Abstract
Heparan sulfate (HS) molecules are ubiquitous in animal tissues where they function as ligands that are dramatically involved in the regulation of the proteins they bind. Of these, chemokines are a family of small proteins with many biological functions. Their well-conserved monomeric structure can associate in various oligomeric forms especially in the presence of HS. Application of protein surface analysis and energy calculations to all known chemokine structures leads to the proposal that four different binding modes are created by the folding and oligomerization of these proteins. So, based on the present state of our knowledge, four different clusters of amino acids should be involved in the recognition process. Our results help to rationalize how unique sequences of HS specifically bind any given chemokine. The conclusions open the route for a rational design of compounds of therapeutical interest that could influence chemokine activity.
Chemokines, derived from chemoattractant cytokine and now comprising more than 50 members, represent a recently identified family of small proteins (8 to 12 kDa in their monomeric form). Depending on the structure of a conserved cysteine-containing motif in the amino-terminal region of the molecule, four subgroups have been characterized and named C, CC, CXC, or CX3C according to the number and spacing of these cysteine residues. Based on the physiological features of these proteins, chemokines also have been classified as inflammatory (or inducible) or homeostatic (or constitutive) (1, 2).
Although the chemotactic effect on leukocytes represents an important activity of the chemokines, it appears that these proteins also control a range of other functions that extend well beyond the regulation of leukocyte migration, including development, angiogenesis, neuronal patterning, hematopoiesis, viral infection, wound healing, and metastasis. The chemokine receptors, 20 of which have been identified, and through which these effects are transduced, are G protein-coupled, seven-helix transmembrane receptors (2). The observation that most chemokines bind to several different receptors and that most chemokine receptors exhibit overlapping specificity raises the question of the specificity of this system.
Glycosaminoglycans (GAGs) are linear polysaccharides present on all animal cell surfaces and in the extracellular matrix, where they are usually found to be attached covalently to core proteins to form the proteoglycan family (3). Each tissue produces specific repertoires of GAGs (4), some of which are known to bind and regulate chemokine activity (5, 6). Several lines of evidence point to the importance of one particular GAG, heparan sulfate (HS), in promoting chemokine activity. First, in vitro, almost all chemokines studied to date appear to bind HS, suggesting that this represents a fundamental aspect of these proteins. Second, the finding that, in vivo, T lymphocytes secrete CC chemokine as a complex with proteoglycans indicates that this form is physiologically relevant (7). Finally, it is known that association of chemokines with HS helps to stabilize concentration gradients along the endothelial surface, providing directional cues for migrating leukocytes. HS may also protect chemokines from proteolytic degradation and induce oligomerization of chemokine, thus promoting local high concentrations in the vicinity of the G-coupled signaling receptors (8). The functional relevance of oligomerization remains controversial, although all chemokines have a clear structural basis for dimerization. Indeed, under conditions required for structural studies, most chemokines have a dimeric or tetrameric organization. Nevertheless, the dissociation constant that characterizes dimer formation is usually below the dissociation constant for receptor binding. Oligomerization appears to be strongly enhanced by HS.
We recently analyzed the HS and heparin binding activity of the chemokine stromal cell-derived factor 1α (SDF-1α) and proposed a structural model of the SDF-1α/heparin complex that rationalizes all of the experimental data (9). Together with the general importance of HS for the biology of chemokines, our results prompted us to extend our modeling study to other members of this family of proteins. Targeting the chemokine/HS interaction clearly offers an approach for the discovery of compounds that modify chemokine activity (10), and we believe that the present study could be of some help for the development of such an approach.
Computational Methods
Calculation of the chemokines Connolly surfaces was performed by using coordinates taken from the Protein Data Bank (11) and the molcad program (12) from the sybyl package. The coordinates of the extended conformation of heparin were taken from the NMR-derived structures (13) deposited in the Protein Data Bank (ID code 1HPN). A variety of chain conformation also was generated for an 18-mer, taking a variety of conformation at each glycosidic linkage. Docking procedures use the grid program (14) to predict the most favorable anchoring position for a charged sulfate group at the surface of the chemokines. From our library of heparin chain conformations, those that displayed an appropriate shape for fitting of sulfate groups with the grid lowest iso-energy contour were selected. The geometry of each of the complexes was optimized by several cycles of energy minimization. All energy calculations were performed with the Tripos force field (15) together with energy parameters especially derived for carbohydrates (16) and sulfated derivatives (17). Details on the different steps of the computational approaches described here are available as supporting information on the PNAS web site, www.pnas.org, and have been described previously in the modeling study of SDF-1α/heparin complex (9).
Results
Comparison of the Oligomerization Modes of Chemokines.
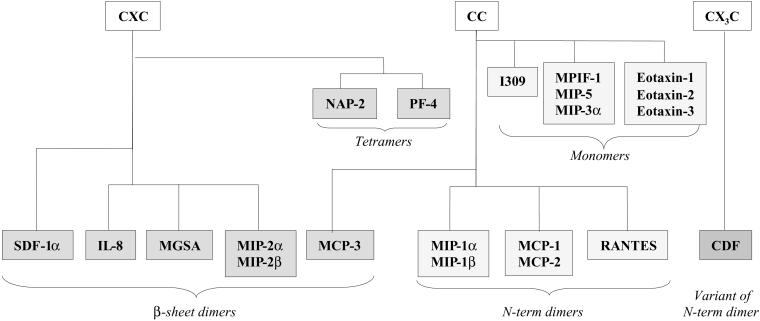
Among the 37 three-dimensional structures of chemokines in the Protein Data Bank (11) (see Table 2, which is published as supporting information on the PNAS web site), only 25% correspond to a monomeric state of the protein, the others being dimers, with the exception of platelet factor-4 (PF-4) and neutrophil-activating peptide-2 (NAP-2), which have been observed as tetrameric structures. The typical chemokine monomer is organized around a triple-stranded antiparallel β-sheet [β(1), β(2), and β(3)] overlaid by a C-terminal α-helix α(C). The N-terminal region consists of a more or less disordered extended strand β(N). In all cases the N terminus is anchored to the rest of the molecule by disulfide bridges involving the two cysteine residues that are characteristic of the CC, CXC, or CX3C motif. From these monomer templates, several oligomerization modes are observed, as shown in Fig. 1.
Figure 1.
Classification of chemokines with known three-dimensional structure as a function of their quaternary structure.
Dimerization through association of the β-sheets is observed for all chemokines of the CXC family. The dimer interface arises from antiparallel association of the first β-strand, creating a larger β-sheet. On one face of this sheet, the two helices run exactly antiparallel. Monocyte chemoattractant protein (MCP)-3, a member of the CC family of chemokines (18), also adopts this particular quaternary structure referred to hereafter as β-sheet dimer.
Dimerization by the N-terminal strands associating in antiparallel mode is observed in most members of the CC chemokine family. In all cases the dimeric structure is rather extended and the helices are located at the two extremities.
A variant of N-terminal strand dimerization mode has been observed in the recently determined structure of the chemokine domain of fractalin (19), the only member of the CX3C chemokine family. Nevertheless, because of the different arrangement in the disulfide bridge, the N-terminal strand is closer to the protein core, and chemokine domain of fractalkine (CDF) forms a more compact dimer than the CC chemokines.
Tetramerization by association of the two first types of dimer described above is characteristic of PF-4 and NAP-2 (20, 21).
The oligomerization modes summarized in Fig. 1 offer an image of our current knowledge and the experimental methods used for the characterization. NMR experiments (which are generally run in dilute solution) are less likely than x-ray crystallography to characterize oligomeric structures. Nevertheless, in more than 50% of the reported NMR structures, the chemokines were associated in dimer or tetramer (compared with more than 90% for the x-ray structures).
Chemokines and Heparin Binding: Analysis of the Accessible Surfaces.
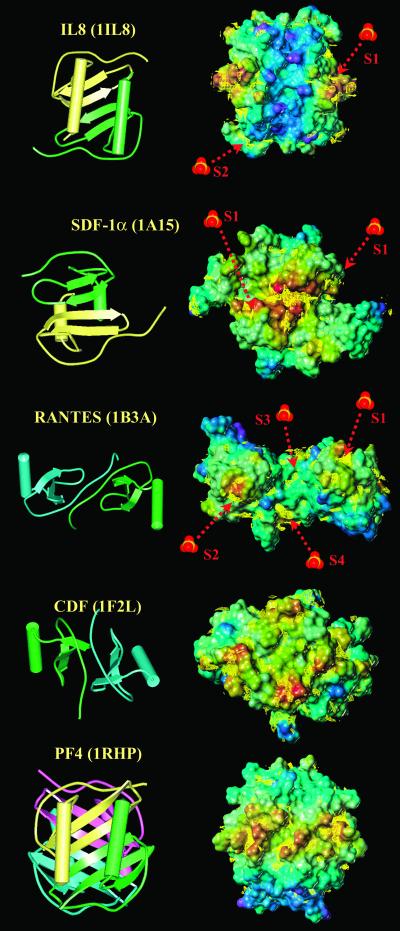
Several studies have been performed on the binding of GAGs by chemokines. They all are known to bind heparin with the exception of macrophage inflammatory protein (MIP)-5, I309, and several eotaxins for which no data are available. By using molcad software (12), the accessible surfaces of 12 chemokines have been computed and color-coded according to electrostatic potential, and some are displayed in Fig. 2.
Figure 2.
(Left) Ribbon and tube representation of selected chemokines available in the Protein Data Bank. (Right) Connolly surface of the chemokine dimers or tetramers as calculated with the molcad program. The surfaces are color-coded according to the electrostatic potential from blue (negative values) to red (positive values). Iso-energy contour for best interaction of a charge negative oxygen (sulfate probe) as calculated with the grid program have been superimposed as yellow lines on the Connolly surface. Localization of sulfate ions cocrystallized with the chemokines have been indicated by a red arrow.
In most CXC chemokine monomers, such as IL-8, a positively charged area of the protein surface is created by the C-terminal α-helix together with the loop connecting the extended N-terminal strand region with the first β-strand of the sheet. All CXC chemokines display the same positive area on the surface of the α-helices, except SDF-1α whose dimer is very different: the most positive area is on the opposite side, i.e., on the back of the β-sheet formed by the dimer association.
For the chemokines of the CC family (with the exception of MCP-3), the most positive area consists of two bulbous regions created by the rather long loop connecting the third β-strand, β(3), and the C-terminal α-helix. When the two loops are not too far away, as in MIP-1α and RANTES (regulated upon activation, normal T cell expressed and secreted), an almost continuous positively charged surface is created. The more dense dimerization mode of the sole CX3C family member (CDF) does not allow these loops to be exposed. In this particular dimer the most positively charged surface is on the opposite side, on the flat area created by the antiparallel association of the two N-terminal strands.
As for the tetramers, in common with IL-8 and other CC chemokines, they share the arrangement of positively charged amino acids at the surface of the C-terminal α-helices. The two IL-8-type dimers associate in such a way that the positively charged areas form a continuous ring around the globular-shaped tetramer.
Prediction of Surface Areas with High Affinity for Sulfate Groups.
To gain more detailed insight into the binding site for sulfated GAGs, the grid program was run by using a negatively charged oxygen as the probe for a sulfate group. The iso-energy contours, corresponding to the lower energy of interaction (between −4.5 to −6 kcal/mol, depending on the proteins), are superimposed on the Connolly surfaces in Fig. 2. The prediction of binding sites for sulfate groups could be validated because sulfate ions, originating from the crystallization buffer, sometimes are observed in crystal structures of proteins, as with IL-8 (22), SDF-1α (23), MCP-1 (24), and RANTES (25). Comparisons between the observed position for sulfate ions and iso-contours for the lowest interaction energy values are displayed in Fig. 2. In all cases, the crystallographic localization of the sulfate ions corresponds to a predicted binding site. In most cases, the binding occurs in a positively charged area of the protein surface.
Chemokines and Heparin Binding: Modeling of the Complexes.
The predicted binding site for sulfate ions, together with the location and shape of the positively charged area of the protein surface, provide the basis for docking heparin fragments onto representative members of each class of chemokine. In each case, several heparin fragments were selected from a library of randomly generated conformations. Different possible binding modes were considered, and several cycles of geometry optimization were performed (Table 3, which is published as supporting information on the PNAS web site). General findings are as follows:
All binding modes were independent of the orientation of the heparin chain because of the 2-fold symmetry of all chemokine dimers studied here.
A large majority of interactions are caused by salt bridges between heparin acidic group (carboxylate and sulfate) and protein basic side chains. Few hydrogen bonds are also predicted to occur.
There is no ambiguity for the lowest energy geometry of the interaction, i.e., orientation of the heparin chain at the protein surface, the energy gap between the best orientation and the second one always being several tens of kcal/mol. However, there could be some ambiguity for the translation of the heparin chain on the protein surface. There is little energy difference between the best docking mode and the one resulting from the translation of one monosaccharide along the chain (carboxylate and sulfate of iduronic acid replacing the sulfate from GlcNS,6S).
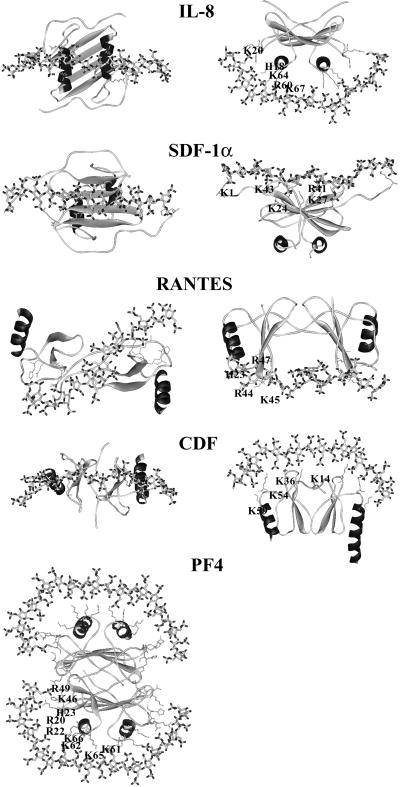
Only the lowest energy complexes are displayed in Fig. 3.
Figure 3.
Representation of the lowest energy models of several chemokine/heparin fragment interactions. The proteins are represented by ribbons except for the side chains of the basic amino acids directly involved in polysaccharide binding. The heparin molecule is represented by sticks. Hydrogen atoms are not displayed.
When binding to IL-8, heparin is predicted to adopt a curved shape that allows the establishment of an electrostatic interaction with two peptide sequences that are rich in basic amino acids: the C-terminal α-helix (R60, K64, K67, and R568) and the loop connecting the elongated N-terminal region with the first β-strand (H18 and K20). This model, in which the polysaccharide runs perpendicular to the two α-helices and bridges the two monomers, is in agreement with the prediction made by Wade using another docking strategy (26). Previously, a different model was proposed in which the heparin chain adopts a horseshoe shape and runs parallel to both helices (27). Our docking study predicts that the PF-4 tetramer binds HS in a similar way to IL-8. The positively charged ring around the PF-4 tetramer (Fig. 2) allows for the docking of two heparin oligosaccharides, each being perpendicular to a pair of C-terminal α-helices. In addition to the two binding regions present on IL-8, the loop between the β-strands β(2) and β(3) also displays a basic character (R46 and R49) and may further extend the length of the putative binding site. The model proposed in Fig. 3 is similar to the predicted binding mode of heparin in bovine PF-4 (28). Because of the particular ring shape of the binding area, HS oligomers could bind in different ways, as long as they mimic the ring fragment, allowing for several possibilities of binding.
The SDF-1α/heparin complex has been previously modeled and discussed (9), and it is presented here only for comparison with the other chemokines. In this model, the polysaccharide adopts a straight and extended shape and interacts with the β-sheet formed at the dimer interface. The participating basic amino acids belong to the β(1) (K24 and K27) and β(2) (R41 and K43) strands with an additional stabilization caused by the N-terminal lysine. Therefore, when viewing the molecules along the helix axes (Fig. 3 Left), the binding regions of SDF-1 and IL-8 are located on the two opposite sides of the dimer.
In the RANTES/heparin complex the polysaccharide is predicted to adopt an S shape that allows it to interact with the two positively charged bulbous regions present at the surface of the protein (Fig. 2). On each monomer this particular region is created by a cluster of basic amino acids (R44, K45, and R47) located in the loop connecting the two strands β(2) and β(3). An additional contact with heparin is established by H23 located in the loop connecting β(N) and β(1).
In the CDF model the binding site consists of a flat positive region made up of a large cluster of basic amino acids: K54 of the loop between β(3) and α(C) together with K59 at the beginning of this helix, K36 from the loop connecting β(1) and β(2), and K14 that is located at the end of the first-strand β(N). As displayed in Fig. 3 the putative HS binding areas of RANTES and CDF are located on the two opposite site of the molecule.
Discussion
Proposal of Four Docking Modes for the Interaction between Chemokines and HS.
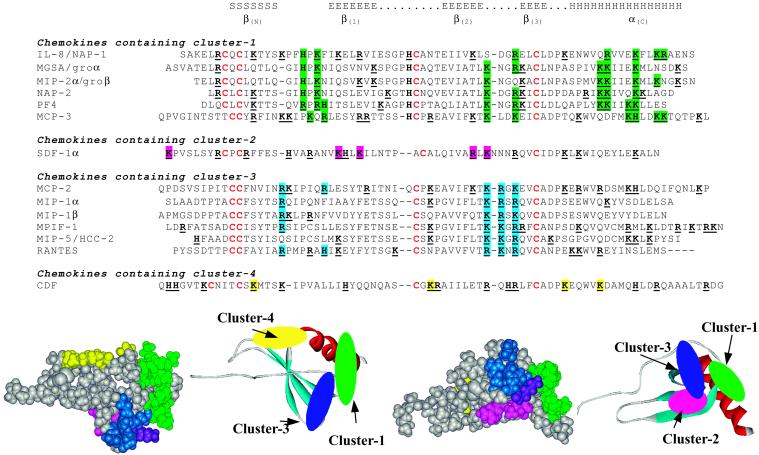
From the present modeling study, and in agreement with modeling data published previously (26, 28), four different docking modes are proposed for the interaction between chemokine dimers and heparin or HS: one mode for each of the CC and CX3C types of chemokines and two different modes within the CXC family. In each case, different clusters of basic amino acids can be defined as an arrangement of amino acids at the protein surface that provides efficient binding of HS. Fig. 4 displays a sequence alignment deduced from the superimposition of three-dimensional structures. The chemokines have been classified as a function of the occurrence of each type of cluster and the amino acids implicated have been colored.
Figure 4.
(Upper) Alignment of chemokine sequences based on the three-dimensional structure fit. Amino acids that are proposed to be involved in HS binding have been colored. (Lower) Space fill and ribbon representation of a chimeric chemokine protein that consists of poly-Ala chain with all basic amino acids participating in the cluster. Color coding is as follows: green, cluster 1; magenta, cluster 2; blue, cluster 3; and yellow, cluster 4.
Cluster 1 is characteristic of IL-8 and has also been proposed for melanoma growth stimulating activity (MGSA), MIP-2α, and NAP-2. The main characteristic of this binding site concerns two α-helices separated by a gap. Because basic amino acids are present on one side of the C-terminal helix and on contiguous turns, i.e., separated by about 3.5 aa, the peptide signature consists of (B)Bxx(x/B)BxxB(B) in the C-terminal region. The loop L(βN-β1) also participates in the binding and exhibits a conserved HxK motif. This mode of binding is also very likely to occur in the NAP-2 and PF-4 tetramer.
Cluster 2 has been observed only in SDF-1 where it forms a crevasse at the interface between the β-strands. It is the only binding site in this study to have a concave shape and to accommodate heparin or HS, having a fully extended conformation. Two basic amino acids in both β(1) and β(2) strands characterize this binding site but, because there is only one member in the family at the present time, this cannot be defined as a characteristic motif.
Cluster 3 is observed in all CXC chemokines with the exception of the MCP that may represent a family with different characteristics. It mainly involves the loop between β(2) and β(3) strands with a BBXB conserved motif. As in the case of site 1, in the dimer there is a gap between the two loops that the heparin chain has to bridge or fit into. Basic amino acids located at the beginning and the end of the loop connecting β(N) and β(1) also participate in the establishment of such a cluster.
Cluster 4 is based on the recent CDF crystal structure (19) and comprises a flat area made up of different loops. Two loops are involved, the ones between β(1) and β(2) and between β(2) and β(3).
The MCP chemokines are more difficult to describe in terms of the characterization of clusters. The three MCP chemokines share about 60% amino acid identity and putatively have very different heparin binding sites. MCP-3, which makes a CC type of dimer (18), displays a cluster-1 arrangement of basic amino acids, albeit with some degeneration. MCP-2 forms a N-terminal dimer (29) and its sequence contains the signature motif of cluster-3. MCP-1 forms the same dimer but no characteristic signature can be identified in the amino acid sequence.
To visualize the location of the clusters on the chemokine surface a chimera protein has been built by using the monomer of IL-8 as a template. All of the amino acids have been mutated to Ala except for the four cysteine residues involved in disulfide bridges. The 15 amino acids that belong to the clusters all have been mutated to His, Lys, or Arg, depending on the most frequent occurrence. The corresponding protein surface is displayed in Fig. 4 together with the location of the clusters in a ribbon representation of the monomer. Except for cluster 2 and cluster 3 that share one amino acid (a Lys colored in violet), the clusters are not contiguous and are scattered on different faces of the monomer.
Validation of the Modeling Study: Comparison of the Theoretical Complexes with Biochemical Data.
In the absence of crystal structures of complexes between chemokines and heparin or HS fragments, other experimental methods can be used to identify the amino acids involved in the interaction. Although site-directed mutagenesis is commonly used, direct NMR observation of the proton resonances of the side chains that are perturbed upon heparin binding also has proven to be a powerful method (30).
The amino acids that have been experimentally identified as being involved in heparin binding are listed in Table 1. There is an excellent agreement between the models derived from the present study and the experimental data, validating the clusters proposed here. Some discrepancies may be observed at the edge of clusters: mutation of Arg-68 of IL-8 caused a large decrease in affinity for heparin (31) whereas, in our study, this amino acid is included in cluster 1, but at the edge. The only point of disagreement that arises is found between our modeling and some experimental data obtained with RANTES (32). However, recent site-directed mutagenesis analysis (33) clearly established the role of Arg-44, Lys-45, and, more particularly, Arg-47 for binding heparin, in perfect agreement with the present study.
Table 1.
Known mutations of chemokines that affect the binding of HS
| Chemokine | Mutated amino acids | Chain length | Reference |
|---|---|---|---|
| Containing cluster-1 | |||
| Il-8 | D52 to S72 (including K54, R60, K64, K67, R68) | (38) | |
| Il-8 | K20, R60, K64, K67, R68 | (31) | |
| Il-8 | dp24 (Il-8 dimer) | (27) | |
| PF-4 | L59 to S70 (including K61, K62, K65, K66) | (39) | |
| PF-4 | R20, R22, H23, T25, K46, R49 | (30) | |
| PF-4 | R20, R22 | dp12 | (40) |
| PF-4 | dp20 of CS | (41) | |
| PF-4 | dp40 (PF-4 tetramer) | (42) | |
| Containing cluster-2 | |||
| SDF-1α | K1, K24, K27, R41, K43 | dp14 | (9, 43) |
| Containing cluster-3 | |||
| MIP-1α | K45, R46 | (44) | |
| MIP-1α | R18, K45, R46, R48 (no detectable role for K61) | (45) | |
| MIP-1β | R18, K45, R46 (no detectable role for K19, R22, K48) | (46) | |
| RANTES | 55 to 66 | (32) | |
| R44, R47 (no detectable role for K55, K56, R59) | (33, 47) |
Furthermore, from the present model, a synthetic HS analog consisting of two hexasaccharides linked by a spacer through their reducing end and therefore presenting a 2-fold symmetry that fits the symmetry and the organization of the protein dimer has been prepared (R. Vives and H.L.-J., unpublished work). This molecule has a several-fold enhanced affinity for RANTES, compared with the equivalent heparin dodecamer, validating our model and its use to design chemokine ligands.
Functional and Biological Implications of the Proposed Models.
In target cells, chemokines bind to and activate seven-helix transmembrane domain receptors that are coupled to G proteins (10). It is well established that the N-terminal region of chemokines is critical for biological activity (34). In most cases mutations that prevent HS binding do not appear to modify receptor binding. The present study shows that, in most cases, the HS-interacting surface is found on the C-terminal α-helix or in loops facing the N-terminal domain, and thus is distinct from the receptor-interacting surface. Nevertheless, recent studies have shown that receptor binding and receptor activation are not necessarily connected. For example, SDF-1 or RANTES in which the N-terminal domain was modified still bound to their cognate receptors, but the signal was not transduced (35, 36). It is possible therefore to consider that binding to GAGs may modulate signaling rather than binding to the receptor itself. This view is supported by a recent study showing that HS supported the Ca2+ mobilization induced by chemokines, whereas the GAG dermatan sulfate inhibits this cellular response (6). It is also known that a number of GAGs in complex with RANTES inhibit HIV replication by interacting with the chemokine receptor CCR5 but fail to induce Ca2+ mobilization (37). Therefore, in addition to the presentation of chemokines, GAGs might play an active part in chemokine function. Finally, it is possible that a single GAG may have different effects on a chemokine depending on which receptor it uses.
Among several questions that arise from these considerations are the following: How can HS bind and regulate so many different chemokines? Does HS simply localize and concentrate active proteins on the cell surface based on affinity considerations, or does it promote critical changes in protein conformation that allow interaction with and/or activation of its signaling receptors?
In this context, we have provided a complete description of the complexes formed by chemokines and HS. It is probable that new binding modes will be characterized in the future, when new structures of chemokines are solved. Our analysis provides clear evidence that, in each case, only a small subset of sulfate groups is involved in complex formation, a point consistent with the emerging view that unique sequences specifically bind a given chemokine. Although the present study would not allow for a very fine view of the GAG sequence optimal for binding (i.e., effect of 2-N-sulfate compared with 6-O-sulfate), it does help in identifying optimal arrangement of sulfate-rich and sulfate-poor portions of HS (S-domains and NAc-domains) that would interact with the clusters. As our comprehension of chemokine-induced receptor activation improves these data should provide a basis for the understanding of how HS may modulate chemokine function. From a therapeutic viewpoint the models we propose should assist with the rational design of HS mimics that can modulate the formation of chemokine-HS complexes.
Supplementary Material
Acknowledgments
We are indebted to Professor W. Mackie (University of Leeds, Leeds, U.K.) for his help in the redaction of the manuscript. This work was supported by a grant from the Agence Nationale de la Recherche contre le SIDA.
Abbreviations
- GAG
glycosaminoglycan
- HS
heparan sulfate
- MCP
monocyte chemoattractant protein
- RANTES
regulated upon activation, normal T cell expressed and secreted
- NAP-2
neutrophil-activating peptide-2
- MIP
macrophage inflammatory protein
- SDF-1α
stromal cell-derived factor 1α
- PF-4
platelet factor 4
- CDF
chemokine domain of fractalkine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cyster J G. Science. 1999;286:2098–2102. doi: 10.1126/science.286.5447.2098. [DOI] [PubMed] [Google Scholar]
- 2.Rossi D, Zlotnik A. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Park P W, Reizes O, Bernfield M. J Biol Chem. 2000;275:29923–29926. doi: 10.1074/jbc.R000008200. [DOI] [PubMed] [Google Scholar]
- 4.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. J Biol Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 5.Kuschert G S, Coulin F, Power C A, Proudfoot A E, Hubbard R E, Hoogewerf A J, Wells T N. Biochemistry. 1999;38:12959–12968. doi: 10.1021/bi990711d. [DOI] [PubMed] [Google Scholar]
- 6.Hirose J, Kawashima H, Yoshie O, Tashiro K, Miyasaka M. J Biol Chem. 2001;267:5228–5234. doi: 10.1074/jbc.M007542200. [DOI] [PubMed] [Google Scholar]
- 7.Wagner L, Yang O O, Garcia-Zepeda E A, Ge Y, Kalams S A, Walker B D, Pasternack M S, Luster A D. Nature (London) 1998;391:908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- 8.Hoogewerf A J, Kuschert G S, Proudfoot A E, Borlat F, Clark-Lewis I, Power C A, Wells T N. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 9.Sadir R, Baleux F, Grosdidier A, Imberty A, Lortat-Jacob H. J Biol Chem. 2001;276:8288–9296. doi: 10.1074/jbc.M008110200. [DOI] [PubMed] [Google Scholar]
- 10.Cascieri M A, Springer M S. Curr Opin Chem Biol. 2000;4:420–427. doi: 10.1016/s1367-5931(00)00113-7. [DOI] [PubMed] [Google Scholar]
- 11.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldherr-Teschner M, Goetze T, Heiden W, Knoblauch M, Vollhardt H, Brickmann J. In: Advances in Scientific Visualization. Post F H, Hin A J S, editors. Heidelberg: Springer; 1992. pp. 58–67. [Google Scholar]
- 13.Mulloy B, Forster M J, Jones C, Davies D B. Biochem J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodford P J. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 15.Clark M, Cramer R D I, van den Opdenbosch N. J Comput Chem. 1989;8:982–1012. [Google Scholar]
- 16.Pérez S, Meyer C, Imberty A. In: Modeling of Biomolecular Structures and Mechanisms. Pullman A, Jortner J, Pullman B, editors. Dordrecht, the Netherlands: Kluwer; 1995. pp. 425–454. [Google Scholar]
- 17.Imberty A, Bettler E, Karababa M, Mazeau K, Petrova P, Pérez S. In: Perspectives in Structural Biology. Vijayan M, Yathindra N, Kolaskar A S, editors. Hyderabad: Indian Academy of Sciences and Universities Press; 1999. pp. 392–409. [Google Scholar]
- 18.Meunier S, Bernassau J M, Guillemot J C, Ferrara P, Darbon H. Biochemistry. 1997;36:4412–4422. doi: 10.1021/bi9627929. [DOI] [PubMed] [Google Scholar]
- 19.Hoover D M, Mizoue L S, Hanel T M, Lubkowski J. J Biol Chem. 2000;275:23187–23193. doi: 10.1074/jbc.M002584200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Chen L, Bancroft D P, Lai C K, Maione T E. Biochemistry. 1994;33:8361–8366. doi: 10.1021/bi00193a025. [DOI] [PubMed] [Google Scholar]
- 21.Malkowski M G, Wu J Y, Lazar J B, Johnson P H, Edwards B F. J Biol Chem. 1995;270:7077–7087. doi: 10.1074/jbc.270.13.7077. [DOI] [PubMed] [Google Scholar]
- 22.Gerber N, Lowman H, Artis D R, Eigenbrot C. Proteins. 2000;38:361–367. [PubMed] [Google Scholar]
- 23.Dealwis C, Fernandez E J, Thompson D A, Simon R J, Siani M A, Lolis E. Proc Natl Acad Sci USA. 1998;95:6941–6946. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubkowski J, Bujacz G, Boque L, Domaille P J, Handel T M, Wlodawer A. Nat Struct Biol. 1997;4:64–69. doi: 10.1038/nsb0197-64. [DOI] [PubMed] [Google Scholar]
- 25.Hoover D M, Shaw J, Gryczynski Z, Proudfoot A E I, Wells T. Protein Pept Lett. 2000;7:73–82. [Google Scholar]
- 26.Bitomsky W, Wade R C. J Am Chem Soc. 1999;121:3004–3013. [Google Scholar]
- 27.Spillmann D, Witt D, Lindahl U. J Biol Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 28.Stuckey J A, St Charles R, Edwards B F. Proteins. 1992;14:277–287. doi: 10.1002/prot.340140213. [DOI] [PubMed] [Google Scholar]
- 29.Blaszczyk J, Coillie E V, Proost P, Van Damme J, Opdenakker G, Bujacz G D, Wang J M, Ji X. Biochemistry. 2000;39:14075–14081. doi: 10.1021/bi0009340. [DOI] [PubMed] [Google Scholar]
- 30.Mayo K H, Ilyina E, Roongta V, Dundas M, Joseph J, Lai C K, Maione T, Daly T J. Biochem J. 1995;312:357–365. doi: 10.1042/bj3120357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuschert G S, Hoogewerf A J, Proudfoot A E, Chung C W, Cooke R M, Hubbard R E, Wells T N, Sanderson P N. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 32.Burns J M, Gallo R C, DeVico A L, Lewis G K. J Exp Med. 1998;188:1917–1927. doi: 10.1084/jem.188.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proudfoot A E, Fritchley S, Borlat F, Shaw J P, Zwahlen C, Trkola A, Clapham P R, Wells T N. J Biol Chem. 2001;276:10620–10626. doi: 10.1074/jbc.M010867200. [DOI] [PubMed] [Google Scholar]
- 34.Clark-Lewis I, Kim K S, Rajarathnam K, Gong J H, Dewald B, Moser B, Baggiolini M, Sykes B D. J Leukocyte Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 35.Crump M P, Gong J H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J L, Baggiolini M, Sykes B D, Clark-Lewis I. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polo S, Nardese V, De Santis C, Arcelloni C, Paroni R, Sironi F, Verani A, Rizzi M, Bolognesi M, Lusso P. Eur J Immunol. 2000;30:3190–3198. doi: 10.1002/1521-4141(200011)30:11<3190::AID-IMMU3190>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Burns J M, Lewis G K, DeVico A L. Proc Natl Acad Sci USA. 1999;96:14499–14504. doi: 10.1073/pnas.96.25.14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb L M, Ehrengruber M U, Clark-Lewis I, Baggiolini M, Rot A. Proc Natl Acad Sci USA. 1993;90:7158–7162. doi: 10.1073/pnas.90.15.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loscalzo J, Melnick B, Handin R I. Arch Biochem Biophys. 1985;240:446–455. doi: 10.1016/0003-9861(85)90049-9. [DOI] [PubMed] [Google Scholar]
- 40.Mikhailov D, Young H C, Linhardt R J, Mayo K H. J Biol Chem. 1999;274:25317–25329. doi: 10.1074/jbc.274.36.25317. [DOI] [PubMed] [Google Scholar]
- 41.Petersen F, Brandt E, Lindahl U, Spillmann D. J Biol Chem. 1999;274:12376–12382. doi: 10.1074/jbc.274.18.12376. [DOI] [PubMed] [Google Scholar]
- 42.Stringer S E, Gallagher J T. J Biol Chem. 1997;272:20508–20514. doi: 10.1074/jbc.272.33.20508. [DOI] [PubMed] [Google Scholar]
- 43.Amara A, Lorthioir O, Valenzuela A, Magerus A, Thelen M, Montes M, Virelizier J L, Delepierre M, Baleux F, Lortat-Jacob H, Arenzana-Seisdedos F. J Biol Chem. 1999;274:23916–23925. doi: 10.1074/jbc.274.34.23916. [DOI] [PubMed] [Google Scholar]
- 44.Graham G J, MacKenzie J, Lowe S, Tsang M L, Weatherbee J A, Issacson A, Medicherla J, Fang F, Wilkinson P C, Pragnell I B. J Biol Chem. 1994;269:4974–4978. [PubMed] [Google Scholar]
- 45.Koopmann W, Krangel M S. J Biol Chem. 1997;272:10103–10109. doi: 10.1074/jbc.272.15.10103. [DOI] [PubMed] [Google Scholar]
- 46.Koopmann W, Ediriwickrema C, Krangel M S. J Immunol. 1999;163:2120–2127. [PubMed] [Google Scholar]
- 47.Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, Vita C. Biochemistry. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.