Abstract
Background
Coinfection with multiple viruses is a common occurrence in the pig industry. Porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) are two significant pathogens, with a notable prevalence of their coinfection observed in clinical settings. However, reports on corresponding combination vaccines against PRRSV and PCV2 are scarce. The capsid (Cap) protein encoded by PCV2 is highly immunogenic, and recombinant Cap expressed in vitro can self-assemble into virus-like particles (VLPs), providing a promising strategy for developing bivalent vaccines against both PRRSV and PCV2.
Method
Three novel nanoparticle vaccines (Cap-DS, Cap-DP, and Cap-DE) were engineered by inserting multiple neutralizing epitopes from PRRSV (GP5B epitope, GP3I epitope, and GP5IV epitope) into the PCV2 Cap protein. These recombinant proteins, produced using a baculovirus expression system, successfully formed VLPs in vitro.
Results
Immunization of BALB/c mice with these nanoparticle vaccines significantly enhanced T-lymphocyte immune responses and elicited high-titer antibodies against both PCV2 and PRRSV. Notably, Cap-DS induced more potent serum neutralizing antibodies against PRRSV compared to Cap-DP and Cap-DE. Piglets immunized with Cap-DS, upon challenge with PRRSV, demonstrated significant protection compared to the PBS-immunized control group.
Conclusions
These findings indicate that Cap-DS is a promising candidate for the development of a bivalent nanoparticle vaccine against PRRSV and PCV2 infections, addressing a significant gap in current swine vaccination strategies.
Graphical Abstract

Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-025-03514-8.
Keywords: Porcine reproductive and respiratory syndrome virus, Porcine circovirus type 2, Virus-like particles, Nanoparticle vaccine, Baculovirus expression
Introduction
In modern swine production, porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) are among the most serious disease threats [1–5]. PRRSV is a single-stranded RNA virus in the family Arteriviridae, causing reproductive failure in pregnant pigs, as well as severe respiratory disease in newborn/weaned piglets and growing pigs [6–11]. Meanwhile, PCV2 is a small single-stranded DNA virus belonging to the Circoviridae family [12, 13]. PCV2 is the main cause of porcine circovirus associated disease (PCVAD), including post weaning multiple system exhaustion syndrome (PMWS), porcine dermatitis, and nephrotic syndrome (PDNS). In pig farms, the serious PRRSV infections are often accompanied by PCV2 and the synergistic effect of these two viruses often leads to more serious consequences for farming [14–17].
Large-scale vaccination serves as an economical and effective strategy to defend against virus infection. At present, commercially available vaccines against PRRSV or PCV2 alone have been widely applied in the clinic. However, PRRSV vaccines are less effective and often fail to provide robust protection against the virulent strains due to two primary factors. First, PRRSV exhibits an extremely high mutation rate, while most existing PRRSV vaccines are developed based on specific strains [18]. Due to the potentially significant differences in antigenic characteristics between newly emerging strains and vaccine strains, these vaccines often struggle to provide effective protection against new variants. Second, safety concerns associated with modified live virus (MLV) vaccines for PRRSV, including the risk of reversion to virulence and potential transplacental transmission, complicate their use in breeding herds [19]. As a result, the traditional PRRSV vaccines frequently fail to achieve the expected efficacy in clinical applications. In contrast, numerous studies have demonstrated that PCV2 Cap protein subunit vaccines have exceptional efficacy, resulting in substantial reductions in infection rates, associated morbidities, and PCVAD incidence in the field [20, 21]. Nevertheless, they cannot completely prevent coinfections with PRRSV. The immune response induced by PCV2 vaccines mainly targets PCV2-specific antigens. PRRSV, equipped with unique immune-evasion mechanisms, can suppress the host’s immune system, thereby weakening the immune responses triggered by PCV2 vaccines. Moreover, PCV2 itself can also cause immunosuppression [22]. This not only impairs the pig’s immune function but also exacerbates the severity of PRRSV infection, creating a vicious cycle [16, 23]. Given the numerous limitations of the current single-pathogen vaccines, the development of a bivalent vaccine targeting both PRRSV and PCV2 is highly warranted.
Virus-like particles (VLPs) are non-replicating viral structures that mimic the natural virus but lack its genetic material, rendering them incapable of causing infection [24]. Composed of viral structural proteins that self-assemble into a particle resembling the virus, VLPs can effectively simulate the natural infection process and elicit a robust immune response. This characteristic has garnered significant attention in virology and vaccine research. VLPs present a host of benefits in the realm of vaccine development [25]. Compared to free, soluble protein subunits, VLPs exhibit a granular structure, which significantly enhances uptake efficiency by antigen-presenting cells (APCs) and facilitates greater accumulation in draining lymph nodes [26]. Furthermore, VLPs can present antigens on their outer surfaces in a regular and systematic manner, which is conducive to a more effective immune response [25]. Additionally, as a vaccine delivery vehicle, VLPs can stimulate CD8⁺ T cells through the cross-presentation mechanism [27]. For instance, VLP-based vaccines for human papillomavirus (HPV) and for hepatitis B virus (HBV) have been successfully developed and widely used, demonstrating their efficacy in disease prevention [28–30]. Moreover, VLPs are currently being explored for COVID-19 vaccine development, showing promising immunogenicity and safety profiles [31, 32]. This advancement underscores the potential of VLPs technology in advancing vaccine innovation.
Research on neutralizing antibody epitopes of PRRSV and PCV2 has been ongoing for many years [33, 34]. Studies indicate that the principal neutralizing epitopes are located within the structural proteins GP5 and GP3 of PRRSV [35–39]. Notably, the amino acid (aa) sequence 37–45 (S37HIQLIYNL45) within the PRRSV GP5 is recognized as a core antigenic site GP5B epitope; the IV epitope of PRRSV GP5 (187–200 aa, T187PLTRVSAERWGRL200) and the I epitope of PRRSV GP3 (61–72 aa, Q61AAAEILEPGKS72) have also been identified as crucial immunogenic sites [40–43]. Overall, significant progress has been made in understanding the neutralizing epitopes of PRRSV; however, the selection of appropriate carriers to effectively present these epitopes remains a central challenge in the development of effective subunit vaccines against PRRSV. PCV2, a non-enveloped virus, has a Cap protein that can self-assemble into VLPs, exhibiting considerable immunogenicity and effectively preventing PCV2 infection [44]. Recent studies have provided in-depth insights into the structural properties of PCV2 Cap protein-derived VLPs, with existing literature reporting their crystal structures. This progress offers valuable foundations for the application of Cap protein-based VLPs in multivalent vaccine development [45–47]. Consequently, this study aims to engineer the Cap protein of PCV2 to maintain the VLP structure and its protective efficacy against PCV2 while also serving as a carrier for PRRSV.
In this study, we designed and evaluated three innovative nanoparticle vaccines (Cap-DP, Cap-DS, and Cap-DE) that integrate multiple neutralizing epitopes of PRRSV into the Cap protein of PCV2. These recombinant proteins were successfully self-assembled into VLPs, eliciting robust immune responses in BALB/c mice. Notably, the Cap-DS vaccine exhibited the most pronounced neutralizing antibody response and protective efficacy against PRRSV challenge in immunized piglets. These findings highlight a promising strategy for the development of effective bivalent vaccines targeting both PRRSV and PCV2, with significant implications for enhancing swine health management.
Materials and methods
Ethics statement
The BALB/c mice and the piglets used in this study were from the Laboratory Animal Center of Huazhong Agricultural University and a commercial PRRSV-free farm, respectively. The animal experiments were approved by the Animal Ethics Committee of the Huazhong Agricultural University under the approval ID number: HZAUMO-2023–0207 (mice), HZAUMO-2025–0024 (mice), HZAUSW-2024–0012 (pigs).
Cells, viruses, peptides and antibodies
Sf9 and High Five (Hi5) insect cells were maintained in SF-SFM and HF502C medium (Womei, Suzhou, China), respectively. These cells were cultured in a shaker at 110 r/min and 27 °C. PK-15 cells and Marc-145 cells were maintained at 37 °C with 5% CO2 in Dulbecco’s modified eagle medium (DMEM) (Hyclone, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Shanghai, China). The PRRSV strain WUH3 (GenBank accession number: HM853673) [48] and PCV2 strain WuHan (GenBank accession number: FJ598044.1) [49] were preserved in our laboratory. The PRRSV GP5B peptide and GP35 peptide (GP3 epitope I and GP5 epitope IV tandem structure) were synthesized by Genscript Biotech Corporation (Nanjing, China) with their purity exceeded 95%. Mouse monoclonal antibodies anti-PCV2 Cap protein, and anti-PRRSV N protein were prepared and purified in our laboratory. Commercial antibodies used in this study included APC/Cyanine7-conjugated anti-mouse CD3, Brilliant Violet 510™-conjugated anti-mouse CD4, PerCP/Cyanine5.5-conjugated anti-mouse CD8a, Brilliant Violet 421™-conjugated anti-mouse IL-4, PE/Cyanine7-conjugated anti-mouse IFN-γ, FITC-conjugated anti-mouse CD44, Brilliant Violet 605™-conjugated anti-mouse CD62L (BioLegend, San Diego, CA, USA), Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (ABclonal, Wuhan, China), and DyLight 488 goat anti-mouse IgG (Abbkine, Wuhan, China).
Protein modeling
The monomeric structures of both the Cap and capsid-derived proteins were predicted using AlphaFold2 [50, 51]. To generate the three-dimensional (3D) structure of VLPs formed by the modified Cap proteins, we employed homology modeling with SWISS-MODEL [52, 53]. The template for this modeling was the previously reported crystal structure of PCV2 Cap protein (PDB ID: 3R0R) [54]. Once the modeling was complete, we downloaded the results as PDB files and imported them into PyMOL (PyMOL Molecular Graphics System, Version 1.3) to create the protein models.
Construction of recombinant baculovirus
The PCV2 Cap gene fragment fused with the PRRSV peptide was synthesized and cloned into pFast-Bac-Dual vector. This recombinant vector was then transformed into the E. coli DH5α strain for amplification. The subsequent experimental procedures followed the guidelines provided in the Bac-to-Bac baculovirus expression system manual (Invitrogen). Briefly, plasmids isolated from the transformed E. coli DH5α strain were re-transformed into the E. coli DH10Bac strain. Shuttle plasmids were identified through blue/white spot screening and extracted. Following this, Sf9 insect cells were transfected with the shuttle plasmids. After 3–4 days, the supernatants from the Sf9 insect cells were harvested, yielding the first generation of recombinant baculovirus.
Indirect immunofluorescence assay (IFA)
Recombinant baculoviruses were inoculated into Sf9 cells cultured in 6-well plates, with uninfected cells serving as controls. The cells were incubated at 27 °C for 24 h. Subsequently, they were fixed with 4% paraformaldehyde for 10 min. The fixative was then discarded, and cooled methanol was added for permeabilization for another 10 min. After washing three times with PBS, the cells were blocked with 5% bovine serum albumin (BSA) at 37 °C for 1 h. After additional washes with PBS, the cells were incubated with a monoclonal antibody anti-PCV2 Cap protein (serving as the primary antibody), followed by incubation with DyLight 488 goat anti-mouse IgG as the secondary antibody. Finally, the cell nuclei were counterstained with 0.01% 4’, 6-diamidino-2-phenylindole (DAPI) at room temperature (RT) for 15 min. After washing with PBS, the cells were observed under an inverted fluorescence microscope (Olympus IX73, Tokyo, Japan).
Recombinant proteins expression and purification
We propagated the primary generation baculovirus in Sf9 cells for 3 passages, then inoculated the fourth passage baculovirus into Hi5 insect cells for recombinant protein expression. After 10 days, the supernatants were collected by centrifugation at 4000 r/min for 5 min. The supernatants were run on a GE HiTrap SP FF 5 mL column after filtration with a 0.45 μm membrane. The column was washed with 50 mM phosphate buffer (pH 6.5) (Buffer A) and then eluted with 50 mM phosphate buffer containing 1.0 M NaCl (pH 6.5) (Buffer B). The final purification step was performed using Superose 6 Increase 10/300 GL gel filtration columns. The purified recombinant proteins were formulated in 20 mM Tris–HCl, 50 mM NaCl, and 10% glycerol (pH 6.5) (Buffer C).
Western blotting
The supernatants of the infected or mock-infected Hi5 insect cells were detected by SDS-PAGE. Then the proteins were transferred to PVDF membranes (Roche, Mannheim, Germany) using a Bio-Rad company (Hercules, CA, USA) protein transfer device. Tris-Buffered Saline with Tween-20 (TBST) containing 5% (w/v) skim milk was used to block the membranes overnight at 4 °C. After three times of washing with TBST, the membranes were incubated with anti-PCV2 Cap protein monoclonal antibody and HRP-conjugated goat anti-mouse IgG successively. The reaction was then visualized using enhanced chemiluminescent reagent (Bio-Rad, Hercules, CA, USA) and analyzed with the ImageLab4.0.1 software.
Dynamic light scattering (DLS)
Whether the recombinant proteins are self-assembled into nanoparticles were determined by DLS. The purified recombinant proteins (10 μg/mL) were measured three times with 13 runs each time at 37 °C. The size distribution of Cap and Capsid-Derived nanoparticles in 10 mM Tris–HCl system were shown.
Transmission electron microscope (TEM)
After negative staining with phosphotungstic acid (pH 7.0), the purified recombinant proteins were observed and photographed using a transmission electron microscope (JEM-1200EX, Tokyo, Japan) at accelerating voltage of 100 kV.
Vaccination and PCV2 challenge in mice
The purified Cap-WT, Cap-DP, Cap-DS and Cap-DE proteins and PBS were emulsified with ISA201 adjuvant (Seppic, Paris, France). 6–8-weeks-old female BALB/c mice were divided into 5 groups and immunized with these proteins (20 μg per mouse) and PBS on days 0 and 21. In addition, three untreated normal mice will be prepared as the control group to measure the serum levels of interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α). Blood samples were collected on days 21 and 35 after the first immunization to detect the antibody levels against PCV2 or PRRSV by ELISA or serum neutralization assay. At 35 days post-primary immunization (dpi), three mice from each group were sacrificed, serum samples were collected for antibody and inflammatory cytokines detection and spleen tissues were collected for lymphocyte separation. The remaining 5 mice in each group were challenged with 2 × 105.0 TCID50 of PCV2 strain WuHan, and euthanized at 14 days post challenge (dpc). Spleen, lung, and kidney tissues were collected for viral load analysis and histopathological examination, while sera were additionally obtained for quantification of viral load.
Preparation and inactivation of PRRSV strain WUH3
This study inactivated the PRRSV strain WUH3 according to the preparation protocol established by Wuhan Keqian Biology Co., Ltd. for PRRS inactivated vaccines. Briefly, PRRSV was propagated in Marc-145 cells and subjected to three freeze–thaw cycles, followed by centrifugation at 4000 × g for 5 min to remove cellular debris. The supernatant was then inactivated with formaldehyde solution at a final concentration of 0.2% (v/v) for 48 h at 37 °C. Complete viral inactivation was confirmed by subsequent inoculation onto Marc-145 cells.
Vaccination and PRRSV challenge in piglets
The purified Cap-DS protein and inactivated PRRSV strain WUH3 were individually emulsified with ISA 201 adjuvant to prepare subunit and inactivated vaccines, respectively. Fifteen 4–5-week-old piglets negative for PRRSV were randomly divided into three groups (n = 5/group). Group 1 received Cap-DS subunit vaccine (200 μg/piglet) through cervical intramuscular injection, Group 2 was administered inactivated PRRSV WUH3 (106.5 TCID50/piglet) via the same route, and Group 3 received adjuvant-emulsified PBS. All groups received prime immunization followed by two booster doses at 14-day intervals (days 0, 14, and 28) with equivalent antigen quantities. Blood samples were collected at 14, 28 and 38 dpi to detect the neutralizing antibody against PRRSV. Then all the piglets were challenged with 2 × 105.0 TCID50 of PRRSV strain WUH3 at 38 dpi. Rectal temperatures of all piglets were recorded daily from 1 to 14 dpc. The survival rate was monitored daily and calculated over the 14 dpc period. Serum samples were collected at 1, 3, 5, 7, 10 and 14 dpc for viral load analysis. All piglets were euthanized at 14 dpc, and lung tissues were collected for viral load analysis and histopathological examination.
Enzyme-linked immunosorbent assay (ELISA)
The Cap protein or synthetic PRRSV GP5B and GP35 epitope peptides were diluted to the optimal coating concentration with 50 mM carbonate buffer (pH 9.6) and then added to the enzyme plate (Jincanhua, Shenzhen, China) and placed at 4 °C overnight. After washing with PBST, blocking solution was added and incubated for 1 h at 37 °C. The tested sera were first diluted to the appropriate multiple, followed by a twofold serial dilution, and incubated for 1 h at 37 °C. HRP-conjugated goat anti-mouse IgG was added after washing with PBST and incubated at 37 °C for 1 h. After washing, TMB (Invitrogen, Carlsbad, CA, USA) was added and incubated in the dark environment for 15 min. The reaction was stopped by adding H2SO4, and the OD value at 630 nm was measured on a microplate reader (BioTek, Winooski, VT, USA).
PRRSV serum neutralization test
The serum neutralization test for PRRSV was conducted as previously described [55]. Serum samples were inactivated at 56 ℃ for 30 min before testing, then twofold serially diluted and mixed with an equal volume of PRRSV strain WUH3 (200 TCID50/0.1 mL). After incubation for 1 h at 37℃, the mixture was added in quadruplicate to the monolayer of Marc-145 cells in the 96-well cell culture plate. After adsorption for 1 h, the supernatants were discarded and DMEM medium containing 2% FBS was then added. The plate was kept at 37 °C with 5% CO2 and the cytopathic effect (CPE) was observed for 5 days. The titers of neutralizing antibody against PRRSV were calculated by the Reed-Muench method.
PCV2 serum neutralization test
The PCV2 serum neutralization test was carried out with 96-well cell culture plate as previously described [56]. Briefly, serum samples were inactivated at 56 ℃ for 30 min before detection, then twofold serially diluted and mixed with an equal volume of PCV2 strain WuHan (200 TCID50/0.1 mL). After incubation for 1 h at 37 ℃, the mixture was added in quadruplicate to monolayer of PK-15 cells in the 96-well cell culture plate. After adsorption for 1 h, the supernatants were discarded and DMEM medium containing 2% FBS was added. The cells were incubated at 37 °C with 5% CO2 for 24 h and then washed with PBS. D-glucosamine (300 mM) was added and incubated at 37 ℃ for 30 min. After washing with PBS again, DMEM medium containing 2% FBS was added for further cultivation for 48 h, then fixed with 4% paraformaldehyde, and permeabilized with methanol. The cell culture plate was blocked with 4% BSA for 1 h at 37 °C, and then incubated with anti-PCV2 Cap protein monoclonal antibody for another 1 h at 37 °C. After then, the cells were incubated with DyLight 488 goat anti-mouse IgG for 1 h at 37 °C. The cells were observed using an inverted fluorescence microscopy (Olympus IX73, Tokyo, Japan). The titers of neutralizing antibody against PCV2 were calculated by the Reed-Muench method.
Lymphocyte proliferation assay
The splenic lymphocytes of mice were isolated and cultured in 96-well plates with RPMI-1640 medium containing 10% FBS at 37 °C with 5% CO2, then stimulated with a mixture of purified proteins (Cap-WT, Cap-DP, Cap-DS, and Cap-DE mixed at a 1:1:1:1 mass ratio) at concentration of 5 µg/mL, and the unstimulated cells were used as negative controls. After 44 h of incubation, CCK-8 reagent (10 μL) (Beyotime, Shanghai, China) was added to each well and then incubated for another 4 h. The OD value at 450 nm was read with a microplate reader (BioTek, Winooski, VT, USA). The stimulation index (SI) was calculated as follows: OD value of the well containing stimulated cells /OD value of the well containing unstimulated cells.
Flow cytometry analysis
The methods for lymphocyte isolation and intracellular cytokine staining were performed in accordance with a previously published study [57]. In brief, the isolated T lymphocytes were seeded into a cell culture plate at a density of 1 × 107 cells per well. Each well was supplemented with a 5 µg/mL protein mixture containing purified Cap-WT, Cap-DP, Cap-DS, and Cap-DE proteins (mixed at a 1:1:1:1 mass ratio) and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Subsequently, Brefeldin A Solution (BioLegend, San Diego, CA, USA) was added to block protein secretion. After centrifugation, the cells were resuspended in Cell Staining Buffer (BioLegend, San Diego, CA, USA) and stained with fluorochrome-conjugated surface antibodies (diluted according to the manufacturer’s protocol for flow cytometry) for 20 min at 4 °C in the dark. Following two washes with Cell Staining Buffer, cells were fixed and permeabilized using Fixation Buffer (BioLegend, San Diego, CA, USA) for 20 min at RT in the dark. The fixed cells were washed twice with Intracellular Staining Perm Wash Buffer (BioLegend, San Diego, CA, USA) and centrifuged at 350 × g for 5 min. For intracellular cytokine detection, cells were incubated with fluorochrome-conjugated anti-IL-4 and anti-IFN-γ antibodies for 20 min at RT in the dark. After the final washes, cells were analyzed on a CytoFLEX LX flow cytometer (Beckman Coulter, Brea, CA, USA), and data were processed using CytExpert software.
Cytokine detection
Serum samples collected at 35 dpi were centrifuged (5,000 r/min, 10 min, 4 °C) and subsequently stored at − 80 °C until further analysis. The concentrations of TNF-α and IL-6 were measured using mouse-specific ELISA kits (Mlbio, Shanghai, China) according to the manufacturer’s instructions. Absorbance was recorded at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
Quantitative analysis of viral loads
The viral loads in the spleens, lungs and serum samples of mice or piglets were detected by real-time quantitative PCR (qPCR). Total DNA/RNA was extracted using FastPure Viral DNA/RNA Mini Kit (Vazyme, Nanjing, China). The extracted PRRSV RNA was reverse-transcribed into cDNA by HiScript III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, Nanjing, China). The qPCR primers and probes of PCV2 and PRRSV were listed in Table S1. The content of PCV2 Cap gene or PRRSV N gene was then detected by qPCR with AceQ Universal U+ Probe Master Mix V2 (Vazyme, Nanjing, China). Standard curves were established with CT values from serially diluted plasmid standards and then the copy numbers of the samples were calculated according to the standard curves.
Cleaved Caspase-3 immunofluorescence assay
Spleen tissues were fixed in 4% paraformaldehyde, dehydrated via graded ethanol, cleared in xylene, and embedded in paraffin. Sections were mounted on slides, baked at 60 °C for 30 min, deparaffinized in xylene, and rehydrated through graded ethanol. Antigen retrieval was performed by heating in citrate buffer under controlled conditions. After PBS washes, sections were blocked with 3% BSA for 30 min, incubated overnight at 4 °C with rabbit anti-mouse Cleaved Caspase-3 antibody (1:200), and stained with CY3-conjugated secondary antibody (1:500) for 50 min at RT. Nuclei were counterstained with DAPI for 10 min. Slides were treated with autofluorescence quencher, rinsed, and mounted with anti-fade medium. Cleaved Caspase-3-positive cells exhibiting red fluorescence were imaged using a fluorescence microscope.
Histopathology and immunohistochemistry (IHC)
Spleen, lung, and kidney tissues of mice and lung tissues of piglets were fixed in 4% paraformaldehyde for 24 h and then embedded in paraffin for sectioning. Histopathological alterations of the sections were observed by hematoxylin–eosin (HE) staining under an optical microscope (Nikon, Tokyo, Japan). IHC was used to detect PCV2- or PRRSV-specific antigens. Briefly, the embedded tissue sections were dewaxed and hydrated, and antigen retrieval was performed by autoclaving. Then sections were subsequently stained immunohistochemically and counterstained. Anti-PCV2 Cap protein monoclonal antibody or Anti-PRRSV N protein monoclonal antibody was used as primary antibodies, while HRP-conjugated goat anti-mouse IgG was used as secondary antibody for IHC detection.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism v8.01 (GraphPad Software) (La Jolla, CA, USA). Data are presented as mean ± SEM in each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Results
Designing PCV2 capsid-derived nanoparticles for the insertion of PRRSV neutralizing epitopes
The Cap protein of PCV2 is a crucial structural component, consisting of 234 amino acid residues. Extensive structural analysis via X-ray crystallography has elucidated that the Cap protein adopts a stable, barrel-like conformation characterized by eight β-sheet strands interconnected through hydrogen bonding (Fig. 1A and B). In its native state, the Cap protein self-assembles into 60-mer spherical VLPs, a process essential for its biological functions (Fig. 1B). Within the Cap protein, six distinct antigenic epitopes have been identified by immunogenic studies (Fig. 1A).
Fig. 1.
Designing PCV2 capsid-derived nanoparticles for the insertion of PRRSV neutralizing epitopes. A Schematic representation of the antigenic epitopes and β-sheet structures within the amino acid sequence of the Cap protein. The six known epitopes (epitopes 1–6) are identified based on previous studies. Light blue arrows (B–I) indicate the eight β-sheets present in the Cap protein. The red pentagram highlights the region containing inserted PRRSV neutralizing epitopes. B Three-dimensional structural schematic of the VLPs assembled from the Cap protein. The left side depicts the overall structure of the VLPs, while the right side illustrates the monomeric form of the Cap protein. Notably, the gray region (85G–88P) and the green region (231L–234 K) are displayed on the surface of the VLPs, highlighting important surface features. Additionally, the pink region (169S–180R) is shown, corresponding to epitope 5, which is characterized as non-neutralizing. C Design schematic of Cap-DS, Cap-DP, and Cap-DE. The gray area denotes the outer surface of the virus-like particles. The regions in which exogenous fragments are inserted are indicated in red (85G–88P), green (231L–234K), and yellow (169S–180R), respectively
Recent studies have highlighted the surface areas of nanoparticles as key platforms for displaying drugs and antigenic epitopes [58, 59]. By analyzing the surface regions, structural framework, and antigenic epitopes of the PCV2 Cap protein, we identified amino acid residues 85–88 and 231–234 of PCV2 Cap protein as promising sites for the introduction of foreign viral epitopes (Fig. 1B). These regions do not overlap with any known antigenic epitopes and are strategically located away from the core of the eight β-sheets (Fig. S1A), making them ideally positioned on the outer, fully exposed surface of the nanoparticles (Fig. S1B). To further explore this potential, we substituted the amino acids at positions 85–88 of Cap protein with PRRSV neutralizing epitope GP5B, fused to a dendritic cell-binding peptide (DCBp, FYPSYHSTPQRP) [60]. The simultaneous incorporation of PRRSV GP35 neutralizing epitope, characterized by a tandem fusion of GP3 epitope I and GP5 epitope IV, at the C-terminus of Cap protein resulted in the creation of a novel engineered nanoparticle, designated as Cap-DS (Fig. 1C). Structural predictions conducted using AlphaFold2 revealed that Cap-DS maintains a tertiary structure closely resembling that of the wild-type Cap protein (Fig. S2). Furthermore, confidence analysis highlighted the structural reliability of Cap-DS, particularly within the core region of the eight β-strands, which exhibited confidence levels exceeding 90% (Fig. S2). These findings collectively suggest that the modifications of Cap protein do not significantly compromise its structural integrity or multimerization, thereby providing preliminary validation for the viability of these alterations. Previous studies have identified a decoy epitope within Cap protein, spanning amino acids 169 to 180, located at the interface between the protein’s inner and outer surfaces[54, 61]. Substituting this decoy epitope with the GP35 epitope presents a promising strategy for eliciting neutralizing antibodies against PRRSV [62, 63]. Based on these results, we substituted the PCV2 decoy epitope with PRRSV GP35 epitope which fused with DCBp, leading to the generation of the novel construct Cap-DP as a reference group. Additionally, we engineered a nanoparticle, Cap-DE, which builds on Cap-DS and incorporates modifications from Cap-DP, aimed to evaluate whether the concomitant introduction of these modifications can further enhance the immunogenicity of the polyvalent vaccine (Fig. 1C). Together, we designed three engineered VLPs to advance bivalent vaccination strategies against PRRSV and PCV2.
Preparation and structural characterization of capsid-derived nanoparticles
We utilized the baculovirus expression system to express wild-type Cap (Cap-WT) and three engineered capsid nanoparticles: Cap-DP, Cap-DS, and Cap-DE. As illustrated in Fig. 2A, a substantial amount of soluble proteins, with molecular weights ranging from 25 to 35 kDa, could be detected in the culture medium of insect cells 10 days post-baculovirus infection. Importantly, no significant degradation bands were observed during incubation at 27 °C over several days, indicating the remarkable stability of these recombinant proteins.
Fig. 2.
Preparation and structural characterization of capsid-derived nanoparticles. A SDS-PAGE and Coomassie Brilliant Blue staining of supernatant from Hi5 insect cell culture post-induction for 10 days. The recombinant Cap proteins exhibit theoretical molecular weights of 27.95 kDa (Cap-WT), 30.75 kDa (Cap-DP), 33.27 kDa (Cap-DS), and 34.61 kDa (Cap-DE). B Size exclusion chromatography (SEC) elution profile of the purified recombinant Cap proteins on a Superose 6 column. C SDS-PAGE and Coomassie Brilliant Blue staining of the purified recombinant Cap proteins. D Western blotting analysis of the purified recombinant Cap proteins using a monoclonal antibody against PCV2 Cap protein. E–H TEM images of the purified recombinant Cap proteins. I–L DLS analysis results of the purified recombinant Cap proteins
Given that the isoelectric points of both wild-type and engineered Cap proteins are above 6.0, we employed cation exchange chromatography as the initial purification step. To isolate proteins capable of self-assembling into VLPs, we followed up with size exclusion chromatography for secondary purification. The target proteins primarily eluted around 6–8 mL (Fig. 2B), achieving a purification yield exceeding 90% (Fig. 2C). The eluted fractions corresponded to proteins with molecular weights greater than 100 kDa, suggesting significant self-assembly of the purified Cap proteins. We further performed western blotting (Fig. 2D) and IFA (Fig. S3) to confirm the presence of specific proteins recognized by monoclonal antibody against PCV2 Cap protein, thereby validating them as products of the target gene expression.
To further assess the capacity of these expressed Cap proteins to form VLPs, we analyzed the size and morphology of the purified fractions using transmission electron microscopy. The results revealed a high density of uniform spherical nanoparticles, averaging approximately 20 nm in diameter, thus confirming the formation of stable VLP structures (Fig. 2E–H). Complementary DLS analysis (Fig. 2I–L) corroborated these findings, indicating that the average size of the majority of purified nanoparticles was indeed around 20 nm. Taken together, our engineered Cap proteins effectively form stable VLPs, while the insect expression system facilitates the efficient production of abundant secreted VLPs.
Antibody responses of capsid-derived nanoparticles vaccines in mice
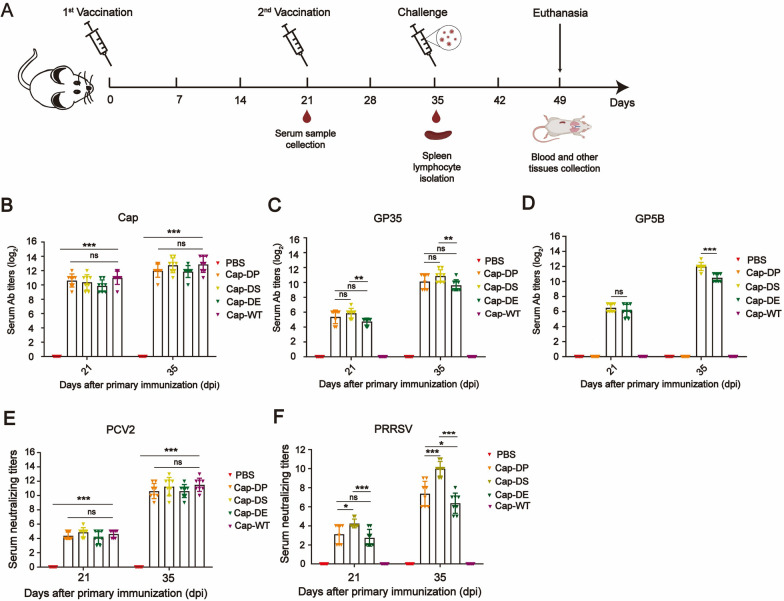
To assess the immunogenicity of the recombinant Cap-DP, Cap-DS, and Cap-DE proteins, mice were intramuscular administered twice at 21-day interval with each protein which was previously formulated with ISA201 adjuvant (Fig. 3A). Serum IgG antibodies against PCV2 Cap protein, PRRSV GP5B, and GP35 antigen peptides were measured using ELISA at 21 and 35 dpi. As shown in Fig. 3B, specific antibodies against the Cap protein were detectable in the immunized groups at 21 dpi, and these levels significantly increased following booster immunization. Notably, the modifications did not compromise the immunogenicity of the Cap protein. Additionally, after booster immunization, elevated levels of antibodies specific to GP35 (Fig. 3C) and GP5B (Fig. 3D) in the immunized groups were observed. These findings demonstrated that the inclusion of GP5B and GP35 epitopes in the Cap protein effectively elicited robust humoral immune responses against PRRSV, while maintaining the protein’s immunogenicity against PCV2.
Fig. 3.
Humoral immune responses induced by capsid-derived vaccines in mice. A Schematic of the immunization and challenge procedures in animal studies, created using BioRender (https://biorender.com). B–D Serum IgG titers against Cap (B), GP35 (C), and GP5B (D) peptides were measured by ELISA at 21 and 35 dpi. E–F Neutralizing antibody titers in sera collected at 21 and 35 dpi were tested against PCV2 (E) and PRRSV (F) using an end-point dilution reduction assay. Data are presented as mean ± SEM for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant
We further evaluated the serum neutralizing antibodies induced by the recombinant Cap-DP, Cap-DS, and Cap-DE vaccines against PCV2 and PRRSV. As illustrated in Fig. 3E, no neutralizing antibodies were detected in the sera of mice from the PBS group after the primary immunization. In contrast, mice immunized with Cap-WT, Cap-DP, Cap-DS, or Cap-DE vaccines exhibited low levels of neutralizing antibodies against PCV2, with titers ranging from 2 to 4. Following booster immunization, there was a significant increase in serum neutralizing antibody levels across all vaccination groups, except for the PBS group, aligning with the ELISA results. Notably, neutralizing antibody titers against PCV2 were comparable among the Cap-DP, Cap-DS, and Cap-DE groups.
For PRRSV, neutralizing antibodies remained undetectable in the sera of mice receiving PBS or Cap-WT protein after two immunizations. However, mice immunized with Cap-DP, Cap-DS, or Cap-DE developed detectable neutralizing antibodies against PRRSV following the second immunization, with titers ranging from 5 to 11 (Fig. 3F). The Cap-DS group exhibited significantly higher neutralizing antibody titers compared to the Cap-DP and Cap-DE groups (Fig. 3F). Additionally, the serum neutralizing antibody titers in the Cap-DP group were significantly higher than those in the Cap-DE group (Fig. 3F). These results support the ELISA findings, indicating that the Cap-DS vaccine elicited robust neutralizing antibody responses against both PCV2 and PRRSV.
Induction of spleen lymphocyte immune responses by capsid-derived vaccines
To evaluate the efficacy of recombinant capsid-derived vaccines in enhancing cellular immunity, we conducted a comprehensive assessment of their impacts on lymphocyte proliferation. Using the CCK-8 assay, we found that after stimulation with the capsid-derived mixed proteins (equimolar mixtures of purified Cap-WT, Cap-DP, Cap-DS, and Cap-DE proteins), the SI of splenic lymphocytes in the capsid-derived vaccine group was significantly higher than that in the PBS control group (Fig. 4A). This finding confirms that the nanoparticle proteins can effectively activate the mouse immune system. Subsequently, we used flow cytometry to evaluate the cytokine secretion of each specific cell type (Fig. S4). The results showed that in the capsid-derived vaccine group, the proportions of CD4⁺ cells secreting interleukin-4 (IL-4) and interferon-γ (IFN-γ) increased significantly (Fig. 4B and C). The simultaneous increase in the levels of IL-4 and IFN-γ secreted by CD4⁺ cells indicates that immunization with the capsid-derived vaccine can induce a balanced Th1 and Th2 immune response. Moreover, we observed that the nanoparticle capsid-derived vaccine significantly enhanced the activation of antigen-specific multifunctional CD8⁺ T cells, as evidenced by a notable increase in the proportion of CD8⁺ T cells that released IFN -γ (Fig. 4D). This result indicates that the capsid-derived vaccine can effectively induce a robust cytotoxic T lymphocyte (CTL) immune response in mice.
Fig. 4.
Cellular immune responses in splenic lymphocytes of mice immunized with capsid-derived vaccines. A Lymphocyte proliferation in response to different immunogens. Splenocytes isolated from immunized mice were stimulated in vitro with equimolar mixtures of purified proteins (Cap-WT, Cap-DP, Cap-DS, and Cap-DE), and the SI was assessed using the CCK-8 assay. B–H Proportions of IFN-γ+CD4+ (B), IL-4+CD4+ (C), IFN-γ+CD8+ (D), Central m.CD4+ (E), Central m.CD8+ (F), Effector m.CD4+ (G), and Effector m.CD8+ (H) T cells in the spleens of mice immunized with PBS or different immunogens. Data are presented as mean ± SEM for each group. *, P < 0.05; ns, not significant
In addition, we evaluated the safety of the vaccine by measuring the levels of IL-6 and TNF-α in the serum of experimental mice at 35 dpi. IL-6 and TNF-α are key cytokines in the body’s inflammatory response, and a significant increase in their levels typically indicates a strong inflammatory reaction [58]. The experimental results showed that there was no significant difference in the levels of IL-6 and TNF-α in the serum of experimental mice compared with healthy mice without treatment (Fig. S5). This finding indicates that although these vaccines can induce a strong cellular immune response, they do not provoke a strong inflammatory reaction in the body, highlighting the advantage of this vaccine in terms of safety.
Finally, we used flow cytometry to evaluate the ability of the capsid-derived vaccine to elicit CD4⁺ and CD8⁺ memory T cells in the spleen, which is crucial for the host’s antiviral response. Although the capsid-derived vaccine did not significantly increase the proportion of CD4⁺ central memory T cells (CD44highCD62Lhigh, Tcm) (Fig. 4E), it did markedly increase the proportion of CD8⁺ central memory T cells (Fig. 4F) and the proportion of CD4⁺ and CD8⁺ effector memory T cells (CD44highCD62Llow, Tem) (Fig. 4G and 4H). This indicates that the capsid-derived nanoparticles may induce a strong immune memory response.
Capsid-derived vaccines could provide effective immunization protect against PCV2 challenge in mice
To evaluate whether the capsid-derived vaccines can provide effective immune-protection against PCV2 infection, mice were challenged with PCV2 at 35 dpi. The viral loads in sera, spleens, lungs and kidneys were quantified by qPCR, with the standard curve for absolute quantification shown in Fig. S6. At 14 dpc, the PBS group exhibited high levels of viral loads in sera, spleens, lungs and kidneys. In contrast, the capsid-derived vaccine groups demonstrated a reduction in viral load, with PCV2 levels in sera, spleens, lungs and kidneys being reduced to approximately 1/100 to 1/10000 of those observed in the PBS group (Fig. 5A–D). Notably, some mice in the capsid-derived vaccines groups showed undetectable levels of PCV2. Consistently, the results of IHC analysis (Fig. 5E–G) also demonstrated that the content of PCV2 in the spleen, kidney, and lung tissues of mice treated with PBS was significantly higher than that in the tissues of mice immunized with the capsid-derived vaccines. Immunofluorescence technique further corroborated these findings. As depicted in Fig. S7, the spleens from the PBS group displayed marked lymphocyte apoptosis. In contrast, spleens from the mice immunized with the capsid-derived vaccines exhibited less tissue lymphocyte apoptosis. Among the three modified capsid-derived vaccines tested, Cap-DS demonstrated the most effective protection, aligning with previous immunogenicity results. Collectively, these data indicate that the recombinant Cap vaccines provide effective protection against PCV2 infection in mice.
Fig. 5.
Protective efficacy of capsid-derived vaccines against PCV2 challenge in mice. Serum, spleen, lung, and kidney samples from mice immunized with various immunogens were collected at 14 dpc. Viral loads in sera (A), spleens (B), lungs (C), and kidneys (D) were quantified by qPCR. Some samples were below the detection threshold of qPCR; “n” in “n/5” denotes the number of samples with positive qPCR results. E–G Tissue samples from the spleens (E), lungs (F), and kidneys (G) were analyzed by IHC for PCV2 presence (bar = 50 μm)
Enhanced immunogenicity and protection from PRRSV in piglets after Cap-DS vaccine administration
The preliminary studies in mice revealed that the Cap-DS vaccine exhibited superior immunogenicity relative to Cap-DP and Cap-DE. Given the constraints on infecting mice with PRRSV, the Cap-DS vaccine was chosen for a comprehensive evaluation of its protective efficacy in piglets. Piglets were intramuscularly administered thrice with 200 μg of Cap-DS, 106.5 TCID50 PRRSV WUH3 inactivated vaccine and PBS at two-week intervals (Fig. 6A). Serum neutralizing antibodies were detectable in all piglets at 28 dpi, with a significant escalation in antibody titers observed by 38 dpi (Fig. 6B). Upon challenge with 2 × 105.0 TCID50 of PRRSV via intramuscular injection, the mean rectal temperature of PBS-treated piglets was significantly higher than that of both Cap-DS and WUH3 groups from 5 to 14 dpc, while the WUH3 group showed elevated temperatures compared with the Cap-DS group between 6 and 11 dpc (Fig. 6C). The viral loads in sera were quantified using qPCR, with the standard curve for absolute quantification presented in Fig. S8. Notably, the serum viral loads in Cap-DS-immunized and WUH3 inactivated vaccine-immunized piglets were substantially diminished in comparison to those in PBS-immunized piglets from 3 to 10 dpc (Fig. 6D). Meanwhile, we measured the PRRSV content in the lung tissues of the surviving pigs at 14 dpc. The results revealed that the viral RNA copies in the tissues were almost all at a relatively low level, ranging from 104 to 105 copies/g (Fig. 6E). In alignment with these findings, the survival rate of Cap-DS-immunized and WUH3 inactivated vaccines-immunized piglets was 80% and 60%, respectively, contrasted with the PBS group, which achieved only a 40% survival rate by 14 dpc (Fig. 6F). The macroscopic examination of nine surviving piglets’ lung morphology following exposure to the highly pathogenic PRRSV is depicted in Fig. S9. Within the PBS-immunized group, the two surviving piglets exhibited consolidated lungs with extensive bilateral hemorrhages, characteristic of severe PRRSV infection. In stark contrast, the four surviving piglets in the Cap-DS-immunized group and three surviving piglets in the WUH3 inactivated vaccines showed only minimal congestion and negligible hemorrhaging, maintaining relatively intact lung morphology. Consistently, histopathological analysis of lung tissues unveiled severe congestion and pronounced interstitial hyperplastic pneumonia in the PBS group, indicative of PRRSV infection. Conversely, the lung tissues of piglets immunized with Cap-DS and WUH3 inactivated vaccines exhibited minimal bleeding and infection (Fig. 6G). IHC analysis corroborated the presence of robust PRRSV signals in the lungs of the two surviving PBS-immunized piglets, while the lungs of the Cap-DS-immunized and WUH3 inactivated piglets displayed negligible PRRSV signals (Fig. S10). Collectively, these results substantiate that the Cap-DS vaccine provides robust and effective protection against PRRSV infection in piglets.
Fig. 6.
Protective efficacy of the Cap-DS vaccine against PRRSV challenge in piglets. A Schematic diagram of the immunization and challenge experimental procedures for piglets. Illustrations were created using BioRender (https://biorender.com). B Neutralizing antibody titers in sera collected at 14, 28, and 38 dpi against PRRSV were assessed using an end-point dilution reduction assay. C Mean rectal temperatures of immunized piglets from 1 to 14 dpc following PRRSV (WUH3 strain) challenge. Red “ + ” symbol indicates deceased piglets. D Viral loads in serum samples collected at 1, 3, 5, 7, 10, and 14 dpc were quantified by qPCR. E Viral loads in lung samples collected at 14 dpc were quantified by qPCR. F Survival curve of piglets within 14 dpc. G Histopathological examination of lung sections following PRRSV challenge (HE staining; bar = 200 μm)
Discussion
PRRSV and PCV2 predominantly target porcine alveolar macrophages (PAMs). Although PCV2 infection alone inflicts minimal damage to PAMs, it significantly compromises the innate immune response, thereby exacerbating PRRSV-induced pathologies. This synergistic impairment presents a formidable challenge in managing coinfected pigs [64, 65]. Traditional inactivated PRRSV vaccines offer only partial protection, while live-attenuated PRRSV vaccines carry risks such as viral dissemination and potential reversion to virulence [66–69]. Furthermore, the high cost and stringent dosage requirements of these vaccines limit their practicality, particularly in cost-sensitive farming environments. In this study, we developed a novel subunit vaccine, Cap-DS, by utilizing the PCV2 Cap protein as a carrier to express the neutralizing epitopes GP5B, GP5IV, and GP3I of PRRSV. This strategy not only addresses the limitations of existing vaccines but also offers a more economical and secure alternative for the porcine industry.
Our study elucidates the rational design of a bivalent vaccine based on three key principles: the selection of regions that minimally disrupt the Cap protein’s tertiary structure, ensuring that the inserted epitopes are prominently displayed on the surface of VLPs, and meticulously avoiding any perturbations to established antigenic sites within the Cap protein. A comprehensive series of in vitro and in vivo experiments consistently supports the effectiveness of this design strategy. Results from TEM and DLS confirm that the engineered capsid-derived proteins have successfully folded into their native tertiary structures. In mouse models, both Cap-DS and Cap-WT elicited comparable levels of antibodies against the Cap protein, providing effective protection against PCV2 infection, which indicates that the insertion of foreign fragments does not compromise the immunogenicity of the Cap protein. Additionally, Cap-DS induced high titers of neutralizing antibodies against PRRSV in both mice and piglets, confirming the effective surface presentation of the foreign epitopes. Given the numerous novel neutralizing epitopes that have been identified for PRRSV, future research could explore these insertion sites for the substitution of other PRRSV epitopes, thereby facilitating the development of PRRSV vaccines.
PCV2 capsid-derived proteins, expressed as soluble proteins through baculovirus expression systems, self-assemble into nanoparticles with diameters ranging from 15 to 30 nm. This nanoscale assembly enhances the delivery efficiency and stability of vaccines, which are crucial for their efficacy [70]. Our study reveals that all three PCV2 capsid-derived protein vaccines elicit effective neutralizing antibody responses. However, compared to Cap-DS, Cap-DP and Cap-DE exhibit diminished immunogenicity. This observation highlights the potential advantage of incorporating a diverse spectrum of neutralizing epitopes on the surface of VLPs to induce immunogenicity. Repetitive insertion of identical epitope types may disrupt conformation, resulting in suboptimal or counterproductive outcomes. Hence, we advocate for future PCV2 modifications to focus on the strategic integration of multiple distinct exogenous epitopes, rather than reiterative insertion of the same epitope type, to maximize the vaccine’s immunogenic potential.
In this study, we also introduced a novel epitope presentation strategy by incorporating a DCBp upstream of the neutralizing epitopes [60, 71]. This modification could enhance antigen recognition by dendritic cells (DCs) and promotes the activation of CD4+ T cells via MHC-II presentation, thereby improving the overall immunogenic profile of the vaccine [72, 73]. In fact, the capsid-derived vaccines effectively stimulated both humoral and cellular immunity, as evidenced by the significant proliferation of splenic lymphocytes in the vaccine groups compared to the PBS group, as shown by the CCK-8 assay. Moreover, the capsid-derived vaccines induced a balanced CD4+ T cell immune response. At 35 dpi, there were notable increases in the proportion of CD4+ T cells secreting Th1 (IFN-γ) and Th2 (IL-4) cytokines. The capsid-derived vaccines also significantly enhanced the activation of antigen-specific multifunctional CD8+ T cells. This was manifested as a remarkable increase in the proportion of CD8+ T cells releasing IFN-γ. Additionally, at 35 dpi, there was no significant difference in the levels of serum inflammatory factors IL-6 and TNF-α between all the vaccinated mice and normal mice, indicating robust yet controlled immune activation. This dual-stimulatory profile suggests that the capsid-derived vaccines can trigger strong immune responses without causing excessive inflammation, marking them as well-balanced immunogens. Flow cytometry further confirmed the enhanced activation of T cell immune memory. The capsid-derived vaccine groups exhibited higher proportions of CD4+ and CD8+ effector memory T cells, supporting their potential as effective and safe vaccine candidates.
In summary, our study presents an innovative subunit vaccine design targeting both PRRSV and PCV2, characterized by the simultaneous incorporation of GP5B and GP35 epitopes into the PCV2 Cap protein. Immunization with Cap-DS in murine and porcine models provided effective protection against viral infections, underscoring its potential as a promising strategy for the prevention and control of PRRSV and PCV2 in clinical settings. This approach not only addresses the limitations of traditional vaccines but also provides a more efficient and cost-effective solution, offering significant benefits for swine health management.
Supplementary Information
Acknowledgements
We would like to thank National Key Laboratory of Agricultural Microbiology Core Facility for assistance during the experiments. This work is supported by the National Key Research and Development Program of China (2022YFD1800801).
Author contribution
SX and LF designed and supervised the research; JM, XX, YZ, WH, JS and XC performed research; JM, XX, YZ, WH and JS analyzed data; JM, XX, SX and LF wrote the manuscript. All authors reviewed the manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Competing interest
The authors declare that there are no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jun Ma and Xun Xiao contributed equally to this work.
References
- 1.Nathues H, Alarcon P, Rushton J, Jolie R, Fiebig K, Jimenez M, et al. Modelling the economic efficiency of using different strategies to control porcine reproductive & respiratory syndrome at herd level. Prev Vet Med. 2018;152:89–102. [DOI] [PubMed] [Google Scholar]
- 2.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. Javma-J Am Vet Med A. 2005;227(3):385–92. [DOI] [PubMed] [Google Scholar]
- 3.Renken C, Nathues C, Swam H, Fiebig K, Weiss C, Eddicks M, et al. Application of an economic calculator to determine the cost of porcine reproductive and respiratory syndrome at farm-level in 21 pig herds in Germany. Porcine Health Manag. 2021;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valdes-Donoso P, Alvarez J, Jarvis LS, Morrison RB, Perez AM. Production losses from an endemic animal disease: porcine reproductive and respiratory syndrome (PRRS) in selected Midwest US sow farms. Front Vet Sci. 2018;5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilalta C, Arruda AG, Tousignant SJP, Valdes-Donoso P, Muellner P, Muellner U, et al. A Review of quantitative tools used to assess the epidemiology of porcine reproductive and respiratory syndrome in U.S. swine farms using Dr. Morrison’s swine health monitoring program data. Front Vet Sci. 2017;4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruedas-Torres I, Sanchez-Carvajal JM, Salguero FJ, Pallares FJ, Carrasco L, Mateu E, et al. The scene of lung pathology during PRRSV-1 infection. Front Vet Sci. 2024;11:1330990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992;4(2):117–26. [DOI] [PubMed] [Google Scholar]
- 8.Bai YZ, Sun Y, Liu YG, Zhang HL, An TQ, Wang Q, et al. Minor envelope proteins from GP2a to GP4 contribute to the spread pattern and yield of type 2 PRRSV in MARC-145 cells. Front Cell Infect Microbiol. 2024;14:1376725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao F, Jiang YF, Li GX, Zhou YJ, Yu LX, Li LW, et al. Porcine reproductive and respiratory syndrome virus expressing E2 of classical swine fever virus protects pigs from a lethal challenge of highly-pathogenic PRRSV and CSFV. Vaccine. 2018;36(23):3269–77. [DOI] [PubMed] [Google Scholar]
- 10.Dokland T. The structural biology of PRRSV. Virus Res. 2010;154(1–2):86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mardassi H, Gonin P, Gagnon CA, Massie B, Dea S. A subset of porcine reproductive and respiratory syndrome virus GP glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J Virol. 1998;72(8):6298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q, Yu X, He D, Ku X, Hong B, Zeng W, et al. Investigation and analysis of etiology associated with porcine respiratory disease complex in China from 2017 to 2021. Front Vet Sci. 2022;9: 960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Zhu H, Zhan C, Chen P, Wu B, Peng Z, et al. Establishment and application of a quadruplex real-time reverse-transcription polymerase chain reaction assay for differentiation of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, porcine circovirus type 3, and streptococcus suis. Microorganisms. 2024;12(3):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Annunzio G, Ostanello F, Muscatello LV, Orioles M, Jacumin N, Tommasini N, et al. Porcine circovirus type 2 and porcine reproductive and respiratory syndrome virus alone or associated are frequent intralesional detected viruses in porcine respiratory disease complex cases in Northern Italy. Front Vet Sci. 2023;10:1234779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drolet R, Larochelle R, Morin M, Delisle B, Magar R. Detection rates of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, and swine influenza virus in porcine proliferative and necrotizing pneumonia. Vet Pathol. 2003;40(2):143–8. [DOI] [PubMed] [Google Scholar]
- 16.Fan PH, Wei YW, Guo LJ, Wu HL, Huang LP, Liu JB, et al. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol J. 2013;10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, et al. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. 2001;38(5):528–39. [DOI] [PubMed] [Google Scholar]
- 18.Kong C, Li D, Hu Y, Gao P, Zhang Y, Zhou L, et al. The genetic variation of porcine reproductive and respiratory syndrome virus replicase protein nsp2 modulates viral virulence and persistence. J Virol. 2023;97(3): e0168922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pileri E, Mateu E. Review on the transmission porcine reproductive and respiratory syndrome virus between pigs and farms and impact on vaccination. Vet Res. 2016;47(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu H, Sun M, Wang N, Zhang S, Deng Z, Xu H, et al. Efficacy comparison in cap VLPs of PCV2 and PCV3 as swine vaccine vehicle. Int J Biol Macromol. 2024;278(Pt 3): 134955. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Xu F, Yuan C, Zhang Y, Ren J, Yue H, et al. Comparison of immune effects of porcine circovirus type 2d (PCV2d) capsid protein expressed by Escherichia coli and baculovirus-insect cells. Vaccine. 2024;42(11):2848–57. [DOI] [PubMed] [Google Scholar]
- 22.Gao YY, Wang Q, Li HW, Zhang S, Zhao J, Bao D, et al. Genomic composition and pathomechanisms of porcine circoviruses: a review. Virulence. 2024;15(1):2439524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park C, Seo HW, Park SJ, Han K, Chae C. Comparison of porcine circovirus type 2 (PCV2)-associated lesions produced by co-infection between two genotypes of PCV2 and two genotypes of porcine reproductive and respiratory syndrome virus. J Gen Virol. 2014;95(Pt 11):2486–94. [DOI] [PubMed] [Google Scholar]
- 24.Morales-Hernandez S, Ugidos-Damboriena N, Lopez-Sagaseta J. Self-assembling protein nanoparticles in the design of vaccines: 2022 update. Vaccines (Basel). 2022;10(9):1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziqi W, Kai C, Costabel U, Xiaoju Z. Nanotechnology-facilitated vaccine development during the coronavirus disease 2019 (COVID-19) pandemic. Exploration (Beijing). 2022;2(5):20210082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon JJ, Suh H, Bershteyn A, Stephan MT, Liu H, Huang B, et al. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat Mater. 2011;10(3):243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Sagaseta J, Malito E, Rappuoli R, Bottomley MJ. Self-assembling protein nanoparticles in the design of vaccines. Comput Struct Biotechnol J. 2016;14:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchooli A, Aghaiypour K, Kiasari BA, Samarbaf-Zadeh A, Ghadiri A, Makvandi M. VLP production from recombinant L1/L2 HPV-16 protein expressed in Pichia pastoris. Protein Pept Lett. 2018;25(8):783–90. [DOI] [PubMed] [Google Scholar]
- 29.Yadav R, Zhai L, Tumban E. Virus-like particle-based L2 vaccines against HPVs: where are we today? Viruses. 2019;12(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roldao A, Mellado MCM, Castilho LR, Carrondo MJT, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines. 2010;9(10):1149–76. [DOI] [PubMed] [Google Scholar]
- 31.Hemmati F, Hemmati-Dinarvand M, Karimzade M, Rutkowska D, Eskandari MH, Khanizadeh S, et al. Plant-derived VLP: a worthy platform to produce vaccine against SARS-CoV-2. Biotechnol Lett. 2022;44(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li RQ, Chang ZJ, Liu HL, Wang YN, Li MH, Chen YL, et al. Double-layered N-S1 protein nanoparticle immunization elicits robust cellular immune and broad antibody responses against SARS-CoV-2. J Nanobiotechnol. 2024;22(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung BK, Kim HR, Lee YH, Jang H, Chang KS. Comparison of immune responses to the PCV2 replicase-capsid and capsid virus-like particle vaccines in mice. J Microbiol Biotechnol. 2019;29(3):482–8. [DOI] [PubMed] [Google Scholar]
- 34.Lin HX, Ma Z, Hou X, Chen L, Fan HJ. Construction and immunogenicity of a recombinant swinepox virus expressing a multi-epitope peptide for porcine reproductive and respiratory syndrome virus (vol 7, pg 43990, 2017). Sci Rep-Uk. 2017;7:46592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Xiao S, Fang L, Yu X, Song Y, Niu C, et al. DNA vaccines co-expressing GP5 and M proteins of porcine reproductive and respiratory syndrome virus (PRRSV) display enhanced immunogenicity. Vaccine. 2006;24(15):2869–79. [DOI] [PubMed] [Google Scholar]
- 36.Ma H, Li X, Li J, Zhao Z, Zhang H, Hao G, et al. Immunization with a recombinant fusion of porcine reproductive and respiratory syndrome virus modified GP5 and ferritin elicits enhanced protective immunity in pigs. Virology. 2021;552:112–20. [DOI] [PubMed] [Google Scholar]
- 37.Ren JQ, Sun WC, Lu HJ, Wen SB, Jing J, Yan FL, et al. Construction and immunogenicity of a DNA vaccine coexpressing GP3 and GP5 of genotype-I porcine reproductive and respiratory syndrome virus. BMC Vet Res. 2014;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu XG, Wang ZS, Zhang Q, Li ZC, Ding L, Li W, et al. Baculovirus as a PRRSV and PCV2 bivalent vaccine vector: Baculovirus virions displaying simultaneously GP5 glycoprotein of PRRSV and capsid protein of PCV2. J Virol Methods. 2012;179(2):359–66. [DOI] [PubMed] [Google Scholar]
- 39.Zhu S, Guo X, Keyes LR, Yang HC, Ge XN. Recombinant encephalomyocarditis viruses elicit neutralizing antibodies against PRRSV and CSFV in mice. PLoS ONE. 2015;10(6): e0129729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piñeyro PE, Kenney SP, Giménez-Lirola LG, Heffron CL, Matzinger SR, Opriessnig T, et al. Expression of antigenic epitopes of porcine reproductive and respiratory syndrome virus (PRRSV) in a modified live-attenuated porcine circovirus type 2 (PCV2) vaccine virus (PCV1-2a) as a potential bivalent vaccine against both PCV2 and PRRSV. Virus Res. 2015;210:154–64. [DOI] [PubMed] [Google Scholar]
- 41.Hu GW, Wang ND, Yu WT, Wang ZF, Zou YW, Zhang Y, et al. Generation and immunogenicity of porcine circovirus type 2 chimeric virus-like particles displaying porcine reproductive and respiratory syndrome virus GP5 epitope B. Vaccine. 2016;34(16):1896–903. [DOI] [PubMed] [Google Scholar]
- 42.Li GP, Liu L, Xu BJ, Hu JX, Kuang HY, Wang X, et al. Displaying epitope B and epitope 7 of porcine reproductive and respiratory syndrome virus on virus like particles of porcine circovirus type 2 provides partial protection to pigs. J Vet Med Sci. 2021;83(8):1263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrowski M, Galeota JA, Jar AM, Platt KB, Osorio FA, Lopez OJ. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol. 2002;76(13):6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang SJ, Bae SM, Lee HJ, Jeong YJ, Lee MA, You SH, et al. Porcine circovirus (PCV) genotype 2d-based virus-like particles (VLPs) Induced broad cross-neutralizing antibodies against diverse genotypes and provided protection in dual-challenge infection of a PCV2d virus and a type 1 porcine reproductive and respiratory syndrome virus (PRRSV). Pathogens. 2021;10(9):1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamazares E, Gutiérrez F, Hidalgo A, Gutiérrez NA, Espinoza FI, Sánchez O, et al. A heterologous viral protein scaffold for chimeric antigen design: an example PCV2 virus vaccine candidate. Viruses-Basel. 2020;12(4):385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Meng XP, Wang SN, Li ZQ, Yang L, Tu LQ, et al. Virus-like particles of recombinant PCV2b carrying FMDV-VP1 epitopes induce both anti-PCV and anti-FMDV antibody responses. Appl Microbiol Biot. 2018;102(24):10541–50. [DOI] [PubMed] [Google Scholar]
- 47.Zhang HW, Qian P, Liu LF, Qian SH, Chen HC, Li XM. Virus-like particles of chimeric recombinant porcine circovirus type 2 as antigen vehicle carrying foreign epitopes. Viruses-Basel. 2014;6(12):4839–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Xiao SB, Wang YW, Xu SS, Jiang YB, Chen HC, et al. Immunogenicity of the highly pathogenic porcine reproductive and respiratory syndrome virus GP5 protein encoded by a synthetic ORF5 gene. Vaccine. 2009;27(13):1957–63. [DOI] [PubMed] [Google Scholar]
- 49.Li DG, Wang J, Xu SG, Cai SX, Ao CJ, Fang LR, et al. Identification and functional analysis of the novel ORF6 protein of porcine circovirus type 2. Vet Res Commun. 2018;42(1):1–10. [DOI] [PubMed] [Google Scholar]
- 50.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, et al. The 2.3-angstrom structure of porcine circovirus 2. J Virol. 2011;85(21):11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue CY, Wang W, Liu QL, Miao ZW, Liu K, Shen HF, et al. Chimeric influenza-virus-like particles containing the porcine reproductive and respiratory syndrome virus GP5 protein and the influenza virus HA and M1 proteins. Arch Virol. 2014;159(11):3043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fort M, Olvera A, Sibila M, Segales J, Mateu E. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs. Vet Microbiol. 2007;125(3–4):244–55. [DOI] [PubMed] [Google Scholar]
- 57.Sun YY, Gao YN, Su TJ, Zhang LJ, Zhou HR, Zhang J, et al. Nanoparticle vaccine triggers interferon-gamma production and confers protective immunity against porcine reproductive and respiratory syndrome virus. ACS Nano. 2025;19(1):852–70. [DOI] [PubMed] [Google Scholar]
- 58.Chen YL, Zhu JH, Wang SQ, Li MH, Sun XK, Liu SY, et al. Modular nano-antigen display platform for pigs induces potent immune responses. ACS Nano. 2024;18(42):29152–77. [DOI] [PubMed] [Google Scholar]
- 59.Yang D, Su MJ, Guo DH, Zhao FY, Wang MJ, Liu JY, et al. Combination of S1-N-terminal and S1-C-terminal domain antigens targeting double receptor-binding domains bolsters protective immunity of a nanoparticle vaccine against porcine epidemic Diarrhea virus. ACS Nano. 2024;18(19):12235–60. [DOI] [PubMed] [Google Scholar]
- 60.Curiel TJ, Morris C, Brumlik M, Landry SJ, Finstad K, Nelson A, et al. Peptides identified through phage display direct immunogenic antigen to dendritic cells. J Immunol. 2004;172(12):7425–31. [DOI] [PubMed] [Google Scholar]
- 61.Trible BR, Kerrigan M, Crossland N, Potter M, Faaberg K, Hesse R, et al. Antibody recognition of porcine circovirus type 2 capsid protein epitopes after vaccination, infection, and disease. Clin Vaccine Immunol. 2011;18(5):749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung BK, Kim HR, Jang H, Chang KS. Replacing the decoy epitope of PCV2 capsid protein with epitopes of GP3 and/or GP5 of PRRSV enhances the immunogenicity of bivalent vaccines in mice. J Virol Methods. 2020;284: 113928. [DOI] [PubMed] [Google Scholar]
- 63.Yu C, Li X, Liu JW, Diao WZ, Zhang LC, Xiao Y, et al. Replacing the decoy epitope of PCV2b capsid protein with a protective epitope enhances efficacy of PCV2b vaccine. Vaccine. 2016;34(50):6358–66. [DOI] [PubMed] [Google Scholar]
- 64.Zhou L, Kang RM, Yu JF, Xie B, Chen CY, Li XY, et al. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-like, Jxa1-like, and Mlv-like strains. Viruses-Basel. 2018;10(10):551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Jiao D, Jing Y, He Y, Han W, Li Z, et al. Genetic characterization and pathogenicity of a novel recombinant PRRSV from lineage 1, 8 and 3 in China failed to infect MARC-145 cells. Microb Pathog. 2022;165: 105469. [DOI] [PubMed] [Google Scholar]
- 66.Binjawadagi B, Dwivedi V, Manickam C, Ouyang K, Wu Y, Lee LJ, et al. Adjuvanted poly(lactic-co-glycolic) acid nanoparticle-entrapped inactivated porcine reproductive and respiratory syndrome virus vaccine elicits cross-protective immune response in pigs. Int J Nanomedicine. 2014;9:679–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Charerntantanakul W. Porcine reproductive and respiratory syndrome virus vaccines: immunogenicity, efficacy and safety aspects. World J Virol. 2012;1(1):23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H, Kim HK, Jung JH, Choi YJ, Kim J, Um CG, et al. The assessment of efficacy of porcine reproductive respiratory syndrome virus inactivated vaccine based on the viral quantity and inactivation methods. Virol J. 2011;8:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roca M, Gimeno M, Bruguera S, Segalés J, Díaz I, Galindo-Cardiel IJ, et al. Effects of challenge with a virulent genotype II strain of porcine reproductive and respiratory syndrome virus on piglets vaccinated with an attenuated genotype I strain vaccine. Vet J. 2012;193(1):92–6. [DOI] [PubMed] [Google Scholar]
- 70.Song JX, Wang MX, Zhou L, Tian PP, Sun ZY, Sun JR, et al. A candidate nanoparticle vaccine comprised of multiple epitopes of the African swine fever virus elicits a robust immune response. J Nanobiotechnol. 2023;21(1):424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang YC, Zhou M, Li YY, Luo ZC, Chen HC, Cui M, et al. Recombinant rabies virus with the glycoprotein fused with a DC-binding peptide is an efficacious rabies vaccine. Oncotarget. 2018;9(1):831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moser M, Murphy KM. Dendritic cell regulation of TH1-TH2 development. Nat Immunol. 2000;1(3):199–205. [DOI] [PubMed] [Google Scholar]
- 73.Quaratino S, Duddy LP, Londei M. Fully competent dendritic cells as inducers of T cell anergy in autoimmunity. Arthritis Res Ther. 2000;97(20):10911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.