doi: 10.1016/s0065-230x(08)60956-2. [DOI] [PubMed] [Google Scholar]
- 57.Stewart N, Hicks G G, Paraskevas F, Mowat M. Evidence for a second cell cycle block at G2/M by p53. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 58.Takahashi K, Suzuki K. DNA synthesis-associated nuclear exclusion of p53 in normal human breast epithelial cells in culture. Oncogene. 1994;9:183–188. [PubMed] [Google Scholar]
- 59.Taylor, W. R., M. L. Agarwal, A. Agarwal, D. W. Stacey, and G. R. Stark. p53 inhibits entry into mitosis when DNA synthesis is blocked. Submitted for publication. [DOI] [PubMed]
- 60.Tevosian S G, Shih H H, Mendelson K G, Sheppard K-A, Paulson K E, Yee A S. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 61.Tishler R B, Calderwood S K, Coleman C N, Price B D. Increases in sequence specific DNA binding by p53 following treatment with chemotherapeutic and DNA damaging agents. Cancer Res. 1993;53:2212–2216. [PubMed] [Google Scholar]
- 62.Tlsty T D. Normal diploid human and rodent cells lack a detectable frequency of gene amplification. Proc Natl Acad Sci USA. 1990;87:3132–3136. doi: 10.1073/pnas.87.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toledo F, Buttin G, Debatisse M. The origin of chromosome rearrangements at early stages of AMPD2 gene amplification in Chinese hamster cells. Curr Biol. 1993;3:255–264. doi: 10.1016/0960-9822(93)90175-n. [DOI] [PubMed] [Google Scholar]
- 64.Vogan K, Bernstein M, Leclerc J M, Brisson L, Brossard J, Brodeur G M, Pelletier J, Gros P. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53:5269–5273. [PubMed] [Google Scholar]
- 65.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 66.White A E, Livanos E M, Tlsty T D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 67.Woodruff K A, Rosenblatt J D, Moore T B, Medzoyan R H, Pai D S, Noland J L, Yamashiro J M, Wada R K. Cell type-specific activity of the N-myc promoter in human neuroblastoma cells is mediated by a downstream silencer. Oncogene. 1995;10:1335–1341. [PubMed] [Google Scholar]
- 68.Wright J A, Smith H S, Watt F M, Hancock M C, Hudson D L, Stark G R. DNA amplification is rare in normal human cells. Proc Natl Acad Sci USA. 1990;87:1791–1795. doi: 10.1073/pnas.87.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 70.Xu L, Meng Y, Wallen R, DePinho R A. Loss of transcriptional attenuation in N-myc is associated with progression towards a more malignant phenotype. Oncogene. 1995;11:1865–1872. [PubMed] [Google Scholar]
- 71.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]