Abstract
The proper assembly of light-harvesting complexes (LHCs) is critical for photosynthesis and requires the biogenesis of light-harvesting chlorophyll a,b-binding proteins (LHCPs) to be coordinated with chlorophyll (Chl) biosynthesis. The mechanism underlying this coordination is not well understood. Here, we show that a conserved molecular chaperone, chloroplast signal recognition particle 43-kDa protein (cpSRP43), provides a molecular thermostat that helps maintain this coordination. cpSRP43 undergoes a conformational rearrangement between a well-folded closed state and a partially disordered open state. Closed cpSRP43 is dedicated to the biogenesis of LHCPs, whereas open cpSRP43 protects multiple Chl biosynthesis enzymes from heat-induced destabilization. Rising temperature shifts cpSRP43 to the open state, enabling it to protect heat-destabilized Chl biosynthesis enzymes. Our results reveal the molecular basis of a posttranslational mechanism for the thermoadaptation of LHC biogenesis. They also demonstrate how an adenosine triphosphate–independent chaperone uses conformational dynamics to switch its activity and client selectivity, thereby adapting to different proteostatic demands under shifting environmental conditions.
A thermo-switchable molecular chaperone helps coordinate light-harvesting complex assembly during photosynthesis.
INTRODUCTION
The capture and conversion of sunlight through photosynthesis provide >99% of the energy used by life on Earth. To optimize the efficiency of this process, photosynthetic organisms evolved antenna-like complexes that funnel photons into photosynthetic reaction centers. In photosynthetic algae and land plants, the antenna for photosystem II is formed by the light-harvesting complexes (LHCs), which comprise the light-harvesting chlorophyll a,b-binding protein (LHCP) family of proteins bound to chlorophyll (Chl) and other photosynthetic pigments (1). LHCPs are integral membrane proteins containing three transmembrane domains and comprise the most abundant family of membrane proteins on Earth (2). These hydrophobic proteins are nuclear encoded, synthesized in the cytosol, and highly prone to misfolding and aggregation in the aqueous environments of the cytosol and stroma during their transport to the thylakoid membrane. Thus, the de novo biogenesis of LHCPs requires effective chaperones and transport machinery that maintain them in a translocation-competent state and mediate their targeted delivery to the thylakoid membrane. The proper folding of LHCPs in the thylakoid membrane further requires the binding of Chls, which are supplied by the tetrapyrrole biosynthesis (TBS) pathway (3–5). Reciprocally, the buildup of free Chl or its biosynthetic precursors in the absence of protein binding leads to excess reactive oxygen species (ROS) that can be cytotoxic (6). Therefore, proper LHC assembly requires the supply of Chl to be precisely coordinated with the de novo biogenesis of LHCPs. The mechanism behind this coordination is not well understood.
The chloroplast signal recognition particle 43-kDa protein (cpSRP43) is a conserved molecular chaperone that coevolved with the LHCs. It participates in both branches of LHC biogenesis: the delivery of LHCPs and the biosynthesis of Chl (7). cpSRP43 is a subunit of the cpSRP responsible for mediating sequence-specific, high-affinity recognition of newly imported LHCPs, effectively protecting them from misfolding and aggregation in the chloroplast stroma (8, 9). This function is carried out by the substrate binding domain (SBD) comprising four ankyrin repeat motifs (ARMs) capped by an N-terminal chromodomain (CD1) (8–10). The other cpSRP subunit, cpSRP54, binds the second chromodomain (CD2) of cpSRP43 via a conserved C-terminal motif (54C) and allosterically enhances cpSRP43’s chaperone activity toward LHCPs (11–15). cpSRP further mediates protein-protein contacts at the thylakoid membrane, via the interaction of cpSRP54 with the chloroplast signal recognition particle receptor cpFtsY and the binding of the third chromodomain (CD3) of cpSRP43 to the Alb3 insertase, forming a dedicated pathway for the membrane transport and insertion of the LHCPs (16–22). Loss of cpSRP43, cpSRP54, or cpFtsY results in growth retardation, weak pigmentation, and reduced LHCP levels in Arabidopsis thaliana, indicating the essential role of the cpSRP pathway in LHC biogenesis (23–25).
In addition to its canonical role in LHCP delivery, more recent work found that cpSRP43 also acts as a chaperone to prevent the heat-induced aggregation of multiple enzymes in the TBS biosynthetic pathway (7, 26). These biochemically detected chaperone activities were corroborated in studies of A. thaliana chaos mutants lacking cpSRP43, which show reduced levels of at least three TBS enzymes including glutamyl tRNA reductase (GluTR), the rate-limiting enzyme in TBS biosynthesis, and GUN4 and CHLH, components of the Mg-chelatase complex (26). This phenotype is more pronounced during heat stress, when all three enzymes were substantially destabilized, and the levels of Chl and multiple Chl precursors were reduced in chaos plants compared to wild-type (WT) plants (26). The participation of cpSRP43 in both LHCP biogenesis and Chl biosynthesis suggests that it could provide a posttranslational mechanism of regulation to balance the levels of LHCPs and Chl during LHC biogenesis (7).
However, the molecular basis underlying the potential regulatory role of cpSRP43 remains elusive. It is unclear how a small, adenosine triphosphate (ATP)–independent chaperone such as cpSRP43 recognizes and protects two distinct types of client proteins, newly synthesized LHCPs and mature TBS enzymes, that differ in size, composition, structure, and folding state. Notably, cpSRP54 enhances the chaperone activity of cpSRP43 toward the LHCPs but represses its activity toward the TBS enzymes (10, 26). This observation indicates that only the apo form of cpSRP43 is capable of TBS protection and suggests that cpSRP43 uses distinct molecular mechanisms to recognize the two different classes of clients. The mechanism of this activity switch is unclear; neither is it clear how cpSRP43 selects between its two classes of client proteins to balance the supply of LHCP and Chl during LHC assembly.
Early crystal structures showed cpSRP43 in a structured closed conformation, in which the helices in all four ARMs are well folded and tightly packed against one another (9). However, nuclear magnetic resonance (NMR) studies detected a second open conformation of cpSRP43 in equilibrium with the closed state (10, 27). In the open state, the two C-terminal ARMs in the cpSRP43 SBD unravel and become partially disordered (10, 27). While we previously proposed and provided evidence that the open state of cpSRP43 allows this chaperone to turn “off” its activity at the thylakoid membrane during LHCP transport (10, 27), the question remains about why a seemingly “inactive” open conformation evolved in cpSRP43.
In this work, we define the role of the different conformational states of cpSRP43 using rational mutations and experimental conditions that shift the conformational equilibrium in this chaperone. Biochemical and biophysical studies showed that closed cpSRP43 specifically binds and protects LHCPs from aggregation, whereas open cpSRP43 is solely responsible for the thermoprotection of TBS enzymes. The closed-to-open transition is strongly temperature dependent and, together with the release of cpSRP54, unleashes open cpSRP43 to stabilize TBS enzymes at elevated temperatures. We propose a model in which the conformational switch of cpSRP43 provides a molecular thermostat that enables photosynthetic organisms to rapidly respond to rising temperature and maintain coordinated LHC biogenesis.
RESULTS
cpSRP43 undergoes a heat-induced conformational change
We previously used 19F-NMR to monitor the conformational dynamics of cpSRP43 (27). The spectra of the cpSRP43 site specifically labeled with an 19F probe, 3-bromo-1,1,1-trifluoroacetone (BTFA), showed two NMR peaks indicating two conformations in slow exchange, which were assigned to a partially disordered open state and a rigidly folded closed state (27). Notably, this conformational change is strongly temperature dependent. The closed state dominated at temperatures below 30°C. The open state became populated between 30° and 40°C and dominated above 40°C (Fig. 1, A and B, red). The heat-induced opening of cpSRP43 could also be detected by circular dichroism (CD), which measures the secondary structure content of proteins (28). The CD spectra of WT cpSRP43 changed substantially between 25° and 55°C, with a loss in molar ellipticity (θ) at 222 nm at 55°C (Fig. 1C), consistent with the partial unfolding of α helices in the C-terminal ARMs of cpSRP43 in the open state (27). This conformational change is reversible, with >80% of helical content restored at 25°C after three heating cycles (Fig. 1C). The temperature dependence of θ222 agreed well with that from 19F-NMR measurements and yielded a mid-transition temperature (Tm) of 35°C (Fig. 1, B and D, red, and Table 1). These results show that elevated temperature induces the closed-to-open transition of cpSRP43.
Fig. 1. Identification of mutations that stabilize the open or closed state of cpSRP43.
(A) 19F-NMR spectra of BTFA-labeled WT cpSRP43 at the indicated temperatures. The open (red “O”) and closed (green “C”) state peaks were assigned in (27). Spectra were collected in CD buffer with 50 mM NaCl. ppm, parts per million. (B) The fraction of cpSRP43 variants in the open state at different temperatures determined by 19F-NMR. Data were from the spectra shown in (A) and fig. S1. “*” denotes the data collected after the completion of one heating cycle, where 10 to 20% of cpSRP43 did not return to the closed state. (C) CD spectra of WT cpSRP43 at 25°C (solid) and 55°C (dashed). Data were collected in CD buffer with 50 mM NaCl and repeated for three heat cycles (dark to light red). Values represent mean residue ellipticity ± SE from n = 3 technical replicates. (D) Temperature dependence of helical content for R189L, WT, DE3NQ, and DE6NQ cpSRP43, measured as per-residue molar ellipticity (θ) at 222 nm by CD in CD buffer with 50 mM NaCl. Representative data from three technical replicates are shown. Values represent mean residue ellipticity ± SD. The lines are fits of the data to Eq. 2, and the obtained Tm values from n = 3 technical replicates are summarized in Table 1. (E and F) Closed state structure of the SBD of A. thaliana cpSRP43 [Protein Data Bank (PDB): 3DEP (9)] showing the hydrogen bonding interactions of R189 (E) or the seven closely spaced acidic residues (F). Sequence conservation of these acidic residues across 1000 land plants is shown in WebLogo [(F), top right] (51). The charge-neutralizing mutations are indicated [(F), bottom right].
Table 1. Summary of the Tm values of the closed-to-open conformational change for indicated cpSRP43 variants under different conditions.
Values are presented as mean fitted values ± SE from n = 3 technical replicates.
| [Urea] (M) | FL or SBD | [NaCl] (mM) | R189L | WT | DE3NQ | DE6NQ |
|---|---|---|---|---|---|---|
| 0 | FL | 0 | <25* | 28.2 ± 0.41 | 39.1 ± 0.45 | 41.3 ± 0.76 |
| 50 | <25* | 34.6 ± 0.41 | 42.0 ± 0.62 | 42.0 ± 0.57 | ||
| 100 | <25* | 37.2 ± 0.30 | 43.4 ± 0.07 | 42.9 ± 0.49 | ||
| 200 | 25.7 ± 0.58 | 39.9 ± 0.23 | 44.8 ± 0.21 | 43.4 ± 0.26 | ||
| FL + L18† | 200 | 38.4 ± 0.90 | ||||
| FL + 54C† | 200 | 35.2 ± 0.45 | ||||
| SBD | 200 | 29.9 ± 0.62 | 42.0 ± 0.19 | 47.2 ± 0.39 | ||
| 0.5 | FL | 200 | <25* | 37.5 ± 0.38 | 42.7 ± 0.24 | |
| 1 | 200 | <25* | 34.0 ± 0.46 | 40.1 ± 0.50 |
*Limits shown when the Tm is below the lowest temperature tested (25°C) and not measurable.
†Includes excess L18 or 54C peptide.
To rigorously define the function of cpSRP43 in its different conformations, we sought conservative mutations that would bias the chaperone toward either conformational state. To favor the open state, we introduced an R189L mutation that disrupts two hydrogen bonds in the closed state structure of cpSRP43 (Fig. 1E) (9). 19F-NMR and CD measurements showed that the open state is much more populated in R189L compared to WT cpSRP43 (figs. S1 and S2). The estimated Tm for opening is 10° to 15°C for R189L, more than 20°C lower than that for WT cpSRP43 (Fig. 1, B and D, blue). Thus, most of R189L is in the open state at room temperature (25°C) under physiological salt (50 mM NaCl) conditions. The 19F-NMR chemical shifts of open and closed R189L are the same as those of WT cpSRP43 (fig. S1). In addition, under conditions that stabilize the closed state, including low temperature (17°C), higher salt (see fig. S6 later), and the presence of the FDPLGL-containing L18 peptide derived from the cpSRP43 recognition motif in LHCP, the 1H,15N–heteronuclear single-quantum coherence (HSQC) spectra of WT cpSRP43 and R189L overlaid well (fig. S3). Only residues near the FDPLGL binding site or the site of mutation showed large chemical shift perturbations. These observations strongly suggest that the R189L mutation shifts the conformational equilibrium of cpSRP43 toward the open state, without affecting the closed state structure.
To drive cpSRP43 toward the closed state, we noted that the SBD has an acidic surface with seven evolutionarily conserved Asp and Glu residues in close proximity (Fig. 1F), generating electrostatic repulsion that would destabilize compact folding in the SBD. Charge-neutralizing mutations at these residues in variants DE3NQ (D154/187N and E221Q) and DE6NQ (D154/185/187/219N and E221/223Q) are predicted to stabilize the closed state by 3.4 and 3.9 kcal, respectively, at physiological salt concentration [Eqs. 3A and 3B in Materials and Methods (29) and fig. S4, A and B]. Consistent with these predictions, the Tm value for the closed-to-open transition was shifted to 42°C for DE3NQ and DE6NQ, ~7°C higher than that for WT cpSRP43 (Fig. 1, B and D; Table 1; and figs. S1 and S2, green and purple). The CD spectra and 19F-NMR chemical shifts of these mutants were the same as WT cpSRP43 at temperatures far below and above their respective Tm values (figs. S1 and S2). Except for residues near the mutated sites, most peaks in the 15N-HSQC spectra of DE3NQ match the assignments for closed cpSRP43 (fig. S3) (10), providing additional evidence that the mutations do not affect the closed state structure. Thus, the charge-neutralizing mutations shift the conformational equilibrium of cpSRP43 toward the closed state.
To independently verify the effects of the mutations on cpSRP43 conformation, we took advantage of the fact that the partially disordered open state is favored by low doses of chemical denaturants (27). CD and 19F-NMR measurements showed that, compared to WT and R189L cpSRP43, the opening of DE3NQ is more resistant to urea (Table 1 and fig. S5). This provides further evidence for the strong stabilization of the closed state by charge-neutralizing mutations on the acidic surface of cpSRP43.
Last, we explored the effect of salt to further tune the conformation of cpSRP43. Charge screening at higher ionic strength reduces the electrostatic repulsion at the acidic surface of cpSRP43 and is predicted to stabilize the closed state by ~2.4 kcal/mol at 200 mM NaCl relative to 0 mM NaCl (fig. S4C). CD measurements showed that this was indeed the case: The closed state was greatly stabilized at higher salt concentrations in WT cpSRP43 and R189L (Table 1 and fig. S6). In contrast, the salt dependence was much smaller with DE3NQ and negligible in DE6NQ (fig. S6), as predicted (fig. S4C). Thus, the open-to-closed rearrangement can also be tuned by salt concentration in addition to temperature.
Together, the results in this section show that the open-to-closed transition of cpSRP43 can be regulated by diverse environmental factors including temperature, ionic strength, and low doses of denaturants. We successfully isolated rational mutations that modulate this rearrangement, with R189L favoring the open state and DE3NQ and DE6NQ hyperstabilizing the closed state, without altering the structure of either conformation.
Closed cpSRP43 chaperones LHCP
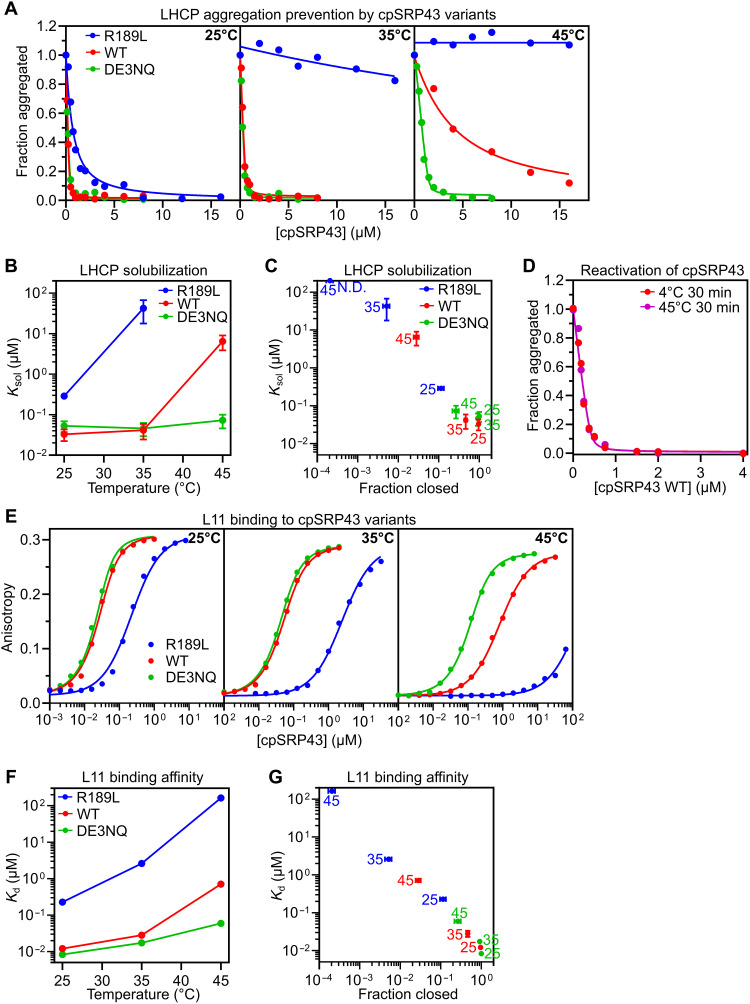
Using the mutations and experimental conditions that drive cpSRP43 toward different conformations, we aimed to define the activity of each conformational state. We first tested the chaperone activity toward its canonical substrate, newly synthesized LHCPs, using an established assay that detects large aggregates based on turbidity at 360 nm (8). Dilution of urea-denatured LHCP into buffer led to rapid and extensive aggregation, whereas dilution into buffer containing WT cpSRP43 resulted in quantitative, dose-dependent reductions in LHCP aggregation (Fig. 2A). The chaperone concentration dependence of LHCP solubilization yields solubilization constants (Ksol), which represent apparent dissociation constants (Kd) of the chaperone for LHCP as it competes with the aggregation of the latter. R189L was less effective at chaperoning LHCP compared to WT and DE3NQ at 25°C, with a >10-fold increase in Ksol (Fig. 2, A and B, and Table 2). This defect was more pronounced at 35° and 45°C, at which little to no solubilization of LHCP was observed even at 16 μM R189L. In contrast, WT cpSRP43 and DE3NQ remained effective at solubilizing LHCP at 35°C. The high chaperone activity of DE3NQ persisted even at 45°C, at which WT cpSRP43 displayed weaker chaperone activity, consistent with the higher stability of the closed state in DE3NQ than WT cpSRP43 (Fig. 2, A and B, and Table 2). The activity of the cpSRP43 variants toward LHCP across different temperatures correlated with the fraction of chaperone in the closed state from CD measurements (Fig. 2C). These data provide definitive evidence that closed cpSRP43 mediates the chaperoning of LHCP.
Fig. 2. Closed cpSRP43 mediates chaperone activity toward the LHCPs.
(A) Turbidity assays to measure the protection of LHCP by increasing concentrations of cpSRP43 variants at 25°C (left), 35°C (middle), and 45°C (right) in CD buffer with 50 mM NaCl. Optical density at 360 nm (A360) was normalized to that without cpSRP43. Representative data from three technical replicates are shown. Lines are fits of the data to Eq. 4A. (B and C) Obtained Ksol values for LHCP plotted as a function of temperature (B) or fraction of chaperone in the closed state (C). Values represent mean fitted values ± SE of n = 3 technical replicates. (D) Turbidity assays of reactivated WT cpSRP43 heated at 45°C for 30 min and then returned to ice (violet) or unheated (red) carried out toward LHCP at 25°C as in (A). A360 was normalized to that without cpSRP43. Representative data from three technical replicates are shown. Lines are fits of the data to Eq. 4A. (E) Equilibrium titrations to measure the binding of the indicated cpSRP43 variants to HiLyte Fluor 488–labeled L11 peptide at 25°C (left), 35°C (middle), and 45°C (right) in CD buffer with 50 mM NaCl. Representative data from three technical replicates are shown. Lines are fits of the data to Eq. 6A. (F and G) Obtained dissociation constant (Kd) values for L11 peptide binding were plotted as a function of temperature (F) or fraction of chaperone in the closed state (G). Values represent mean fitted values ± SE of n = 3 technical replicates. The numbers next to the data points in (C) and (G) indicate the temperatures at which the measurement was made. The fraction of chaperone in the closed state was from the measurements in Fig. 1D.
Table 2. Summary of the interaction of cpSRP43 variants with LHCP, the L11 peptide, and 54C peptide.
All values were measured at 50 mM NaCl and are presented as mean fitted values ± SE from n = 3 technical replicates.
| Ksol values (μM) for LHCP | ||||
|---|---|---|---|---|
| T (°C) | R189L | WT | DE3NQ | |
| 45 | N.D.* | 6.5 ± 3 | 0.073 ± 0.03 | |
| 35 | 42 ± 25 | 0.042 ± 0.02 | 0.046 ± 0.02 | |
| 25 | 0.29 ± 0.03 | 0.033 ± 0.01 | 0.053 ± 0.02 | |
| T (°C) | WT (unheated) | WT (reactivated) | ||
| 25 | 0.020 ± 0.003 | 0.018 ± 0.005 | ||
| Kd values (μM) for L11 binding to cpSRP43 variants | ||||
| T (°C) | R189L | WT | DE3NQ | |
| 45 | 160 ± 16 | 0.71 ± 0.03 | 0.059 ± 0.006 | |
| 35 | 2.6 ± 0.1 | 0.028 ± 0.005 | 0.017 ± 0.002 | |
| 25 | 0.23 ± 0.01 | 0.012 ± 0.001 | 0.0083 ± 0.0008 | |
| Kd values (μM) for 54C binding to cpSRP43 variants | ||||
| T (°C) | R189L | WT | DE3NQ | |
| 45 | 6.7 ± 0.5 | 5.0 ± 0.1 | 3.3 ± 0.2 | |
| 35 | 1.2 ± 0.1 | 0.23 ± 0.02 | 0.38 ± 0.03 | |
| 25 | 0.16 ± 0.03 | 0.036 ± 0.003 | 0.083 ± 0.02 | |
*Not detectable.
Recognition and protection of LHCP by cpSRP43 require a sequence-specific interaction with the conserved FDPLGL motif in a stromal loop of LHCPs (9–11, 30, 31). To test whether high-affinity recognition of this motif also occurs exclusively in closed cpSRP43, we measured the binding affinity of the cpSRP43 variants for an L11 peptide containing the FDPLGL motif based on cpSRP43-induced anisotropy change of HiLyte Fluor 488–labeled L11 (Fig. 2E). The effects of cpSRP43 mutations and temperature on L11 binding affinity mirrored those on LHCP solubilization by this chaperone (Fig. 2, E to G, and Table 2). The equilibrium Kd for L11 binding across the chaperone variants and temperatures also strongly correlated with the fraction of chaperone in the closed state (Fig. 2G). Thus, the closed state of cpSRP43 specifically recognizes the FDPLGL motif in LHCP.
We further asked whether the chaperone activity of cpSRP43 toward LHCP is reversible following a return from heat-induced opening. To this end, we subjected WT cpSRP43 to a 30-min heat treatment at 45°C, well above its Tm at 35°C. Following its return to 25°C, cpSRP43 remained highly active in mediating LHCP solubilization following the heat treatment, with Ksol values indistinguishable from those of WT cpSRP43 without the heat treatment (Fig. 2D and Table 2).
Collectively, these results show that the closed state of cpSRP43 is responsible for the specific recognition of LHCPs and for protecting this family of membrane proteins from aggregation, whereas the open state is not. This activity is diminished upon heat-induced transition of cpSRP43 to the open conformation but is fully reversible after the heat stress.
Open cpSRP43 mediates thermoprotection of TBS enzymes
We next determined how the chaperone activity of cpSRP43 toward its second class of clients, the TBS enzymes, depends on its conformational state. Mature, folded GUN4, a model TBS enzyme substrate for cpSRP43 (26), aggregates rapidly upon heat treatment as monitored by increased turbidity at 360 nm (fig. S7). The aggregation time courses of GUN4 fit well to first-order kinetics, and the observed rate constant of aggregation was independent of GUN4 concentration (fig. S7, A and B, and table S1). As protein misfolding/unfolding is concentration independent whereas aggregation is strongly concentration dependent, these results indicate that GUN4 undergoes rate-limiting misfolding, followed by rapid aggregation under heat stress. The misfolding and aggregation of GUN4 is accelerated by rising temperature, plateauing at a rate constant of 0.12 min−1, or a half-time of 5.6 min, above 42°C (fig. S7, C and D, and table S1).
To assess the ability of cpSRP43 variants to prevent heat-induced GUN4 aggregation, we chose a condition at the midpoint of GUN4 aggregation kinetics (5 min at 42°C). Under these conditions, the open state population varied substantially across the cpSRP43 mutants: 99% for R189L, 67% for WT, and 28% for DE3NQ (Fig. 1D). The cpSRP43-mediated thermoprotection of GUN4 was strongest for mutant R189L, which is primarily in the open state under these conditions, and lowest for mutant DE3NQ, which is hyperstabilized in the closed state (Fig. 3A and Table 3). The same trend was observed for the thermoprotection of a second TBS enzyme, GluTR, by the cpSRP43 variants (Fig. 3B and Table 3).
Fig. 3. Open cpSRP43 mediates the thermoprotection of TBS enzymes.
(A) Turbidity assays to measure the heat-induced aggregation of 10 μΜ GUN4 and its protection by the indicated cpSRP43 variants in the low-salt (LS) buffer with 50 (left), 100 (middle), and 200 (right) mM NaCl. A360 was normalized to that without cpSRP43. Representative data from three technical replicates are shown. The lines are fits of the data to Eq. 4B. (B) Same as (A), except with 2.7 μM GluTR as the client protein. (C and D) The fitted Ksol values from the data in (A) (see Materials and Methods) are plotted as a function of NaCl concentration (C) and fraction of chaperone in the closed state (D). The numbers next to the data indicate the NaCl concentration at which the measurements were made. Values represent mean fitted values ± SE of n = 3 technical replicates.
Table 3. Summary of the chaperone activities of cpSRP43 variants toward TBS enzymes.
| Ksol values (μM) for GUN4* | ||||
|---|---|---|---|---|
| NaCl (mM) | FL or SBD | R189L | WT | DE3NQ |
| 50 | FL | 0.28 ± 0.1 | 0.89 ± 0.3 | 3.3 ± 0.09 |
| 50 | SBD | 3.4 ± 1.0 | 11.6 ± 2.5 | N.D.† |
| 100 | FL | 0.37 ± 0.03 | 1.2 ± 0.07 | 12.6 ± 0.3 |
| 200 | FL | 0.31 ± 0.1 | 2.8 ± 0.08 | 37.0 ± 2.6 |
| Ksol values (μM) for GluTR* | ||||
| NaCl (mM) | FL or SBD | R189L | WT | DE3NQ |
| 200 | FL | 0.18 ± 0.1 | 0.47 ± 0.1 | 1.4 ± 0.4 |
| 200 | SBD | 1.9 ± 0.9 | 12.4 ± 2.2 | N.D.† |
*Values were measured at 42°C and are shown as mean fitted values ± SE from n = 3 technical replicates.
†Not detectable.
To further tune the conformational equilibrium of cpSRP43 at constant temperature, we increased ionic strength, which reduces the open state population (fig. S6). WT cpSRP43 and DE3NQ showed reduced thermoprotection of GUN4 at higher ionic strength, with a 5- to 10-fold rise in Ksol values at 200 mM NaCl compared to 50 mM NaCl (Fig. 3, A and C, and Table 3). This is consistent with the diminished open state population at higher salt in these chaperone variants (fig. S6). In contrast, R189L, which predominantly populates the open state at 42°C even at 200 mM NaCl, retained effective GUN4 protection (Fig. 3, A and C, blue, and Table 3). We note that heat-induced aggregation of GUN4 is slower at higher ionic strength (fig. S7, E and F); hence, the reduced protection of GUN4 by WT cpSRP43 and DE3NQ under these conditions cannot be attributed to more aggressive GUN4 aggregation. The Ksol values for cpSRP43 variants at different salt concentrations correlated with the population of the chaperone in the open state observed by CD (Fig. 3D). Thus, while the open state of cpSRP43 lacks activity toward the LHCPs, it is responsible for protecting mature TBS enzymes from heat-induced misfolding and aggregation. In contrast, the closed state does not recognize and protect this second class of client proteins.
cpSRP54 and C-terminal chromodomains directly regulate TBS thermoprotection
We previously found that binding of the other subunit of cpSRP, cpSRP54, weakens with rising temperature [Fig. 4, A and B, red; (26)], which correlated with its release from cpSRP43 upon heat stress in A. thaliana. However, cpSRP54 also stabilizes the closed state of cpSRP43 (Table 1 and fig. S8) (10), the population of which diminishes at elevated temperatures. This raises the question: Is the heat-induced release of cpSRP54 from cpSRP43 due to the intrinsic temperature sensitivity of their interaction or to the opening of cpSRP43 at high temperature? To distinguish between these models, we leveraged the set of cpSRP43 variants that open at different temperatures. cpSRP54 binding was measured on the basis of changes in the fluorescence anisotropy of a fluorescein-labeled 54C peptide bearing the cpSRP43-binding motif of cpSRP54 (13). WT cpSRP43 and DE3NQ bound 54C with two- to fivefold higher affinities than R189L, consistent with the higher population of open state in R189L and the preference of cpSRP54 for the closed state. However, the interaction of all three variants with 54C displayed a strong temperature dependence that was largely independent of their conformational equilibrium (Fig. 4, A and B, and Table 2). For example, DE3NQ is predominantly closed between 25° and 35°C, over which its 54C binding weakened fourfold. Similarly, R189L is exclusively open between 35° and 45°C, over which its 54C binding weakened fivefold (Fig. 4, A and B, and Table 2). These results show that the intrinsic temperature sensitivity of the cpSRP43-cpSRP54 interaction is primarily responsible for the disassembly of cpSRP at high temperature, whereas the conformational state of the cpSRP43 SBD contributes modestly.
Fig. 4. cpSRP54 and C-terminal chromodomains modulate TBS thermoprotection by open cpSRP43.
(A) Equilibrium titrations to measure the binding affinity of the 54C peptide for cpSRP43 variants at rising temperatures. Reactions were carried out in CD buffer with 50 mM NaCl. Representative data from three technical replicates are shown. The lines are fits of the data to Eqs. 6A and 6B. (B) The obtained mean Kd values ± SE from n = 3 technical replicates are plotted as a function of temperature. (C) Temperature dependence of the helical content for R189L, WT, and DE3NQ cpSRP43 (SBD), measured as per-residue molar ellipticity (θ) at 222 nm by CD in CD buffer with 200 mM NaCl and fit as in Fig. 1D. Representative data from three technical replicates are shown. Values represent mean residue ellipticity ± SD of the signal over 10 s. (D and E) Comparison of the Ksol values between FL and SBD constructs in the thermoprotection of GUN4 (D) and GluTR (E) for cpSRP43 variants. Ksol values shown are from fits of the data in Fig. 3 (A and B) and fig. S9 (B and C) to Eq. 4B for n = 3 technical replicates.
While the cpSRP43 SBD is necessary and sufficient to chaperone the LHCPs (10), at least one of the C-terminal chromodomains is further required for cpSRP43 to effectively protect TBS enzymes (26). However, the presence of CD2 also biases cpSRP43 toward the open state (10). To distinguish whether the requirement for CD2 is due to its effect on SBD conformation or to a direct interaction with TBS proteins, we examined the impact of deleting CD2 and CD3. For R189L, both the full-length (FL) chaperone and the SBD are predominantly open at 42°C (Fig. 4C, Table 1, and fig. S9A). If CD2CD3 exerts its effect by enhancing the open state population, then its deletion should have no impact on the activity of R189L. In contrast to this prediction, R189L (SBD) showed >10-fold reduced chaperone activity toward GUN4 (Fig. 4D, Table 3, and fig. S9B, blue) and GluTR (Fig. 4E, Table 3, and fig. S9C, blue) compared to FL-R189L, indicating a direct involvement of CD2 and/or CD3 in the thermoprotection of TBS enzymes. In addition, the effect of CD2CD3 deletion on the chaperone activity toward TBS enzymes was consistent across the cpSRP43 variants irrespective of their conformational equilibria (Fig. 4, D and E; Table 3; and fig. S9, B and C). Therefore, the C-terminal chromodomains of cpSRP43 directly participate in the recognition and protection of TBS enzymes under heat stress.
Open cpSRP43 protects TBS enzymes during heat stress in plants
In previous work, we showed that cpSRP43 is necessary for the maintenance of both LHCP and TBS levels in plants, especially during heat stress (7, 26). To assess how the conformational state of cpSRP43 affects its TBS protection activity in planta, we introduced the set of cpSRP43 mutants that lies at different points of the open-to-closed conformational equilibrium into the chaos line of Arabidopsis plants lacking genomically expressed cpSRP43 (32). We constructed transgenic plants in which the expression of WT, R189L (RL), DE3NQ (DE3), or DE6NQ (DE6) cpSRP43 is driven by the endogenous cpSRP43 promoter. Genotyping identified two individual lines expressing each variant with WT-like steady-state levels of cpSRP43 for further investigation.
In contrast to the slow growth and pale pigmentation phenotype of chaos plants, the growth and pigmentation of all four transgenic lines expressing cpSRP43 variants were comparable to the WT plant Landsberg-0 (Ler-0; Fig. 5A and fig. S10), indicating that all cpSRP43 variants rescued the defects in chloroplast development upon cpSRP43 loss. In addition, the steady-state levels of Chl and Chl precursors, such as Mg-porphyrins and protochlorophyllide, were restored to WT-like levels in the transgenic lines expressing cpSRP43 variants (Fig. 5, B to D). The steady-state levels of cpSRP43 clients, LHCa1 and GUN4, and the cpSRP43-binding partner cpSRP54 were also comparable across the cpSRP43 transgenic lines and similar to those in Ler-0 at room temperature (Fig. 5E). Therefore, under normal growth conditions, neither the RL nor DE3/6NQ mutations affect the growth of plant seedlings, Chl synthesis, and steady-state levels of cpSRP43 clients. These results are consistent with biochemical data, as the conformational defect of R189L can be effectively rescued by the binding of LHCP and cpSRP54 [Table 1 and fig. S8; (10)], and DE3NQ and DE6NQ are highly effective in chaperoning newly synthesized LHCPs (Fig. 2).
Fig. 5. Characterization of transgenic cpSRP43 Arabidopsis lines.
(A) Representative images of 18-day-old seedlings of WT (Ler-0), chaos, and transgenic lines expressing the indicated cpSRP43 variants using the endogenous cpSRP43 promoter. Seedlings were grown under normal conditions. Scale bars, 0.5 cm. (B to D) Steady-state levels of Chl (B) and Chl precursors including Mg-porphyrins (C) and protochlorophyllide [PChlide; (D)] in 18-day-old seedlings shown in (A). MgP, Mg-protoporphyrin IX; MgPMME, MgP monomethylester; FW, fresh weight. (E) Steady-state levels of cpSRP43, cpSRP54, and GUN4 in 18-day-old seedlings shown in (A) were analyzed by immunoblotting using the indicated antibodies. The Ponceau S–stained large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RbcL) and actin is shown as loading control.
We next tested the effect of the cpSRP43 conformational variants on the stability of model TBS proteins under short-term heat stress conditions using these transgenic lines. We conducted 0.5- to 1-hour heat treatments (42°C) on seedlings of the transgenic lines expressing WT and cpSRP43 variants. The translation elongation inhibitor, cycloheximide (CHX), was added before heat treatment to block new protein synthesis and thus measure the half-life of only the existing pool of proteins. The level of LHCa1 remained the same in the WT and transgenic lines during the heat treatment, indicating that LHCPs are stable during short-term heat stress (Fig. 6 and fig. S11). In contrast, the level of two model TBS enzymes, GUN4 and CHLH, decreased quickly over the course of heat treatment in transgenic plants expressing WT cpSRP43 (Fig. 6 and fig. S11). This indicates that these enzymes are thermolabile and subject to rapid degradation during heat stress, in agreement with biochemical data (fig. S7). Compared to the WT transgenic line, GUN4 was substantially stabilized in the cpSRP43 RL line during the heat treatment, whereas it was similarly destabilized in the DE3 and DE6 transgenic plants (Fig. 6 and fig. S11). The same effect of cpSRP43 mutations was observed with CHLH (Fig. 6 and fig. S11). The stabilization of TBS enzymes by mutant cpSRP43 R189L, which promotes the open conformation, provides in vivo evidence that the open state of cpSRP43 is more effective at protecting TBS clients from misfolding and degradation during short-term heat stress.
Fig. 6. Open cpSRP43 stabilizes the levels of TBS enzymes in planta during short-term heat stress.
(A) Steady-state levels of the indicated proteins in 10-day-old transgenic seedlings before and after 0.5 and 1 hour of heat treatment at 42°C in the presence of CHX were analyzed by immunoblotting using the indicated antibodies. The Ponceau S–stained RbcL and actin are shown as loading controls. (B and C) Semiquantitative analysis with the ImageJ software of the immunoblots in (A). The relative amounts of GUN4 (B) and CHLH (C) were normalized to their levels before exposure to elevated temperature (0 hours at 42°C), with actin serving as the loading control for quantification. An independent biological replicate is shown in fig. S11.
DISCUSSION
The biogenesis of LHCs is a rate-limiting step in biomass production from solar energy by photosynthetic organisms and presents multiple challenges to the cellular proteostasis network. A particular challenge is the requirement for precise coordination between the biogenesis of LHCPs, mediated by the cpSRP pathway, and the supply of Chl molecules, synthesized by the TBS pathway. Inadequate balance between the two branches leads to the accumulation of aggregated LHCPs or toxic ROS, generating proteostatic or phototoxic stress (6). This balance must be further maintained under different environmental conditions, such as varying light intensity and temperature during the day-night cycle and across seasons. Our previous work uncovered chaperone activities of cpSRP43 toward client proteins in both the LHCP biogenesis and Chl synthesis pathways, but how it coordinates the two pathways was unclear. This work elucidates the molecular basis of this coordination and posits cpSRP43 as a molecular thermostat, potentially helping photosynthetic organisms balance the supply of Chl with LHCP biogenesis at elevated temperatures.
Our previous biophysical studies demonstrated the presence of two conformations of cpSRP43 at equilibrium, a structured closed state and a partially disordered open state. Here, we found that this conformational change provides the fundamental mechanism that explains both the dual chaperone activity of cpSRP43 and its regulation by environmental factors. Using rational mutations and conditions that tune this conformational equilibrium, we show that hyperstabilizing cpSRP43 in the closed state enhanced chaperone activity toward LHCP but abolished its activity toward two TBS enzymes (Figs. 2 and 3). The opposite was observed with chaperone variants that populate the open conformation (Figs. 2 and 3). The results of biochemical studies were buttressed by in vivo data showing that R189L, in which the open state is more populated, more effectively stabilized TBS proteins compared to WT or closed-state–stabilized cpSRP43 variants in plants under heat stress (Fig. 6). These results provide definitive evidence that each conformational state in cpSRP43 is used to handle a distinct class of client proteins. Closed cpSRP43 is dedicated to the chaperoning and membrane transport of newly synthesized and imported LHCPs, whereas open cpSRP43 protects mature TBS enzymes from heat stress.
The conformational change in cpSRP43 is exquisitely sensitive to temperature. Free cpSRP43 undergoes the closed-to-open transition over a narrow temperature range, 30° to 40°C, under low-salt conditions. In vivo, this conformational change likely occurs at a higher temperature range due to the stabilization of closed cpSRP43 by LHCP and cpSRP54 (Table 1 and fig. S8). The observation that mutant R189L better stabilized TBS enzymes than WT cpSRP43 in plants subjected to a 42°C treatment (Fig. 6) suggests that the open conformation is only partially populated by WT cpSRP43 under these conditions in vivo. Notably, this temperature-dependent transition of cpSRP43 correlates with the acceleration in the misfolding and aggregation of TBS proteins with rising temperature, which plateaus at a rate of ~0.1 min−1 for GUN4 above 40°C (fig. S7). Heat-induced destabilization of TBS enzymes was supported by in planta data, which showed that GUN4 and CHLH are degraded within an hour when WT plants were subjected to heat stress at 42°C (Fig. 6). This contrasts with the stability of LHCP during heat stress (Fig. 6), suggesting that the demand for TBS protection outweighs that for de novo LHCP generation under these conditions. Thus, the temperature-dependent conformational switch enables cpSRP43 to rapidly sense and respond to these altered demands, providing short-term thermoprotection for TBS enzymes precisely when this activity is needed.
In addition to the conformational change in the cpSRP43 SBD, the C-terminal chromodomains of this chaperone and its binding partner cpSRP54 further modulate chaperone activity toward the TBS enzymes. While the C-terminal chromodomains were previously shown to favor the opening of cpSRP43, the results here provide evidence that they directly contribute to the binding and protection of TBS enzymes. On the other hand, cpSRP54 favors the closed conformation of cpSRP43, inhibits its TBS protection activity, and is released from this chaperone at elevated temperatures. We find that the strong temperature dependence of the cpSRP54-cpSRP43 interaction occurs largely independently of its effect on the open-to-closed transition in the SBD and likely provides a second molecular mechanism to sense rising temperature. Together with the opening of the cpSRP43 SBD, the release of cpSRP54 enforces the switch in the client preference of cpSRP43 from the LHCPs to the TBS enzymes at elevated temperatures.
We propose a model for how the dual chaperone functions of cpSRP43, enabled by its conformational switch, help coordinate Chl synthesis with LHCP biogenesis for plants experiencing elevated temperature (Fig. 7). At normal temperature (<35°C for A. thaliana), cpSRP43 is predominantly in the closed state and tightly bound to cpSRP54, providing a dedicated pathway for the protected transport and insertion of newly imported LHCPs at the thylakoid membrane. Under these conditions, TBS enzymes are stably folded and mediate the biosynthesis of Chl that is integral to LHCP folding and assembly. At higher temperatures, TBS enzymes are increasingly misfolded, aggregated, and susceptible to proteolytic degradation. Under these conditions, an increasing population of cpSRP43 is released from cpSRP54 and switches to the open conformation, which stabilizes TBS enzymes. This allows plants to rapidly respond to heat stress and maintain the balance of Chl synthesis with LHCP biogenesis.
Fig. 7. Model for cpSRP43’s dual chaperone activities.
Under normal conditions, cpSRP43 primarily populates the closed conformation in which its ARMs in the SBD are tightly folded and further stabilized by interactions with cpSRP54 (left). Closed cpSRP43 confers high-affinity recognition and protection of newly imported LHCP but has no activity toward TBS enzymes. At elevated temperatures, a fraction of cpSRP43 transitions to the open conformation in which the C-terminal ARMs become partially disordered (right). Open cpSRP43 does not recognize LHCP but effectively protects mature TBS enzymes from misfolding, aggregation, and resulting proteolytic degradation. cpSRP54, which inhibits the TBS protection activity of cpSRP43, also dissociates from the chaperone at elevated temperatures. These changes unleash the thermoprotection activity of cpSRP43 under conditions where this activity is needed.
We speculate that this mechanism is most relevant during peak sunlight hours in the early afternoon, especially in summertime, during which leaf temperature can rise to 35°C and above (33). The transition of a fraction of cpSRP43 to the open conformation under these conditions provides plants short-term thermoprotection during this modest daily heat stress. Further, the expression of TBS enzymes is transcriptionally up-regulated after sunrise (34); the increased abundance of these enzymes during daylight could shift a fraction of cpSRP43 to the open state, which recognizes the TBS clients, via the law of mass action. These hypotheses remain to be tested in vivo. In addition, the physiological role of this mechanism for photosynthetic organisms experiencing long-term or extreme heat stress conditions remains unclear. Last, our biochemical data suggest that other factors, such as salt concentration and low doses of denaturants, extensively modulate the conformational transition of cpSRP43; whether this conformational change could also respond to additional environmental stresses, such as salinity, remains to be tested.
Our results here provide a paradigm in which a molecular chaperone completely alters its activity and client specificity in response to a change in environmental conditions. While temperature (35, 36), pH (37–40), and oxidative stress (41, 42) have all been reported to induce conformational changes in various small, ATP-independent chaperones, in most cases, these chaperones switch between an inactive “storage” mode and an active client-binding mode in response to environmental stress. A notable exception is DegP in bacteria (43) and its eukaryotic homolog HtrA2 in the mitochondrial intermembrane space (44, 45), which acts as a chaperone at low temperature and a protease for the same clients at high temperature. cpSRP43 is unique in that a single chaperone harbors two distinct conformational states that are each active toward different clients. By regulating the population of its two conformations over a broad range of physiological temperatures, cpSRP43 posttranslationally balances the levels of its clients to meet the changing needs for coordinated LHC assembly during light capture. Given that its change in activity arises from an order-to-disorder conformational transition, cpSRP43 also provides an excellent model to understand the mechanisms by which structured versus disordered chaperones specialize in the recognition and protection of distinct types of client proteins.
MATERIALS AND METHODS
Protein expression and purification
Mutations of cpSRP43 were constructed using the QuikChange mutagenesis procedure (Stratagene) according to the manufacturer’s instructions. WT and mutant cpSRP43, LHCP, GUN4, and GluTR were overexpressed and purified as previously described (7, 26, 46).
Circular dichroism
Spectra were collected on an Aviv model 410 CD spectrophotometer in 1-mm-thick quartz cuvettes containing 300 μl of 10 μM cpSRP43 in CD buffer [20 mM Na2HPO4 (pH 7.4)] with 0, 50, 100, or 200 mM NaCl and supplemented with 0.5 or 1 M urea where indicated. Spectra were acquired with 5 s averaging in 1-nm increments from 250 to 200 nm at either 25° or 55°C. Thermal melts were acquired by increasing the temperature in 1°C increments from 25° to 55°C and recording the ellipticity at 222 nm averaged over 10 s. Raw ellipticity at 222 nm in instrument units (millidegrees) was converted to per-residue molar ellipticity (θ) using Eq. 1
| (1) |
in which n is the number of amino acids in the protein (329 for FL and 222 for SBD), c is the concentration in molar (1 × 10−5 M or 10 μM), and d is the pathlength in millimeters (1 mm). The temperature dependence of θ222 was fit to Eq. 2
| (2) |
in which θmax is the molar ellipticity of the open state, Δθ is the difference in molar ellipticity between the open and closed states, m is the slope of the sigmoid, Tm is the transition temperature at which the open and closed states are equally populated, and T is the temperature of the measurement.
Free energy calculation of the effects of mutations and salt
Distances between all pairs of negatively charged residues on the acidic surface (D152, D154, D185, D187, D219, E221, and E223) were calculated in ChimeraX based on the structure of closed cpSRP43 SBD [Protein Data Bank (PDB): 3DEP (9)]. The coulombic contribution to the free energy of the closed state (ΔGelectric) of each pair was computed using Eqs. 3A and 3B (29) and summed over all pairs.
| (3A) |
| (3B) |
In Eqs. 3A and 3B, e is the charge of an electron; z1 and z2 are the charges of the two residue pairs; ε0, εH2O, and ε are the permittivity of free space in a vacuum, in water (estimated to be 78.5), and at the protein surface in water (estimated to be 4), respectively; κ is the inverse of Debye length (λd) as defined in Eq. 3B; I is the ionic strength of buffer and is 0, 0.1, 0.15, and 0.25 for buffers with 0, 50, 100, and 200 mM NaCl, respectively; and T is the temperature in kelvin (29).
Chaperone activity
The chaperone activity of cpSRP43 toward LHCP was measured as previously described (8, 30). cpSRP43 solution was clarified by ultracentrifugation in a TLA-100 rotor (Beckman Coulter) at 100,000 rpm for 30 min at 4°C before the experiment. LHCP aggregation was initiated by the addition of 2 μl of 50 μM LHCP denatured in urea buffer [8 M urea, 10 mM tris, and 100 mM Na2HPO4 (pH 8.0)] to 100 μl of CD buffer with 50 mM NaCl and containing indicated concentrations of cpSRP43 and/or urea. Samples were incubated at 25°, 35°, or 45°C in a water bath for 5 min, followed by 5 min on ice. The endpoint optical density was recorded at 360 nm on an ultraviolet-visible spectrometer (Beckman Coulter) after equilibrium had been reached (5 min). The fraction of aggregated LHCP or TBS enzyme client at equilibrium, f, was determined as a function of cpSRP43 concentration ([cpSRP43]) and fit to Eq. 4A
| (4A) |
in which [client] is the concentration of client protein fit to a shared value across all data and Ksol is the apparent solubilization constant.
Thermoprotection of TBS enzymes by cpSRP43 was measured as previously described (26). Purified, folded recombinant GUN4 and GluTR were exchanged into the light scattering (LS) buffer [50 mM Hepes (pH 7.4)] with specified NaCl concentration and centrifuged for 30 min at 100,000 rpm (TLA-100, Beckman Coulter). Unless otherwise stated, 10 μM GUN4 or 2.7 μM GluTR was mixed with an equal volume of cpSRP43 at indicated concentrations in the same buffer at 4°C. The chaperone-client mixtures were incubated in a water bath at 42°C for 5 min and placed on ice for 5 min. The optical density at 360 nm (A360) was recorded, normalized to that of the client without cpSRP43, and plotted as a function of cpSRP43 concentration. For TBS enzymes, the optical density increases at low cpSRP43 concentrations before it decreases when cpSRP43 concentration was further raised; this is attributed to a model in which each cpSRP43 interacts with multiple TBS proteins when the latter is in large molar excess, thus increasing oligomer size. This effect is described by incorporating a correction term to f as described in Eq. 4B
| (4B) |
in which f is defined in Eq. 4A and Kagg describes the apparent interaction constant of cpSRP43 with TBS oligomers. For [cpSRP43] >> Kagg, Eq. 4B reduces to Eq. 4A.
GUN4 aggregation kinetics
Purified GUN4 was centrifuged for 30 min at 100,000 rpm (TLA-100, Beckman Coulter) and diluted to 1 to 10 μM in the LS buffer with indicated NaCl concentrations. Proteins were incubated in a water bath set between 30° to 46°C for 1 to 60 min and then placed on ice for 5 min. The endpoint A360nm was recorded. Kinetic data were fit to a single exponential function (Eq. 5)
| (5) |
in which Afinal is the optical density at 360 nm when the reaction reached equilibrium, k is the misfolding rate constant, and t is the incubation time at elevated temperatures.
Fluorescence anisotropy
The binding of cpSRP43 to HiLyte Fluor 488–labeled L11 peptide (GSFDPLGLADD) or to fluorescein-labeled 54C peptide (QKQKAPPGTARRKRKAC) was measured on the basis of fluorescence anisotropy, as previously described (10). Measurements were performed in CD buffer with 50 mM NaCl at specified temperatures on a Fluorolog 3-22 spectrofluorometer (HORIBA), using 100 nM labeled peptide and the indicated concentrations of cpSRP43. The samples were excited at 500 nm, and fluorescence anisotropy was recorded at 527 nm. The observed anisotropy value (Aobs) as a function of cpSRP43 concentration ([cpSRP43]) was fit to Eq. 6A
| (6A) |
in which A0 is the anisotropy value of the peptide alone, ΔA is the change in anisotropy at saturating concentrations of cpSRP43, [pep] is the peptide concentration, and Kd is the equilibrium dissociation constant for the interaction between cpSRP43 and peptide. Where noted, data are reported as the fraction bound, calculated for each Aobs based on Eq. 6B
| (6B) |
BTFA labeling
cpSRP43 variants containing a single C150 (D150C, C175A, and C297S) were reduced with 4 mM dithiothreitol (DTT) and exchanged into degassed labeling buffer [50 mM Hepes, 300 mM NaCl, 1 mM EDTA, and 10% glycerol (pH 7.4)]. cpSRP43 (50 μM) was labeled with 2 mM BTFA (Sigma-Aldrich) at room temperature for 2 hours. Labeled cpSRP43 was purified on a HiPrep (Cytiva) column in CD buffer with 50 mM NaCl, and 10% D2O was added to the final sample. The final concentrations of BTFA-labeled cpSRP43 were 35 to 60 μM.
NMR spectroscopy
19F-NMR spectra were acquired in CD buffer with 50 mM NaCl at the indicated temperatures on a Bruker Avance 600 spectrometer equipped with a 5-mm QCI 1H/19F/13C/15N quadruple resonance cryoprobe with a single-axis z-gradient. Where specified, urea or 54C or L18 (VDPLYPGGSFDPLGLADD) peptides were added at the indicated concentrations. NMR data were processed with NMRPipe (47). Peaks were analyzed in MestReNova with peak deconvolutions fit to a generalized Lorentzian using simulated annealing (48). Peak areas were used to determine the relative population of closed and open states.
TROSY HSQC spectra of 2H,15N-labeled cpSRP43 (WT, R189L, or DE3NQ) were recorded on an 800-MHz Bruker Avance spectrometer equipped with a 5-mm TCI 1H/13C/15N triple resonance cryoprobe with a single-axis z-gradient. NMR spectra were acquired at 17°C in NMR buffer [50 mM Na2HPO4 (pH 6.5) and 150 mM NaCl] supplemented with 10% (v/v) D2O. NMR data were processed with NMRPipe (47). Data were visualized and analyzed with NMRviewJ (49). Assignments were transferred from (10), and spectra were normalized to the average intensity of the flexible N terminus (1 to 30).
Plant materials and growth conditions
The A. thaliana WT ecotype Ler-0, chaos (cpsrp43 null mutant), and the various T3 generation transgenic lines of cpSRP43 variants were used in this study. Under standard plant growth conditions, Arabidopsis seeds were sown in soil after a 2-day stratification in darkness at 4°C, and germinating young seedlings were grown under standard long-day conditions (16 hours light/8 hours dark, 100 μmol of photons m−2 s−1, 22°C, and 70% relative humidity). For plants used for heat-shock treatment, seeds were sterilized with 15% (v/v) bleach for 5 min and sown on half-strength Murashige and Skoog (MS) medium supplemented with 0.8% (w/v) agar and 1% (w/v) sucrose. After stratification in darkness at 4°C for 3 days, the plates were placed in a growth chamber with a long-daylight cycle (16 hours light/8 hours dark, 100 μmol photons m−2 s−1, 22°C, and 70% relative humidity). Ten-day-old seedlings were transferred to six-well plates containing half-strength MS medium (without agar) for equilibration overnight. Then, CHX, an inhibitor of protein biosynthesis, was added to the overnight culture and placed into a plant growth chamber prewarmed at 42°C for short-term heat-shock treatment.
Protein extraction and Western blot analysis
Rosette leaves from soil or seedlings from 1/2 MS medium were harvested and immediately frozen and ground in liquid nitrogen. Total leaf protein was extracted from the frozen plant material into 2× Laemmli buffer containing 100 mM tris-HCl (pH 6.8), 4% (w/v) SDS, and 20% (v/v) glycerol by heating the samples for 20 min at 70°C. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). All samples in 2× Laemmli buffer were diluted to the same protein concentration, supplemented with 100 mM DTT, and incubated at 70°C for 5 min. Aliquots (10 to 15 μg) of protein were subjected to SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes (Cytiva), and probed with specific antibodies.
Antibodies against GUN4 (PHY1691; 1:1000) and CHLH (PHY1691; 1:1000) were purchased from PhytoAB (CA); those for LHCa1 (AS01005; 1:2000) and actin (AS132640; 1:1000) were purchased from Agrisera (Vännäs). Antibodies against cpSRP43 (1:1000) and cpSRP54 (1:1000) were donated by D. Schünemann (Ruhr University Bochum). Immunoblotting signals visualized by Clarity Western ECL (Bio-Rad) were detected with the ChemiDoc Imaging System (Bio-Rad).
HPLC analyses of Chl and Chl precursors
Rosette leaves (30 to 50 mg) were harvested in light and weighed to determine the fresh weight of seedlings grown under standard plant growth conditions. Chl and Chl precursors were extracted from frozen seedling powders using the ice-cold pigment-extraction buffer [acetone:0.2 M NH4OH; 9:1 (v/v)] at −20°C for at least 30 min. After centrifugation (14,000g for 15 min at 4°C), the supernatant was subjected to high-performance liquid chromatography (HPLC) for the analyses of Chl and Chl precursors. HPLC analyses were conducted using the Agilent 1260 Infinity II Prime LC system equipped with a diode array and fluorescence detectors (Agilent Technologies), essentially as previously described (50). The Agilent PC-bound OpenLab CDS CS Workstation was used for HPLC data analyses.
Statistical analysis
Gaussian (normal) distribution was used as the statistical model for error estimates. Statistics are described for all reported values in the figure legends.
Acknowledgments
We thank members of the Shan group for comments on the manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Funding: This work was supported by the Department of Energy [DOE.DE-SC0020661 (S.S. and P.E.W.)], the National Natural Science Foundation of China [Excellent Young Scientists Fund for Hong Kong and Macau, 32322089 (P.W.)], the Research Grant Council of Hong Kong [27118022, 17107223, and 17109224 (P.W.)], the State Key Laboratory of Agrobiotechnology at the Chinese University of Hong Kong [the ITC Funding (P.W.)], and the University of Hong Kong [66620211115918 and the startup fund (P.W.)].
Author contributions: A.R.S.: Writing—original draft, conceptualization, investigation, validation, formal analysis, and visualization. G.K.: Investigation and methodology. C.Z.: Investigation, methodology, validation, and formal analysis. P.W.: Writing—original draft, investigation, methodology, resources, funding acquisition, validation, formal analysis, visualization, supervision, and project administration. P.E.W.: Writing—review and editing, methodology, resources, funding acquisition, validation, supervision, and project administration. S.S.: Conceptualization, writing—review and editing, resources, funding acquisition, validation, supervision, and project administration.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
REFERENCES AND NOTES
- 1.Jarvis P., López-Juez E., Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Leutwiler L. S., Meyerowitz E. M., Tobin E. M., Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 14, 4051–4064 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plumley G. F., Schmidt G. W., Light-harvesting chlorophyll a/b complexes: Interdependent pigment synthesis and protein assembly. Plant Cell 7, 689–704 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dall’Osto L., Bressan M., Bassi R., Biogenesis of light harvesting proteins. Biochim. Biophys. Acta Bioenerg. 1847, 861–871 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Paulsen H., Finkenzeller B., Kühlein N., Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur. J. Biochem. 215, 809–816 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Apel K., Hirt H., Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Wang P., Liang F.-C., Wittmann D., Siegel A., Shan S., Grimm B., Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 115, E3588–E3596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaru-Ampornpan P., Shen K., Lam V. Q., Ali M., Doniach S., Jia T. Z., Shan S., ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat. Struct. Mol. Biol. 17, 696–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stengel K. F., Holdermann I., Cain P., Robinson C., Wild K., Sinning I., Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321, 253–256 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Liang F.-C., Kroon G., McAvoy C. Z., Chi C., Wright P. E., Shan S., Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc. Natl. Acad. Sci. U.S.A. 113, E1615–E1624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cain P., Holdermann I., Sinning I., Johnson A. E., Robinson C., Binding of chloroplast signal recognition particle to a thylakoid membrane protein substrate in aqueous solution and delineation of the cpSRP43-substrate interaction domain. Biochem. J. 437, 149–155 (2011). [DOI] [PubMed] [Google Scholar]
- 12.McAvoy C. Z., Siegel A., Piszkiewicz S., Miaou E., Yu M., Nguyen T., Moradian A., Sweredoski M. J., Hess S., Shan S., Two distinct sites of client protein interaction with the chaperone cpSRP43. J. Biol. Chem. 293, 8861–8873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdermann I., Meyer N. H., Round A., Wild K., Sattler M., Sinning I., Chromodomains read the arginine code of post-translational targeting. Nat. Struct. Mol. Biol. 19, 260–263 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Gao F., Kight A. D., Henderson R., Jayanthi S., Patel P., Murchison M., Sharma P., Goforth R. L., Kumar T. K. S., Henry R. L., Heyes C. D., Regulation of structural dynamics within a signal recognition particle promotes binding of protein targeting substrates. J. Biol. Chem. 290, 15462–15474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathir K. M., Rajalingam D., Sivaraja V., Kight A., Goforth R. L., Yu C., Henry R., Kumar T. K. S., Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43. J. Mol. Biol. 381, 49–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu C.-J., Schuenemann D., Hoffman N. E., Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274, 27219–27224 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Moore M., Harrison M. S., Peterson E. C., Henry R., Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529–1532 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Falk S., Ravaud S., Koch J., Sinning I., The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dünschede B., Bals T., Funke S., Schünemann D., Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J. Biol. Chem. 286, 35187–35195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis N. E., Marty N. J., Kathir K. M., Rajalingam D., Kight A. D., Daily A., Kumar T. K. S., Henry R. L., Goforth R. L., A dynamic cpSRP43-albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J. Biol. Chem. 285, 34220–34230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichacker L. A., Henry R., Function of a chloroplast SRP in thylakoid protein export. Biochim. Biophys. Acta Mol. Cell Res. 1541, 120–134 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Schuenemann D., Gupta S., Persello-Cartieaux F., Klimyuk V. I., Jones J. D. G., Nussaume L., Hoffman N. E., A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 10312–10316 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimyuk V. I., Persello-Cartieaux F., Havaux M., Contard-David P., Schuenemann D., Meiherhoff K., Gouet P., Jones J. D., Hoffman N. E., Nussaume L., A chromodomain protein encoded by the arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Henry R., Yuan J., Cline K., Hoffman N. E., A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 92, 3789–3793 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzvetkova-Chevolleau T., Hutin C., Noël L. D., Goforth R., Carde J.-P., Caffarri S., Sinning I., Groves M., Teulon J.-M., Hoffman N. E., Henry R., Havaux M., Nussaume L., Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19, 1635–1648 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji S., Siegel A., Shan S., Grimm B., Wang P., Chloroplast SRP43 autonomously protects chlorophyll biosynthesis proteins against heat shock. Nat. Plants 7, 1420–1432 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel A., McAvoy C. Z., Lam V., Liang F.-C., Kroon G., Miaou E., Griffin P., Wright P. E., Shan S.-O., A disorder-to-order transition activates an ATP-independent membrane protein chaperone. J. Mol. Biol. 432, 166708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenfield N. J., Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K. K., Fitch C. A., B. García-Moreno E., Distance dependence and salt sensitivity of pairwise, coulombic interactions in a protein. Protein Sci. 11, 1004–1016 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaru-Ampornpan P., Liang F.-C., Nisthal A., Nguyen T. X., Wang P., Shen K., Mayo S. L., Shan S., Mechanism of an ATP-independent protein disaggregase: II. Distinct molecular interactions drive multiple steps during aggregate assembly. J. Biol. Chem. 288, 13431–13445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu C. J., Peterson E. C., Henry R., Hoffman N. E., The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J. Biol. Chem. 275, 13187–13190 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Amin P., Sy D. A. C., Pilgrim M. L., Parry D. H., Nussaume L., Hoffman N. E., Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121, 61–70 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaletz S. T., Weiser M. D., McDowell N. G., Zhou J., Kaspari M., Helliker B. R., Enquist B. J., The energetic and carbon economic origins of leaf thermoregulation. Nat. Plants 2, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto F., Obayashi T., Sasaki-Sekimoto Y., Ohta H., Takamiya K., Masuda T., Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 135, 2379–2391 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franzmann T. M., Menhorn P., Walter S., Buchner J., Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol. Cell 29, 207–216 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Stengel F., Baldwin A. J., Painter A. J., Jaya N., Basha E., Kay L. E., Vierling E., Robinson C. V., Benesch J. L. P., Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. U.S.A. 107, 2007–2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foit L., George J. S., Zhang B. W., Brooks C. L., Bardwell J. C. A., Chaperone activation by unfolding. Proc. Natl. Acad. Sci. U.S.A. 110, E1254–E1262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal P., Tse E., Borst A. J., Delbecq S. P., Shi L., Southworth D. R., Klevit R. E., A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. eLife 4, e07304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clouser A. F., Klevit R. E., pH-dependent structural modulation is conserved in the human small heat shock protein HSBP1. Cell Stress Chaperones 22, 569–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleckenstein T., Kastenmüller A., Stein M. L., Peters C., Daake M., Krause M., Weinfurtner D., Haslbeck M., Weinkauf S., Groll M., Buchner J., The chaperone activity of the developmental small heat shock protein Sip1 Is regulated by pH-dependent conformational changes. Mol. Cell 58, 1067–1078 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Groitl B., Horowitz S., Makepeace K. A. T., Petrotchenko E. V., Borchers C. H., Reichmann D., Bardwell J. C. A., Jakob U., Protein unfolding as a switch from self-recognition to high-affinity client binding. Nat. Commun. 7, 10357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter J., Ilbert M., Graf P. C. F., Özcelik D., Jakob U., Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T., Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Zurawa-Janicka D., Jarzab M., Polit A., Skorko-Glonek J., Lesner A., Gitlin A., Gieldon A., Ciarkowski J., Glaza P., Lubomska A., Lipinska B., Temperature-induced changes of HtrA2(Omi) protease activity and structure. Cell Stress Chaperones 18, 35–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyama Y., Harkness R. W., Kay L. E., Structural basis of protein substrate processing by human mitochondrial high-temperature requirement A2 protease. Proc. Natl. Acad. Sci. U.S.A. 119, e2203172119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaru-Ampornpan P., Chandrasekar S., Shan S., Efficient interaction between two GTPases allows the chloroplast SRP pathway to bypass the requirement for an SRP RNA. Mol. Biol. Cell 18, 2636–2645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Willcott M. R., MestRe Nova. J. Am. Chem. Soc. 131, 13180–13180 (2009). [Google Scholar]
- 49.Johnson B. A., Blevins R. A., NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Fu D., Zhou H., Grimm B., Wang P., The BCM1-EGY1 module balances chlorophyll biosynthesis and breakdown to confer chlorophyll homeostasis in land plants. Mol. Plant 18, 76–94 (2025). [DOI] [PubMed] [Google Scholar]
- 51.One Thousand Plant Transcriptomes Initiative , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1