Abstract
Eukaryotic DNA replication requires the previous formation of a prereplication complex containing the ATPase Cdc6 and the minichromosome maintenance (Mcm) complex. Although considerable insight has been gained from in vitro studies and yeast genetics, the functional analysis of replication proteins in intact mammalian cells has been lacking. We have made use of adenoviral vectors to express normal and mutant forms of Cdc6 in quiescent mammalian cells to assess function. We demonstrate that Cdc6 expression alone is sufficient to induce a stable association of endogenous Mcm proteins with chromatin in serum-deprived cells where cyclin-dependent kinase (cdk) activity is low. Moreover, endogenous Cdc6 is sufficient to load Mcm proteins onto chromatin in the absence of cdk activity in p21-arrested cells. Cdc6 synergizes with physiological levels of cyclin E/Cdk2 to induce semiconservative DNA replication in quiescent cells whereas cyclin A/Cdk2 is unable to collaborate with Cdc6. Cdc6 that cannot be phosphorylated by cdks is fully capable of inducing Mcm chromatin association and replication. Mutation of the Cdc6 ATP-binding site severely impairs the ability of Cdc6 to induce Mcm chromatin loading and reduces its ability to induce replication. Nevertheless, the ATPase domain of Cdc6 in the absence of the noncatalytic amino terminus is not sufficient for either Mcm chromatin loading or DNA replication, indicating a requirement for this domain of Cdc6.
The initiation of DNA replication in eukaryotic cells requires the assembly of multiple proteins at origins together with the action of protein kinases. Among the proteins known to assemble at origins before the initiation of DNA synthesis are the origin recognition complex, Cdc6, and the minichromosome maintenance (Mcm) complex. Together, these constitute a “prereplication” complex to which additional factors bind once cyclin-dependent kinases (cdks) become active. Once all of the necessary proteins have assembled at the origin, additional phosphorylation events trigger initiation (for reviews see refs. 1–5).
Mutational alteration in yeast (6–8) or immunodepletion of Xenopus laevis oocytes (9) has demonstrated that Cdc6 and each origin recognition complex and MCM subunit plays a unique and essential role in replication. Cdc6 is thought to act primarily through recruitment of the Mcm complex to origins because mutations in yeast cdc6 or immunodepletion of X. laevis Cdc6 lead to a loss of Mcm origin association (9–14). Although homologs of virtually all of the proteins implicated in DNA replication initiation have been identified in mammals, the analysis of these counterparts has been hampered by the lack of in vivo experimental systems. In addition, many of the regulatory mechanisms that target replication factors may not be strictly conserved across species. For instance, phosphorylation of Saccharomyces cerevisiae Cdc6 (and the corresponding Schizosaccharomyces pombe Cdc18 protein) by cdks leads to proteolytic degradation (15–18), whereas in X. laevis oocyte extracts and in mammalian cells, cdk phosphorylation of Cdc6 leads to export of the protein from the nucleus to the cytoplasm (19–21). Furthermore, the peak expression time of the yeast Cdc6 homologs appears to be early G1 whereas human Cdc6 is specifically degraded in G1 and accumulates to highest levels in G2/M (22, 23). For these reasons, it is clearly important to extend the investigation of the function and regulation of mammalian Cdc6 in its native setting. We have made use of recombinant adenovirus vectors to examine Cdc6 function in intact quiescent mammalian cells. Adenovirus-mediated expression is a particularly powerful tool because of the ability of the virus to infect an entire population of cells under both growing and quiescent conditions. Using this approach, we provide evidence that Cdc6 is not only necessary for Mcm chromatin association but is sufficient to induce endogenous Mcm chromatin loading in serum-deprived cells. We also show that Cdc6 synergizes with limiting amounts of cyclin E/Cdk2 to induce semiconservative DNA replication, and we use this system to begin to identify domains of Cdc6 critical for this function.
Materials and Methods
Cells and Viruses.
REF52 cells were grown in DMEM and brought to quiescence as described (24). Infections were carried out in DMEM plus 25 mM Hepes, pH 7.5 for 75 min at 37°C in 20 μl/cm2. Adenoviral stocks were purified and maintained as described (25). The Ad-CycE, Ad-Cdk2 (containing a hemagglutinin epitope tag), Ad-p21, and Ad-E2F2 viruses have been described (24, 26). Ad-CycA, Ad-Cdc6Myc, and its corresponding mutants were generated by the method of He et al. (27). The details of these constructions are available on request. Viral titers were determined as described (25) by using anti-72-kDa Abs (generous gift of A. Levine, Princeton Univ.). REF52 cells are less efficiently infected by adenovirus than 293 cells; thus, a working viral titer was determined by comparing flow cytometric analysis of 293 cells vs. REF52 cells by using multiple green fluorescent protein (GFP)-expressing viruses.
Measurements of DNA Synthesis.
Cells were labeled with 50 μM BrdUrd for 20–24 h followed by anti-BrdUrd immunofluorescence as described (28). A total of at least 300 nuclei taken from multiple fields chosen at random were counted for each sample. Density shift assays were performed as follows: Cells were labeled with 65 μM BrdUrd and 4 μCi/ml [3H]thymidine (DuPont/NEN) for 24 h before harvest. Nuclei were prepared by hypotonic lysis in 10 mM Hepes (pH 7.5), 10 mM KCl, 0.05% Nonidet P-40, and 10 mM EDTA. DNA was extracted by digestion with 0.3 mg/ml Pronase (Roche Molecular Biochemicals) and 25 μg/ml RNaseA (Roche Molecular Biochemicals), 1.5% Sarkosyl in 50 mM Tris (pH 8.0), 100 mM NaCl, and 25 mM EDTA. Cesium chloride (CsCl) solution was added directly to the samples and adjusted with water to a refractive index of 1.4035. Samples were centrifuged in a type 65 rotor at 40,000 rpm (25°C) for at least 44 h; fractions were collected from the bottoms of the tubes and analyzed for refractive index. Newly synthesized DNA was detected by spotting 150-μl aliquots of the gradient fractions onto glass fiber filters, precipitation with cold 5% trichloroacetic acid, and then 95% EtOH followed by scintillation counting.
Abs.
Anti-hemagglutinin (y-11), anti-Cdc6 (sc-9964), and anti-cyclin E Ab (M-20) were purchased from Santa Cruz Biotechnology. Anti-Mcm2 (BM28) was purchased from Transduction Laboratories, Lexington, KY. Anti-BrdUrd Ab was purchased from Amersham Pharmacia. Rabbit polyclonal anti-Mcm antiserum has been described (24). This antiserum recognizes endogenous Mcm3 with high affinity and endogenous Mcm5 and/or Mcm7 with lower affinity (these proteins migrate identically by SDS/PAGE). In all experiments protein concentrations were determined, and equal amounts of protein were subjected to SDS/PAGE and confirmed by Ponceau S staining of the immobilized proteins.
Chromatin-Binding Assays.
REF52 cells (4 × 105 cells) were harvested by trypsinization and lysed in 1 ml of CSK buffer (28) containing 0.5% Triton X-100, 1 mM ATP, 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 2 μg/ml pepstatin A, 10 μg/ml leupeptin, 1 mM sodium orthovanadate, 25 mM β-glycerol phosphate, and 5 μg/ml phosvitin. Lysates were incubated on ice for 20 min and then centrifuged at 300 × g for 5 min at 4°C. Chromatin pellets were washed with 1 ml of lysis buffer for 5 min on ice and centrifuged again. For nuclease digestions, chromatin pellets were resuspended in lysis buffer plus 1 mM CaCl2 and 30 units of micrococcal nuclease (Roche Molecular Biochemicals) and then incubated at 37°C for 10 min followed by chilling on ice and centrifugation. Immune-complex kinase assays were performed as described (29).
Results
Cdc6 Induces Mcm Chromatin Association in Quiescent Cells.
To explore the function of mammalian Cdc6, we constructed an adenoviral vector that expresses human Cdc6 under the control of the constitutive cytomegalovirus promoter. To distinguish the ectopic Cdc6 from the endogenous protein, five copies of the myc epitope tag were engineered at the 5′ end of the cdc6 cDNA. An additional vector producing native Cdc6 also was constructed and has properties identical to the tagged Cdc6 in the assays we have used (data not shown).
Numerous studies have demonstrated that Cdc6 is necessary for the Mcm complex to associate with origins of DNA replication and that Mcm origin association is required for cells to enter S phase (9, 14, 30–33). Mammalian cells deprived of serum lack Cdc6 and are incompetent to initiate DNA synthesis presumably because, in part, of the fact that Mcm proteins are not associated with chromosomes (34, 35). We wanted to determine whether ectopic expression of Cdc6 in serum-deprived mammalian cells is sufficient to induce the association of endogenous Mcm proteins with chromatin.
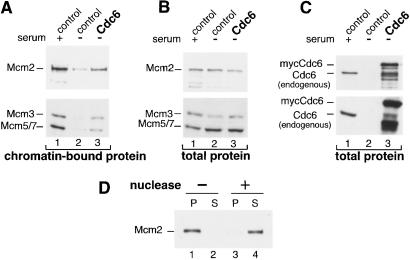
REF52 cells were brought to quiescence by serum deprivation for 48 h and then infected with a control adenovirus expressing GFP (Fig. 1C, lanes 1 and 2) or mycCdc6 (Ad-Cdc6Myc) (Fig. 1C, lane 3). One sample of control-infected cells was induced with serum for 18 h (the time when they typically enter S phase); the other samples were returned to starvation medium. After infection (24 h), all of the cells were harvested and lysed with Triton X-100 in the presence of ATP. Lysates were subjected to low-speed centrifugation, and the pellets were washed to obtain a chromatin-enriched pellet. The proteins were solubilized and separated by SDS/PAGE followed by immunoblot analysis for the endogenous Mcm proteins and for the endogenous and ectopically expressed Cdc6 proteins. As shown in Fig. 1A, the endogenous Mcm2, Mcm3, and Mcm5/7 proteins are associated with the chromatin pellet in serum-stimulated cells but not in quiescent cells (lanes 1 and 2). Strikingly, expression of Cdc6 alone is sufficient to induce a significant amount of these Mcm proteins to become chromatin-associated (Fig. 1A, lane 3). Expression of Cdc6 had no effect on endogenous Mcm protein levels (Fig. 1B). For the sake of simplicity, Mcm2 was used as a marker for Mcm chromatin association in subsequent analyses.
Figure 1.
Expression of Cdc6 in quiescent mammalian cells is sufficient to induce Mcm chromatin association. (A) Chromatin-bound endogenous Mcm proteins. Quiescent REF52 cells were infected with either control virus expressing GFP or mycCdc6 [multiplicity of infection (moi) of 15 focus-forming units (ffu)/cell each]. Cells were then serum-stimulated for 18 h (lane 1) or returned to starvation medium (lanes 2 and 3). Cells were harvested 24 h after infection and fractionated to obtain a chromatin-enriched pellet as described in Materials and Methods. Equal amounts of total protein were analyzed by immunoblotting with anti-Mcm2, or in a separate experiment, an anti-Mcm Ab that recognizes endogenous Mcm3 and Mcm5 and/or Mcm7. (B) Expression of endogenous Mcm proteins. Immunoblot analysis of total cell extracts from A before fractionation. (C) Expression of endogenous and ectopic Cdc6. The same immunoblots as B probed with anti-Cdc6 Ab. (D) Nuclease releases Mcm2 protein from the chromatin pellet. Chromatin-bound proteins prepared as in A were treated with buffer alone (lanes 1 and 2) or with micrococcal nuclease (lanes 3 and 4) and further separated into pellet (P) and supernatant (S) fractions. Identical results were obtained when Mcm3 and Mcm5/7 proteins were examined (data not shown).
To confirm that the presence of Mcm2 in the pellet fraction was caused by association with DNA, a duplicate sample of serum-starved cells expressing Cdc6 was lysed as before but then divided into halves. One half was incubated in lysis buffer, and the other half was incubated in the same buffer plus micrococcal nuclease. The samples were centrifuged again to obtain pellet and supernatant fractions. As shown in Fig. 1D, nuclease treatment nearly quantitatively released Mcm2 into the supernatant, further indicating that Cdc6 induces association of Mcm proteins with chromatin.
Cdc6 Collaborates with Cyclin E/Cdk2 to Induce Semiconservative DNA Replication.
Origins of DNA replication have not been adequately characterized in vertebrate organisms to allow us to determine whether Cdc6 induces the association of mammalian Mcm proteins with presumptive origins. To provide evidence that the observed Mcm chromatin loading was authentic, we wanted to demonstrate that expression of Cdc6 in quiescent cells stimulates DNA synthesis. We infected serum-deprived REF52 cells with the adenovirus expressing Cdc6 and measured the nuclear incorporation of BrdUrd by indirect immunofluorescence. As shown in Fig. 2A even at the highest multiplicities tested, Cdc6 alone could not induce DNA synthesis.
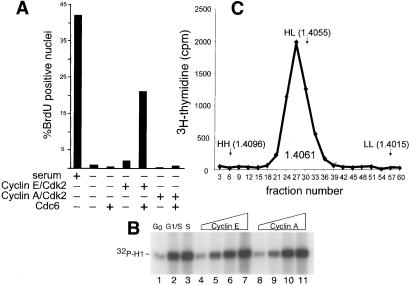
Figure 2.
DNA replication is synergistically induced by coexpression of Cdc6 with cyclin E/Cdk2. (A) Quiescent REF52 cells were infected with Ad-GFP (serum and −, moi of 16 ffu/cell). Ad-Cdc6Myc (moi of 6 ffu/cell), Ad-CycE plus Ad-Cdk2 (moi of 5 ffu/cell each), or Ad-CycA (moi of 1 ffu/cell) plus Ad-Cdk2 (moi of 5 ffu/cell). Cells were subsequently labeled with BrdUrd for 24 h and then stained with anti-BrdUrd Ab and 4′,6-diamidino-2-phenylindole. At least 300 nuclei from randomly chosen microscopic fields were scored for BrdUrd incorporation. (B) Histone H1 kinase activity produced from adenoviral vectors in quiescent cells. Quiescent REF52 cells were infected with Ad-Cdk2 plus control virus (moi of 20 ffu/cell each, lanes 1–3), increasing amounts of Ad-Cdk2 plus Ad-CycE (moi of 2.5, 5, 10, or 20 ffu/cell, lanes 4–7) or Ad-Cdk2 (moi of 2.4, 5, 10, or 20 ffu/cell) plus Ad-CycA (moi of 0.5, 1, 2, or 4 ffu/cell). Control cells were held in starvation medium (lane 1) or stimulated with serum in the presence of hydroxurea for 20 h (lane 2) or released from the arrest for 6 h (lane 3). Cells expressing cyclin E or cyclin A were kept in starvation medium. Cdk2 histone H1 kinase activity was determined by immune-complex kinase assay. (C) DNA synthesis induced by Cdc6 plus cyclin E/Cdk2 in quiescent cells reflects authentic DNA replication. Quiescent REF52 cells were infected with Ad-Cdc6Myc plus Ad-CycE/Ad-Cdk2 as in A. Cells were labeled with BrdUrd and [3H]thymidine for 24 h after infection and then lysed and fractionated by CsCl density gradients as described in Materials and Methods. The refractive index of each fraction was measured, and DNA synthesis was detected by scintillation counting. The positions of DNA taken from quiescent control cells (LL), or cells that had undergone one (HL) or multiple rounds (HH) of S phase in response to serum are marked by arrows, and the refractive index of the peak of DNA synthesis is indicated.
We have previously shown that expression of E2F in conjunction with limiting amounts of cyclin E/Cdk2 leads to a complete round of DNA replication in serum-deprived REF52 cells (24). Because Cdc6 is one of many DNA replication activities controlled by E2F (36–38), we tested whether Cdc6 could collaborate with cyclin E/Cdk2 to induce DNA synthesis. When very low doses of cyclin E and Cdk2 were used to infect serum-deprived cells, very little BrdUrd incorporation was observed by indirect immunofluorescence (Fig. 2A). However, coexpression of Cdc6 with cyclin E/Cdk2 resulted in a 10-fold increase in the number of BrdUrd-positive nuclei. The synergistic effect of coexpression of Cyc6 with cyclin E/Cdk2 was consistently observed in multiple experiments; a representative experiment in shown in Fig. 2A. DNA synthesis was not observed when Cdc6 was expressed with a catalytically inactive form of Cdk2 (data not shown).
In contrast to the effects of cyclin E/Cdk2 plus Cdc6, cyclin A/Cdk2 did not collaborate with Cdc6 to induce DNA synthesis at any viral dose (Fig. 2A). This defect was not caused by a deficiency in cyclin A-dependent kinase activity as shown by the assays in Fig. 2B. Quiescent REF52 cells were infected with Cdk2 virus plus control virus (Fig. 2B, lanes 1–3) or with increasing doses of viruses expressing either cyclin E (Fig. 2B, lanes 4–7) or cyclin A (Fig. 2B, lanes 8–11). Cells were harvested 24 h after infection, and the Cdk2 protein was immunoprecipitated from cell lysates and analyzed for histone H1 kinase activity. Quantification of the phosphorylated histone band indicated that these doses of cyclin E and cyclin A viruses stimulated identical amounts of Cdk2 activity. We further note that the amount of cyclin E/Cdk2 and cyclin A/Cdk2 activity present in cells analyzed in Fig. 2A is slightly less than that seen in serum-stimulated cells at G1/S (Fig. 2B, lane 2) or S/G2 (Fig. 2B, lane 3), and thus the kinase activity is not overproduced. We therefore conclude that Cdc6 specifically collaborates with cyclin E/Cdk2 and not cyclin A/Cdk2 to induce DNA synthesis.
Based on flow-cytometric analysis, Cdc6 plus cyclin E/Cdk2 did not induce quiescent cells to accumulate a G2 DNA content (data not shown.) This result is consistent with our earlier report that adenovirus-mediated expression of E2F alone in quiescent REF52 cells induces significant incorporation of BrdUrd and yet is insufficient to stimulate a complete round of DNA replication (24). In fact, a subpopulation of BrdUrd-positive nuclei from Cdc6 and cyclin E/Cdk2-expressing cells displayed a punctate rather than uniform staining pattern similar to very early S phase cells (J.G.C., unpublished observations). To confirm that the BrdUrd incorporation reflected authentic DNA replication, we performed CsCl density-shift experiments. Quiescent REF52 cells were infected with viruses expressing both Cdc6 and cyclin E/Cdk2 as before and were labeled with both BrdUrd and [3H]thymidine. DNA preparations from these cells were fractionated on a CsCl gradient as described in Materials and Methods. In multiple experiments, all of the incorporated [3H]thymidine incorporated banded at a density of 1.405–1.406, which is the density of the normally replicated (HL) control DNA (Fig. 2C). Furthermore, the method for preparing DNA for these experiments produced large fragments (>15 kb) as determined by agarose gel electrophoresis (data not shown). Because the newly replicated DNA quantitatively shifted to the HL position of the gradient, it appears that DNA synthesis proceeded for at least several kilobases. Therefore, despite the fact that the DNA synthesis was inefficient, we conclude that Cdc6 loading of Mcm proteins is a precursor to semiconservative DNA replication, even in quiescent cells.
Prereplication Complex Formation Occurs in the Absence of cdk Activity.
Human Cdc6 contains three consensus cdk phosphorylation sites within an amino terminal domain, and several studies have demonstrated that phosphorylation of Cdc6, most likely by cyclin A/Cdk2, leads to its export from the nucleus to the cytoplasm, presumably to prevent reinitiation of replication in G2 (19–21). In addition, Herbig et al. (39) and Jiang et al. (21) also have suggested that phosphorylation of Cdc6 may play a positive role in Cdc6 function because overproduction or microinjection of a mutationally altered Cdc6 that cannot be phosphorylated by cdks blocked DNA replication. In contrast, Petersen et al. (20) and Pelizon et al. (19) performed similar experiments but observed no consequences of the lack of Cdc6 phosphorylation other than a failure to relocalize Cdc6 during S phase.
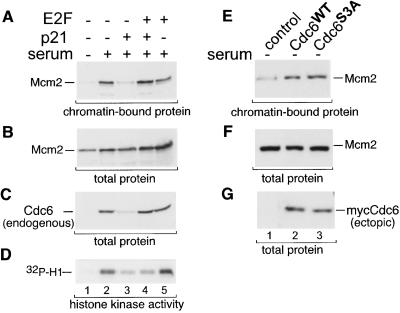
In the experiment shown in Fig. 1, Cdc6 is expressed under conditions in which endogenous cdk activity is very low, and Cdc6 expression by this method does not induce Cdk2 activity (J.G.C., unpublished results). It thus seemed that Cdc6 was not being phosphorylated in quiescent cells, and yet Cdc6 could induce Mcm chromatin association. To directly assess the role of Cdc6 phosphorylation in the loading of Mcm proteins, we infected quiescent REF52 cells with control virus or an adenovirus expressing the p21 cdk inhibitor and then stimulated the cells by serum addition in the presence of aphidicolin to block cells in early S phase. As shown in Fig. 3A, expression of p21 prevented Mcm2 chromatin association (lane 3) and also reduced the Cdk2-associated histone H1 kinase activity (Fig. 3D, lane 3). There was no effect of p21 expression on Mcm2 expression (Fig. 3B, lane 3). However, endogenous Cdc6 expression also was inhibited (Fig. 3C, lane 3) because of the fact that the cdc6 gene is under control of the cdk/Rb/E2F pathway and cdk activity was effectively blocked. (No ectopic Cdc6 was used for this experiment.) To bypass the need for cyclin E/Cdk2 activity to express the endogenous cdc6 gene, we coinfected cells with viruses expressing both p21 and E2F2. Under these conditions, Cdc6 expression was maintained (Fig. 3C, lane 4), although Cdk2 activity was still low (Fig. 3D, lane 4). Importantly, Mcm2 chromatin association also was maintained at levels equal to those seen in serum-stimulated cells with full Cdk2 activity (Fig. 3A, compare lanes 2 and 4). This result suggests that Cdc6 has the ability to induce Mcm chromatin loading independent of cdk activity.
Figure 3.
Cdk activity is required for Cdc6 expression but not Mcm chromatin loading. (A) Chromatin-bound Mcm2 protein. Quiescent REF52 cells were infected with control virus (moi of 5 ffu/cell), or viruses expressing p21 (moi of 5 ffu/cell) or E2F2 (moi of 2 ffu/cell) as indicated, and then returned to starvation medium (lane 1) or stimulated with serum in the presence of 6 μM aphidicolin (lanes 2–5). Chromatin-enriched fractions were prepared as in Fig. 1A. (B) Expression of endogenous Mcm2. Immunoblot analysis of total cell extracts from A before fractionation. (C) Expression of endogenous Cdc6 protein. The same immunoblot as in B probed with anti-Cdc6 Ab. (D) Histone H1 phosphorylation. Endogenous Cdk2 was immunoprecipitated from the same extracts in A–C, and the immunoprecipitates were analyzed for histone H1 kinase activity.
To unequivocally establish that cdk-mediated phosphorylation of Cdc6 is not required for Mcm chromatin loading, we constructed a mutationally altered form of Cdc6 in which the three consensus cdk phosphorylation sites (S54, S74, and S106) have been changed to alanines. Using indirect immunofluorescence, we confirmed previous findings that this mutant form of Cdc6 (Cdc6S3A) is not exported from the nucleus during G2 whereas normal Cdc6 is relocalized from the nucleus to the cytoplasm after S phase (data not shown). Cdc6S3A was expressed from an adenoviral vector at a level similar to normal Cdc6 (Fig. 3G, lanes 2 and 3) and showed a similar ability to induce the chromatin association of Mcm2 (Fig. 3F, lanes 2 and 3). Therefore Cdc6 does not require phosphorylation by cdks to perform its function in Mcm chromatin loading.
The ATPase Domain of Cdc6 Is Required for Prereplication Complex Formation and DNA Replication.
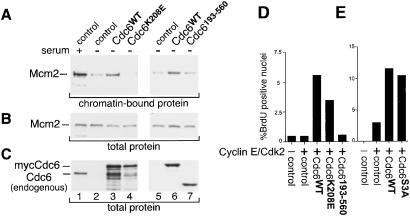
The carboxyl-terminal two-thirds of the Cdc6 protein consists of an evolutionarily conserved ATPase domain. Mutational alterations in this domain that inhibit ATP binding are unable to complement a cdc6 null mutant in yeast (10, 11, 40). Furthermore, microinjection of purified Cdc6 protein that is unable to bind or hydrolyze ATP blocks serum-stimulated DNA replication in mammalian cells (41). We wanted to determine whether ATP binding is required for the human Cdc6 protein to induce Mcm chromatin loading in quiescent cells. To do so, we constructed an additional adenovirus vector that produces Cdc6 in which the ATP-binding domain, also termed the Walker A motif, has been altered (Cdc6K208E). A similarly constructed mutation was confirmed to have the predicted biochemical properties (41). As shown in Fig. 4A (lane 4), Cdc6 that is unable to bind ATP (Cdc6K208E) also is severely impaired in the ability to load Mcm proteins on chromatin in quiescent cells. This result is consistent with previous findings in S. cerevisiae and S. pombe that a similar mutation is unable to complement a cdc6 deletion or load Mcm proteins onto origins.
Figure 4.
The catalytic domain of Cdc6 is required but not sufficient for Mcm chromatin binding and DNA replication. (A) Chromatin-bound endogenous Mcm2 protein. Quiescent REF52 cells were infected at an moi of 15 ffu/cell with either control virus (lanes 1, 2, and 5), virus expressing wild-type mycCdc6 (lanes 3 and 6), Cdc6 that cannot bind ATP (Cdc6K208E, lane 4), or Cdc6 lacking the noncatalytic amino terminus (Cdc6193–560, lane 7). Cells were harvested and chromatin-enriched pellets were prepared as in Fig. 1A. (B) Expression of endogenous Mcm2 in whole-cell extracts before fractionation. (C) Expression of Cdc6. The same samples as in B probed with anti-Cdc6 (Left) or with anti-myc Ab (Right). (D) Stimulation of DNA replication by wild-type and mutationally altered Cdc6 in collaboration with cyclin E/Cdk2. Quiescent REF52 cells were infected with the same Cdc6 viruses as in A plus viruses expressing cyclin E and Cdk2 (moi of 5 ffu/cell each) as indicated. The control is virus expressing GFP. Nuclear BrdUrd incorporation was evaluated as in Fig. 2A.
Sequences in the noncatalytic amino-terminal domain of Cdc6 are required for proteolytic degradation (23) and cdk-dependent nuclear export (19–21). We constructed an adenovirus that expresses myc-tagged Cdc6 lacking the first 192 amino acids (Cdc6193–560). Because this deletion also removed residues required for nuclear localization, a synthetic nuclear localization signal was fused between the tags and amino acid 193 of Cdc6, and the nuclear localization of this protein was confirmed by indirect immunofluorescence (data not shown). As shown in Fig. 4A (lane 7), expression of the amino-terminal deletion mutant failed to induce Mcm chromatin association. We thus conclude that both the ATPase domain of Cdc6 and the amino-terminal domain are required for Mcm chromatin loading.
Given that Cdc6 can stimulate DNA replication if the cells also are expressing cyclin E/Cdk2, we tested whether the mutant forms of Cdc6 that had been analyzed for Mcm chromatin loading also were functional in this assay. We expressed similar amounts of normal Cdc6, Cdc6K208E, Cdc6193–560, and Cdc6S3A in serum-deprived cells coexpressing cyclin E/Cdk2. Once again, expression of normal Cdc6 stimulated DNA replication in collaboration with cyclin E/Cdk2. However, Cdc6K208E, which was severely impaired in inducing Mcm chromatin loading, also was reproducibly reduced in the ability to induce DNA replication. Cdc6K208E consistently produced 63.5% (±3.1%) of the replication-inducing activity of normal Cdc6. In contrast, Cdc6193–560 was entirely unable to induce DNA replication even to the partial level observed with Cdc6K208E, indicating that the catalytic domain of Cdc6 alone is nonfunctional for both Mcm chromatin loading and replication initiation. Consistent with the chromatin loading results shown in Fig. 3D, Cdc6S3A was capable of collaborating with cyclin E/Cdk2 to the same extent as normal Cdc6 (Fig. 4E). Taken together, these results point to an essential replication activity in the amino-terminal domain of Cdc6 and an important role for nucleotide binding by Cdc6, but phosphorylation of Cdc6 itself is not likely to function in replication initiation.
Discussion
We have taken advantage of the ability of adenovirus vectors to transduce quiescent cells to assess the role of Cdc6 in the formation of DNA replication initiation complexes. Because Cdc6 is not expressed in quiescent cells, we can evaluate the function of mutant forms of Cdc6 in the absence of confounding contributions of the endogenous Cdc6 protein. In a sense, the lack of Cdc6 is complemented by the ectopically expressed Cdc6 in a manner analogous to true genetic complementation. In particular, the effects of Cdc6 expression on endogenous Mcm proteins can be determined in the entire population of cells because virtually all of the cells express the Cdc6 transgene. Because the cells are infected after they are brought to quiescence, we have bypassed any possible effects of transgene expression on exit from the cell cycle, a critical aspect of the system with regard to expression of cyclin E/Cdk2 and cyclin A/Cdk2 that would be difficult to address by transient transfection. This experimental system has the ability to delineate the molecular functions of replication components such as Cdc6 and provides the basis for future study.
Mammalian Cdc6 Induces Mcm Chromatin Association.
We have demonstrated that expression of Cdc6 in quiescent mammalian cells is sufficient to induce significant chromatin association of endogenous Mcm proteins. We also provide evidence that phosphorylation of Cdc6 is not required for Mcm chromatin loading. Previous studies have offered conflicting data on this point with some investigators reporting no consequences (19, 20) whereas others report a dominant interfering effect and suggest that phosphorylation of Cdc6 is required for origin firing (21, 39). Possible explanations for the discrepancies between these studies include differences in the form of Cdc6 that is overproduced, particularly when it is expressed as a fusion protein, or the extent to which Cdc6 is overexpressed. Alternatively, the alteration of the target residues to alanines may have an effect on the structure of Cdc6 independent of phosphorylation. We have shown that expression of unphosphorylatable Cdc6, Cdc6S3A, at a low level from an adenoviral vector, was fully capable of chromatin recruitment of Mcm proteins as well as induction of DNA replication. Moreover, we also have shown that wild-type, endogenous Cdc6 induced by E2F in the presence of a cdk inhibitor is also capable of recruiting Mcm proteins to chromatin, arguing against aberrant effects of a mutant or from gross overexpression. Arata and coworkers (35) also found that Mcm proteins are associated with chromatin in cells treated with the cdk inhibitor, butyrolactone, supporting the conclusion that phosphorylation of Cdc6 was not required for Mcm chromatin loading. Because numerous studies have shown that phosphorylation of Cdc6 leads to its nuclear export, we favor a model in which unphosphorylated Cdc6 is functional in promoting Mcm chromatin association and that cdk phosphorylation of Cdc6 is primarily a mode of negative regulation.
We also provide evidence that nucleotide binding by human Cdc6 is required for Cdc6 to induce Mcm chromatin association, as shown by the fact that the Walker A mutant form of Cdc6 (Cdc6K208E) is impaired in the induction of Mcm chromatin loading. Furthermore, although the amino-terminal noncatalytic domain of Cdc6 is not strongly conserved between higher and lower eukaryotes, we have demonstrated a strict requirement for this region in chromatin loading of Mcm proteins and induction of DNA replication. A recent study (23) has identified sequences within the amino-terminal domain of Cdc6 that are necessary for ubiquitin-mediated proteolysis. In addition, the target residues for cdk-mediated nuclear export of Cdc6 also are located in this region. Both modes of regulation most likely represent negative control of Cdc6; our results point to additional positive functions for this domain. We have not directly assessed nucleotide binding by the human Cdc6193–560 described herein. However, in a recent study, the atomic structure of the Pyrobaculum aerophilum Cdc6 homolog, which lacks this amino-terminal extension entirely, was described (42). P. aerophilum Cdc6, cocrystallized with nucleotide, suggesting that Cdc6193–560 also may be capable of binding ATP/ADP.
Cdc6 Induces Semiconservative DNA Replication in Collaboration with Cyclin E/Cdk2.
A wealth of evidence has accumulated over the years that protein kinase activity is required for the initiation of DNA replication (1, 2, 43). It is thus not surprising that expression of Cdc6 in the absence of cdk activity is unable to induce origin firing. Strikingly, however, Cdc6 expression is sufficient to stimulate DNA synthesis in cooperation with limiting amounts of cyclin E/Cdk2 activity. Despite the fact that the DNA replication induced by Cdc6 plus cyclin E/Cdk2 is limited, analysis of newly synthesized DNA confirmed that the replication was semiconservative and resulted in DNA fragments that were several kilobases in length. We suspect that under our experimental conditions, a small number of origins initiated replication, but because of the low levels of other replication activities such as Orc1, Mcm2–7, Cdc45, the Cdc7/Dbf4 kinase, polymerase, and nucleotide precursors coupled with limiting amounts of cdk activity, replication forks could not efficiently elongate.
Other investigators have reported that Cdc6 overexpression in serum-stimulated cells shortens the G1 phase from 16 to 15 h (23); a similar acceleration was observed when recombinant Cdc6 was added to a cell-free replication assay (34). Hateboer et al. (36) also observed that coexpression of Cdc6 with cyclin E/Cdk2 in proliferating cells caused a decrease in the G1 population and an increase in the number of cells in S phase. However, in all of these assays the effects of added Cdc6 were modest because of the presence of considerable amounts of endogenous Cdc6. Our results using serum-deprived cells provides a greater dynamic range and a more robust analysis. Most importantly, these data show that coexpression of Cdc6 with cyclin E/Cdk2 induces authentic DNA replication and thus serves as a useful system for future study.
Dual Roles for Cyclin E/Cdk2 in the Initiation of DNA Replication.
The best-characterized substrates of cyclin E/Cdk2 are the retinoblastoma family proteins, Rb, p130, and p107. Phosphorylation of Rb by cyclin D/Cdk4 and cyclin E/Cdk2 dissociates Rb from E2F and allows the induction of E2F target genes (44, 45). The synergy between low-level cyclin E/Cdk2 expression and Cdc6 shown in Fig. 2 is only seen when cyclin E/Cdk2 activity is low enough to induce endogenous cdc6 expression minimally (J.G.C., unpublished observations). Thus one function of cyclin E/Cdk2 in the induction of S phase is its well-documented role in transcriptional control of E2F target genes such as cdc6.
However, we demonstrate that the role of cyclin E/Cdk2 in Mcm chromatin loading is restricted to its function in E2F-dependent transcriptional control of the cdc6 gene because expression of Cdc6 in the absence of cdk activity (either by ectopic expression or by induction of the endogenous gene by E2F) bypasses the need for cdk activity in Mcm chromatin loading. Cyclin E/Cdk2 activity is not required for prereplication complex formation as long as Cdc6 is produced.
Although the ATP-binding mutant Cdc6K208E was defective for inducing Mcm chromatin association, this mutant was not entirely defective for inducing DNA replication when coexpressed with cyclin E/Cdk2. The underlying basis for this quantitative discrepancy is not clear but may simply reflect the fact that these two assays (Mcm loading and BrdUrd incorporation) are quite different and need not necessarily show a precise quantitative relationship. For instance, it is possible that a low level of chromatin inducing activity is sufficient to achieve substantial DNA replication. In addition, the chromatin-binding assay may be considerably more stringent in that it measures the level of Mcm proteins stably associated with chromatin at the time of lysis, whereas the replication assay reflects the sum total of BrdUrd incorporation over the course of many hours.
These assays also have demonstrated a role for the amino-terminal domain of Cdc6 that is independent of ATP binding. One possible role is suggested from recent data that has shown Cdc6 to recruit the cyclin E/Cdk2 complex to chromatin via motifs found in the Cdc6 amino-terminal region (46). It may be that Cdc6K208E is fully capable of recruiting cyclin E/Cdk2, particularly in the experiments in which the kinase is coexpressed.
Clearly, cyclin E/Cdk2 plays additional roles in replication initiation downstream of Mcm chromatin loading because Cdc6-mediated Mcm chromatin loading is not sufficient for replication without cyclin E/Cdk2, and Cdk2 activity is still required for initiation in X. laevis extracts in which transcriptional control is not important. At least one of those functions is likely to be the loading of the Cdc45 protein onto the newly formed prereplication complex (35, 47, 48), although the precise mechanism of this aspect of cyclin E/Cdk2 function remains to be elucidated.
Acknowledgments
We thank B. Vogelstein, S. Williams, A. Levine, X.-F. Wang, and H. Kimura for generous gifts of reagents, C. Newlon for advice and helpful discussions, N. Turner for technical support, and K. Culler for assistance in the preparation of the manuscript. J.G.C. was supported by an American Cancer Society postdoctoral fellowship (PF-4465), C.-H. P. was a Leukemia Society of America Fellow, T.W.B. was supported by an American Cancer Society postdoctoral fellowship (PF-99–111-01-CCG), and J.R.N. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviatons: moi, multiplicity of infection; Mcm, minichromosome maintenance; ffu, focus-forming units; GFP, green fluorescent protein, cdk, cyclin-dependent kinase.
References
- 1.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 2.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 3.Newlon C S. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 4.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson A D, Blow J J. Curr Opin Genet Dev. 1999;9:62–68. doi: 10.1016/s0959-437x(99)80009-4. [DOI] [PubMed] [Google Scholar]
- 6.Labib K, Tercero J A, Diffley J F X. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 7.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 8.Yan Y, Merchant A M, Tye B K. Genes Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- 9.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Feng L, Hu Y, Huang S H, Reynolds C P, Wu L, Jong A Y. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- 11.Weinreich M, Liang C, Stillman B. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins G, Diffley F X. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 14.Donovan S, Harwood J, Drury L S, Diffley J F X. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jallepalli P V, Kelly T J. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 16.Baum B, Nishitani H, Yanow S, Nurse P. EMBO J. 1998;17:5689–5698. doi: 10.1093/emboj/17.19.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drury L S, Perkins G, Diffley J F. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez M, Calzada A, Bueno A. J Cell Sci. 1999;112:2381–2390. doi: 10.1242/jcs.112.14.2381. [DOI] [PubMed] [Google Scholar]
- 19.Pelizon C, Madine M A, Romanowski P, Laskey R A. Genes Dev. 2000;14:2526–2533. doi: 10.1101/gad.176300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen B O, Lukas J, Sorensen C S, Bartek J, Helin K. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang W, Wells N J, Hunter T. Proc Natl Acad Sci USA. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendez J, Stillman B. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen B O, Wagener C, Marinoni F, Kramer E R, Melixetian M, Denchi E L, Gieffers C, Matteucci C, Peters J-M, Helin K. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leone G, DeGregori J, Jakoi L, Cook J G, Nevins J R. Proc Natl Acad Sci USA. 1999;96:6626–6631. doi: 10.1073/pnas.96.12.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevins J R, DeGregori J, Jakoi L, Leone G. Methods Enzymol. 1997;283:205–219. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- 26.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He T-C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todorov I T, Attaran A, Kearsey S E. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 30.Romanowski P, Madine M A, Laskey R A. Proc Natl Acad Sci USA. 1996;93:10189–10194. doi: 10.1073/pnas.93.19.10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang C, Stillman B. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thömmes P, Kubota Y, Takisawa H, Blow J J. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 34.Stoeber K, Mills A D, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey R A, Williams G H. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arata Y, Fujita M, Ohtani K, Kijima S, Kato J Y. J Biol Chem. 2000;275:6337–6345. doi: 10.1074/jbc.275.9.6337. [DOI] [PubMed] [Google Scholar]
- 36.Hateboer G, Wobst A, Petersen B O, Cam L L, Vigo E, Sardet C, Helin K. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtani K, Tsujimoto A, Ikeda M, Nakamura M. Oncogene. 1998;17:1777–1785. doi: 10.1038/sj.onc.1202105. [DOI] [PubMed] [Google Scholar]
- 38.Yan Z, DeGregori J, Shohet R V, Leone G, Stillman B, Nevins J R, Williams R S. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbig U, Griffith J W, Fanning E. Mol Biol Cell. 2000;11:4117–4130. doi: 10.1091/mbc.11.12.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeRyckere D, Smith C L, Martin G S. Genetics. 1999;151:1445–1457. doi: 10.1093/genetics/151.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbig U, Marlar C A, Fanning E. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Smith C L, DeRyckere D, DeAngelis K, Martin G S, Berger J M. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 43.Bousett K, Diffley J F X. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 45.Nevins J R. Cell Growth Diff. 1998;9:585–593. [PubMed] [Google Scholar]
- 46.Furstenthal L, Kaiser B K, Swanson C, Jackson P K. J Cell Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 48.Zou L, Stillman B. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]