The characterization of protein-folding mechanisms has dominated the landscape of experimental and theoretical biophysics for several decades. Experiment has moved the accessible time scales for the exploration of folding mechanism from minutes to milliseconds to microseconds and faster (1). Theory too has developed to address both the global character of folding mechanism and kinetics (2, 3) and the microscopic details that underlie this character for specific protein sequences and topologies (4). The advent of continuous-flow rapid-micromixing techniques (5, 6), rapid laser-induced temperature-jump heating methods (7), redox-triggered folding experiments (8), and NMR lineshape analysis (9) is beginning to allow direct examination of the earliest events in protein folding and permitting more direct comparison to theories and models of protein folding (10).
In this issue of PNAS, work from Akiyama et al. (11) describes how continuous-flow rapid-mixing methods are used with small-angle x-ray scattering to probe the early stages of folding of cytochrome c. This work follows earlier studies of this type by Eaton and coworkers (12) probing the time-dependent collapse of cytochrome c and complements recent studies by Akiyama et al. (13) who focused on the extent of helix formation during the folding of this protein. In the analysis of these exciting experiments, folding along progress coordinates describing the formation of secondary (helical) structure and the overall degree of collapse, as measured by the protein radius of gyration (Rg), are described and contrasted with three theoretical models for the folding of proteins. Akiyama et al. aim to examine whether the folding of cytochrome c is driven by nonspecific hydrophobic collapse, as would be consistent with the hydrophobic collapse model (14), secondary structure formation, following more closely a framework model for folding (15), or via a concerted mechanism in which collapse and secondary structure formation are nearly concomitant, as discussed recently based on the results of simulation studies on a helical bundle protein and proposed as a dominant folding mechanism for helical topologies (16, 17).
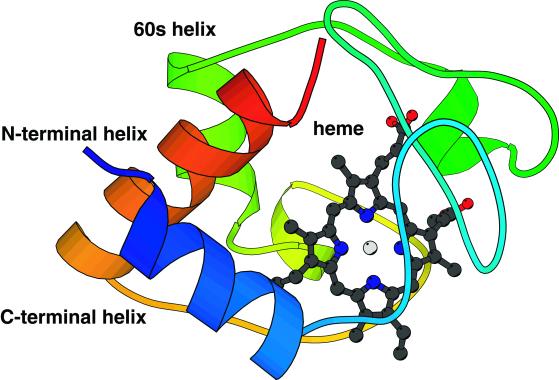
Cytochrome c is comprised of three significant helices and two shorter ones, as well as a small β-sheet (Fig. 1). Complicating the kinetics of folding of cytochrome c is a heme group, which under native conditions is coordinated by a histidine (His-18) and a methionine (Met-80), as well as chemically linked by two cysteine residues. At pH values greater than 7, folding involves a kinetic intermediate arising from the miscoordination of the heme by His-26 or His-33 and this creates an “off-pathway” trap and slows folding to a time scale of tens to hundreds of milliseconds (18). However, folding initiated at low pH values, i.e., from an acid-denatured state near pH 2, and followed after dilution to pH 4.5 by time-resolved circular dichroism (CD) shows relatively rapid folding with at least two distinct phases at 500 μs and 7–15 ms involving stepwise formation of the major helices (13).
Figure 1.
The folded state of cytochrome c is dominated by a three-helical structure, comprised of the N- and C-terminal helices, the “60s” helix, and the significantly hydrophobic heme group. Folding as observed by Akiyama et al. (11) involves initial collapse and helix formation (Rg = 21 Å, fH = 20% native helical content) in less than 160 μs followed by commensurate collapse and helix formation through another intermediate (Rg = 17.7 Å and fH = 70%) emerging on a time scale of 500 μs to native-like helix content and radius of gyration on a time scale of 15 ms.
Akiyama et al. (11) elucidate the kinetic evolution of the collapse coordinate describing cytochrome c folding and observe two intermediates en route to the native state. The first occurs in less than 160 μs (the dead time of the current ultra-fast mixing apparatus) and is shown to have an Rg value 50% larger than the native state (the acid-denatured state is 76% greater than native). Intermediate I also possesses about 20% of the native helix content (13). A second intermediate, with Rg only 29% greater than native and containing 70% of the native helix content, occurs on a time scale of 500 μs. Finally, on a time scale of 15 ms, this intermediate evolves to the native state. Thus, the refolding mechanism for cytochrome c under the conditions of these studies involves the rapid collapse of the polypeptide chain to a conformational ensemble with a radius about 1.5 times the native protein and 20% of its helix content. This initial event is followed by the apparent concomitant formation of helix and chain collapse to a second short-lived intermediate with a majority of the native helix content and 29% less compact than the native state.
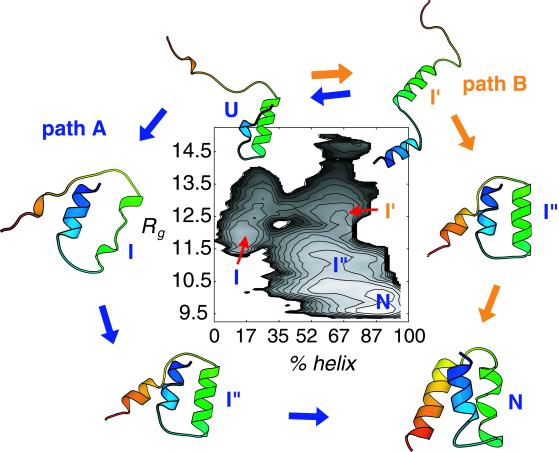
The agreement in general features of the observed mechanism and that from earlier computational folding free-energy studies of a three-helical bundle peptide, a 48-residue version of fragment B of Staphylococcal protein A, is rather remarkable (16, 17). As shown in Fig. 2 (adapted from figure 5B of ref. 17), the studies by C.L.B. and coworkers suggest that this helical protein folds via a mechanism involving two dominant pathways. In one the protein collapses to an ensemble of configurations with ≈17% native helix content and an Rg value 1.3 that of the native protein, and then proceeds to the native state through concomitant helix formation and collapse. An alternative path also observed for this system suggests a more “collision-diffusion” mechanism (19) with significant native helical content present (about 65%) before final collapse to the native state. Although the former mechanism is commensurate with the observations of Akiyama et al. (see figure 5 from ref. 11), the latter seems to be more in line with the mechanism suggested recently for this same three-helical bundle protein from an analysis of NMR lineshapes (9). Both pathways have been observed in other simulation studies (20–22) and suggest that details of solvent conditions, model, and sequence effects may determine which is dominant for a particular experiment.
Figure 2.
Folding of the three-helical bundle fragment B of Staphylococcal protein A progresses from initial collapse of the unfolded states (U) to an intermediate (I) with 17% native helix content and ≈30% expanded from the native state and then to native-like conformations (N) via concomitant collapse and helix formation (I′′) as indicated for path A on the free-energy surface computed by C.L.B. and coworkers. An alternative path also is observed that involves early formation of most of the native helical structure (I′) before significant collapse (path B). The free-energy surface shown here depicts the free-energy cost (contours at 0.5 kcal/mol of increments) for folding vs. progress coordinates for collapse (Rg in Å) and native helical structure formation (fraction of native helical hydrogen bonds). Results are taken from Guo et al. (17). The structures and labels of intermediates shown in this figure are meant only to provide a schematic representation of possible conformations adopted along the folding route for protein A. The protein structural cartoons in both figures were produced by using MOLSCRIPT (24).
The confluence of experiment with theory and computational studies in this example is an exciting harbinger of emerging approaches to understanding protein-folding mechanism. What is apparent from the studies by Akiyama et al. is that examining folding from many different perspectives, i.e., viewpoints as well as progress coordinates, will lead us to more enlightened understanding of folding mechanisms and the interactions that underlie them. Although it may be anticipated that different mechanisms will be at play for different protein topologies (17, 23), it is abundantly clear that examining folding for such systems using many different “rulers” will only enhance our knowledge of folding.
Acknowledgments
I acknowledge financial support from the National Institutes of Health (GM 48807) for the work described here.
Footnotes
See companion article on page 1329.
References
- 1.Eaton W A, Munoz V, Hagen S J, Jas G S, Lapidus L J, Henry E R, Hofrichter J. Annu Rev Biophys Biomol Struct. 2000;29:327–359. doi: 10.1146/annurev.biophys.29.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolynes P, Luthey-Schulten Z, Onuchic J. Chem Biol. 1996;3:425–432. doi: 10.1016/s1074-5521(96)90090-3. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic J N, Socci N D, Luthey-Schulten Z, Wolynes P G. Folding Des. 1996;1:441–450. doi: 10.1016/S1359-0278(96)00060-0. [DOI] [PubMed] [Google Scholar]
- 4.Shea J E, Brooks C L., III Annu Rev Phys Chem. 2001;52:499–535. doi: 10.1146/annurev.physchem.52.1.499. [DOI] [PubMed] [Google Scholar]
- 5.Chan C K, Hu Y, Takahashi S, Rousseau D L, Eaton W A, Hofrichter J. Proc Natl Acad Sci USA. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi S, Yeh S R, Das T K, Chan C K, Gottfried D S, Rousseau D L. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 7.Callender R H, Dyer R B, Gilmanshin R, Woodruff W H. Annu Rev Phys Chem. 1998;49:173–202. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 8.Pascher T. Biochemistry. 2001;40:5812–5820. doi: 10.1021/bi0026223. [DOI] [PubMed] [Google Scholar]
- 9.Myers J K, Oas T G. Nat Struct Biol. 2001;8:552–558. doi: 10.1038/88626. [DOI] [PubMed] [Google Scholar]
- 10.Yeh S R, Rousseau D L. Nat Struct Biol. 2000;7:443–445. doi: 10.1038/75831. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama S, Takahashi S, Kimura T, Oishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack L, Tate M W, Darnton N C, Knight J B, Gruner S M, Eaton W A, Austin R H. Proc Natl Acad Sci USA. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama S, Takahashi S, Ishimori K, Morishima I. Nat Struct Biol. 2000;7:514–520. doi: 10.1038/75932. [DOI] [PubMed] [Google Scholar]
- 14.Dill K A. Biochemistry. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 15.Kim P S, Baldwin R L. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- 16.Boczko E M, Brooks C L., III Science. 1995;269:393–396. doi: 10.1126/science.7618103. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, Brooks C L, III, Boczko E M. Proc Natl Acad Sci USA. 1997;94:10161–10166. doi: 10.1073/pnas.94.19.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colon W, Wakem L P, Sherman F, Roder H. Biochemistry. 1997;36:12535–12541. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 19.Karplus M, Weaver D L. Protein Sci. 1994;3:650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolinski A, Skolnick J. Proteins. 1994;18:353–366. doi: 10.1002/prot.340180406. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Karplus M. Nature (London) 1999;401:400–403. doi: 10.1038/43937. [DOI] [PubMed] [Google Scholar]
- 22.Alonso D O, Daggett V. Proc Natl Acad Sci USA. 2000;97:133–138. doi: 10.1073/pnas.97.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plaxco K W, Simons K T, Baker D. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]