Abstract
Dual inactivation of PTEN and INK4a/ARF tumor suppressor genes is a common feature observed in a broad spectrum of human cancer types. To validate functional collaboration between these genes in tumor suppression, we examined the biological consequences of Pten and/or Ink4a/Arf deficiency in cells and mice. Relative to single mutant controls, Ink4a/Arf−/−Pten+/− mouse embryonic fibroblast cultures exhibited faster rates of growth in reduced serum, grew to higher saturation densities, produced more colonies upon low density seeding, and showed increased susceptibility to transformation by oncogenic H-Ras. Ink4a/Arf deficiency reduced tumor-free survival and shortened the latency of neoplasias associated with Pten heterozygosity, specifically pheochromocytoma, prostatic intraepithelial neoplasia, and endometrial hyperplasia. Compound mutant mice also exhibited an expanded spectrum of tumor types including melanoma and squamous cell carcinoma. Functional synergy between Ink4a/Arf and Pten manifested most prominently in the development of pheochromocytoma, prompting an analysis of genes and loci implicated in this rare human neoplasm. The classical pheochromocytoma genes Ret, Vhl, and Nf-1 remained intact, a finding consistent with the intersection of these genes with pathways engaged by Pten and Ink4a/Arf. Notably, conventional and array-comparative genomic hybridization revealed frequent loss of distal mouse chromosome 4 in a region syntenic to human chromosome 1p that is implicated in human pheochromocytoma. This study provides genetic evidence of collaboration between Pten and Ink4a/Arf in constraining the growth and oncogenic transformation of cultured cells and in suppressing a wide spectrum of tumors in vivo.
The PTEN (also known as MMAC1 and TEP1) gene encodes a phosphatase that negatively regulates the phosphatidylinositol 3-kinase (PI3K) pathway activity (1, 2). PI3K activates a variety of key signaling proteins such as the Ser/Thr kinase AKT. Activated AKT in turn phosphorylates and modulates the activity of a number of important molecules governing cell cycle control and cell survival including forkhead transcription factors and BAD, among other substrates (1, 2). Consistent with the role of Pten as a signaling antagonist of the PI3K pathway, Pten−/− embryonic stem cells exhibit an increased growth rate and an accelerated G1/S transition. In addition, its reintroduction in PTEN-deficient tumor cells down-modulates AKT activity and induces cell cycle arrest and/or apoptosis (2). In the mouse, Pten nullizygosity leads to early embryonic lethality, whereas Pten heterozygotes survive and develop neoplasia in multiple tissues including lymphoid and epithelial hyperplasias and cancers of the prostate, endometrium, intestine, thyroid, adrenal gland, and breast (3–7).
The Ink4a/Arf gene encodes two distinct tumor suppressors, p16INK4a and p19ARF, that function as regulators of the pRB and p53 pathways, respectively (8). p16INK4a and other members of the INK4 family inhibit G1 cyclin D-dependent kinases 4 and 6, thereby preventing CDK4/6-directed pRB hyperphosphorylation and blocking S phase entry. p19ARF inhibits MDM2-mediated degradation of p53 and plays an important role in the apoptotic elimination of aberrantly cycling cells (8). Mice doubly null for p16INK4a and p19ARF are viable but succumb to lymphomas or sarcomas with median latency of ≈30 weeks (9). Relative to wild-type (wt) control cultures, Ink4a/Arf−/− mouse embryonic fibroblasts (MEFs) grow more rapidly, exhibit a high rate of colony formation and immortalization, and are susceptible to transformation by oncogenic H-Ras alone (9).
The PTEN and INK4a/ARF tumor suppressor genes are among the most frequently inactivated genes in human cancer (10, 11). Loss of PTEN function is common in glioblastoma, melanoma, endometrial carcinoma, prostate adenocarcinoma, renal cell carcinoma, and head and neck squamous carcinoma (10). PTEN mutations have also been detected in sporadic cancers of the breast, thyroid, lung, stomach, and hematopoietic systems (10). In addition, germ-line mutations of PTEN underlie three overlapping human autosomal-dominant hamartoma tumor syndromes: Cowden syndrome, Bannayan–Zonana syndrome, and Lhermitte–Duclos disease (2, 10). A wide spectrum of human cancer types also exhibit INK4a/ARF inactivation by mutation, deletion, or epigenetic silencing, particularly in malignant gliomas, melanoma, head and neck squamous carcinoma, and lymphoblastic leukemia (8, 11, 12). It is notable that dual inactivation of PTEN and INK4a/ARF tumor suppressor genes is encountered in several human cancer types, a mutational profile implying functional collaboration between these tumor suppressors. Indeed, this possible synergy is consistent with RAS activation and Ink4a/Arf loss in melanoma genesis in the mouse (13), coupled with the well established biochemical interactions between RAS and PTEN pathways (14). In this study, we examined the potential collaborative interactions between these prominent tumor suppressors on the cellular and organismal levels.
Materials and Methods
Cellular Assays for Growth and Transformation.
Cellular assays were performed as described previously (9, 15, 18). For growth curves and low density seeding assays, early passage (PD ≤ 9) MEFs were cultured in DMEM containing 4% FCS.
Mutant Mouse Tumor Studies.
The production of Pten and Ink4a/Arf mutant mice has been described elsewhere (7, 9) and maintained on a mixed FVB/n C57BL/6 background. Mice heterozygous for Pten and Ink4a/Arf mutant alleles were intercrossed to generate all of the genotypes analyzed in this study.
Histopathology and Immunohistochemistry.
Normal and tumor tissue samples were processed for immunohistochemical analysis by standard techniques. EPOS anti-neuron-specific enolase (Dako) and with anti-chromagranin A antibody (DiaSorin, Stillwater, MN) were used according to the manufacturer's instructions. Diaminobenzadine was used as the chromogen and hematoxylin as the counterstain. Fontana–Mason and iron staining of tissues were performed according to standard procedures.
DNA Isolation, Slot Blot, Southern Blot, Comparative Genomic Hybridization (CGH), and Array-CGH.
DNA for Slot blot, Southern blot, and array-CGH was prepared by the Purogene DNA isolation system (Gentra Systems) following manufacturer's procedures. DNA for conventional CGH was extracted from paraffin-embedded blocks as described previously (16). Southern blot analysis for the loss of heterozygosity of Ink4a/Arf and Pten genes was done as described elsewhere (7, 9). Slot blot and CGH were performed as described previously (16). The array-CGH experiments were performed by using SpectralChip arrays (Spectral Genomics, Houston) according to manufacturer's protocols.
Results and Discussion
Pten Status Modulates the Growth and Transformation of Ink4a/Arf Null MEF Cultures.
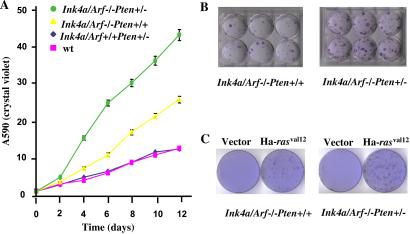
The loss of either Ink4a/Arf or Pten gene function has been shown to exert profound effects on the growth, survival, and oncogenic susceptibility of cultured cells (1, 2, 8, 9, 17). To assess potential cooperative interactions on the cellular level, we examined the compound effects of Pten heterozygosity and Ink4a/Arf nullizygosity. Independently derived early-passage Ink4a/ Arf−/−Pten+/− and Ink4a/Arf−/−Pten+/+ MEFs were found to be indistinguishable with respect to morphology, growth rates, and saturation densities under standard culture conditions (data not shown). However, when grown in reduced serum conditions (4% rather than 10%), Ink4a/Arf−/− Pten+/− MEFs proliferated more rapidly, attained higher saturation densities, and generated 2.5-fold more colonies following low density seeding (P < 0.001) relative to Ink4a/Arf−/−Pten+/+ controls (Fig. 1 A and B). Under the same conditions, no differences in cellular growth rates and colony formation were detected between Ink4a/Arf+/+Pten+/− and wt MEFs (Fig. 1A). Additionally, H-RAS (G12V) transduction generated a 2- to 3-fold increase in the number of transformed foci (foci that emerged more rapidly and attained a larger size) in cultures derived from Ink4a/Arf−/−Pten+/− embryos relative to Ink4a/Arf−/−Pten+/+ controls (Fig. 1C and data not shown, P < 0.001). In contrast, H-RAS (G12V) transduction failed to generate transformed foci in Ink4a/Arf+/+Pten+/− and Ink4a/Arf+/+Pten+/+ MEFs (data not shown). Together, these findings suggest that Pten haploinsufficiency enhances the proliferative potential and oncogenic susceptibility conferred by loss of Ink4a/Arf function; the high resistance of INK4a/Arf−/− cells to apoptotic stimuli (8) does not allow us to exclude a modest improvement in the survival potential brought about by PTEN haploinsufficiency.
Figure 1.
Growth potential of Ink4a/Arf−/−Pten+/+ and Ink/Arf4a−/−Pten+/− MEFs. (A) Growth curve of MEFs. MEFs were plated at 25,000 cells per well in triplicate wells of 12-well plates and cultured for the indicated number of days. Cells were fixed and stained with crystal violet. Adsorption at OD 540 was measured for each time point for individual MEF lines and directly correlates with cell numbers (15). Each symbol represents the mean of triplicate readings of six lines. The standard errors are indicated. (B) Colony formation in MEFs. MEFS were plated at 3,500 cells per 6-well plate, cultured for 12 days, and fixed and stained with crystal violet. Representative plates are shown. (C) Transformation of MEFs. Neoplastic transformation foci of MEFs with Ha-rasVal12. Control plates were transfected with an empty vector.
Impact of Ink4a/Arf Mutation on the Cancer Phenotype of Pten Heterozygous Mice.
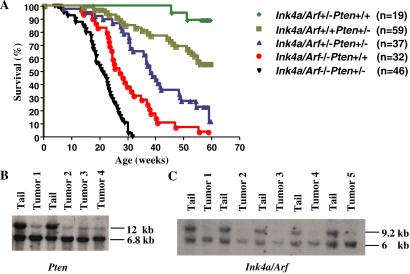
The cell culture-based observations above, along with the co-occurrence of PTEN and INK4a/ARF inactivation in certain human cancer types (2, 8, 10, 11), prompted a detailed analysis of the cancer phenotype of mice mutant for both Pten and Ink4a/Arf. As summarized in Table 1, Ink4a/Arf deficiency increased the penetrance and reduced the latency of cancers associated with Pten heterozygosity. During an observation period of 59 weeks, 100% of the Ink4a/Arf+/+Pten+/+ littermates (n = 23) remained tumor-free, whereas 2 of 19 Ink4a/Arf+/−Pten+/+ mice developed lymphoma or s.c. spindle cell sarcoma at 45 and 51 weeks of age, respectively (data not shown). In line with previous studies (4, 7, 9), we observed mean tumor-free survivals of 28.6 ± 0.7 and 52 ± 1.5 weeks for Ink4a/Arf−/−Pten+/+ and Ink4a/Arf+/+Pten+/− control mice. In contrast, Ink4a/Arf+/−Pten+/− and Ink4a/Arf−/−Pten+/− mice succumbed with shortened latencies of 38 ± 1.1 weeks and 19.2 ± 0.6 weeks, respectively. Notably, all Ink4a/Arf−/− Pten+/− mice died by 32 weeks of age (Fig. 2A).
Table 1.
Tumorigenesis in Ink4a/Arf+/+Pten+/−, Ink4a/Arf−/−Pten+/+, Ink4a/Arf+/−Pten+/−, and Ink4a/Arf−/−Pten+/− mice
| Tumor type | Genotype (numbers)
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Ink4a/Arf+/+Pten+/− (n = 59)
|
Ink4a/Arf−/−Pten+/+ (n = 32)
|
Ink4a/Arf+/−Pten+/− (n = 37)
|
Ink4a/Arf−/−Pten+/− (n = 46)
|
|||||
| No. | Week onset (mean) | No. | Week onset (mean) | No. | Week onset (mean) | No. | Week onset (mean) | |
| Adrenal pheochromocytoma | 14 | 37–59 (42) | 21 | 15–59 (30) | 27 | 7–28 (24) | ||
| Cutaneous melanoma | 4 | 38–52 (46.5) | 3 | 28–31 (29) | ||||
| Peritoneal carcinomatosis/SCC | 3 | 28–39 | 2 | 26, 28 | ||||
| Prostatic hyperplasia | 4 | 36–55 (49) | 6 | 27–57 (40) | 4 | 18–30 (27) | ||
| Endometrial hyperplasia | 10 | 32–57 (46) | 9 | 28–52 (39) | 5 | 22–32 (29) | ||
| Lymphoma | 21 | 2–59 (40) | 11 | 18–56 (35) | 21 | 13–59 (29) | 34 | 7–31 (16) |
| Pulmonary adenocarcinoma | 3 | 39–59 | 2 | 28, 29 | ||||
| Colon adenocarcinoma | 2 | 48, 52 | ||||||
| Pancreatic neuroendocrine tumor | 1 | 18.7 | ||||||
| Thyroid follicular tumor | 2 | 55, 59 | 2 | 37, 49 | ||||
| Breast adenoma | 1 | 55 | 2 | 34, 38 | 2 | 31, 32 | ||
| Malignant germ cell tumor, testis | 2 | 18, 44 | 2 | 7, 25 | ||||
| Undifferentiated sarcoma | 5 | 9–40 | 2 | 19, 23 | ||||
| Angiosarcoma | 1 | 27 | 5 | 14–28 | ||||
| Osteosarcoma | 1 | 13.4 | 1 | 31 | ||||
| Uterine sarcoma | 1 | 52 | 2 | 36, 37 | ||||
Figure 2.
Collaboration of Pten and Ink4a/Arf deficiency in vivo and status of the Ink4a/Arf locus and Pten gene in pheochromocytomas. (A) Kaplan–Meier survival analysis of Ink4a/Arf+/−Pten+/+, Ink4a/Arf−/−Pten+/+, Ink4a/Arf+/+Pten+/−, and Ink4a/Arf and Pten compound mutant mice. Statistically significant differences for pairwise comparison (P < 0.001) were detected between cohorts Ink4a/Arf−/−Pten+/− vs. Ink4a/Arf−/−Pten+/+ and Ink4a/Arf+/−Pten+/− vs. Ink4a/Arf+/+Pten+/−. (B) Southern blot analysis of the Pten gene in pheochromocytomas. Ink4a/Arf+/+Pten+/− mouse tail genomic DNA digested with SacI was the control. The sizes of wt and mutant Pten alleles are 12 and 6.8 kb, respectively. (C) Southern blot of PstI-digested genomic DNA of pheochromocytomas derived from Ink4a/Arf+/−Pten+/− mice or control Ink4a/Arf+/−Pten+/+ mouse tails. The sizes of wt and mutant Ink4a/Arf alleles are 9.2 and 6.0 kb, respectively.
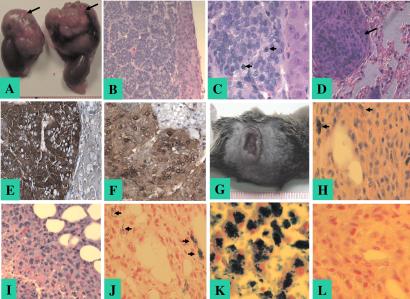
Detailed histopathological surveys of Ink4a/Arf+/+Pten+/− mice confirmed the previously reported spectrum of neoplasms including pheochromocytomas, prostatic intraepithelial neoplasia, endometrial hyperplasia, and breast tumors (3–7). Infiltrating lymphomas were also observed in multiple organs including thymus and spleen as previously described (7); however, our analyses do not distinguish whether these neoplasms represent an aggressive lymphoproliferative disorder or a clonal malignant lymphoma. Specifically, pheochromocytomas, neoplasms derived from chromaffin cells of the adrenal medulla, were observed and measured in size from 2.5 to 10 mm in diameter, with smaller tumors showing compression of the adrenal cortex and larger tumors displaying occasional invasion into the cortex and surrounding adipose tissue. All pheochromocytomas in Ink4a/Arf+/+Pten+/− mice were diagnosed at 36 weeks of age or older (mean age of onset 42 weeks), and the majority of these were unilateral. In contrast, Ink4a/Arf−/−Pten+/− and Ink4a/Arf+/−Pten+/− mice presented with an earlier onset of pheochromocytomas (mean age of onset 24 and 30 weeks, respectively; P < 0.05), with some tumors presenting as early as 7 weeks of age in the Ink4a/Arf−/−Pten+/− cohort. These tumors were often bilateral, larger (5–18 mm), compressed the adrenal cortex, exhibited more mitotic figures (Fig. 3 A–C), and more frequently showed invasion into the surrounding adipose tissue, kidneys, and intestine. Positive anti-chromogranin A and neuron-specific enolase immunoreactivity confirmed the chromaffin origin of these tumors (Fig. 3 E and F). Four of 27 pheochromocytomas arising in Ink4a/Arf−/−Pten+/− mice were metastatic to the lungs (Fig. 3D), whereas no metastasis was observed in the other cohorts. When assayed for tumorigenicity in an SCID explant model, all 10 tested primary pheochromocytoma cell lines derived from Ink4a/Arf−/−Pten+/− or Ink4a/Arf+/−Pten+/− mice readily formed s.c. tumors in SCID mice, whereas only 3 of 8 pheochromocytoma cell lines derived from Ink4a/Arf+/+Pten+/− mice formed tumors (P < 0.05), which grew at a much slower rate (data not shown).
Figure 3.
Histology and immunohistochemistry of pheochromocytoma and cutaneous melanoma from Ink4a/Arf and Pten compound mutant mouse. (A) Gross view of bilateral pattern of pheochromocytoma. The tumors (arrows) arise in the adrenal medulla superior to each kidney. Each unit of the ruler represents 1 mm. (B) Hematoxylin and eosin (H&E) staining of pheochromocytoma (original magnification ×20), showing compressed cortex and classic nested pattern similar to human pheochromocytomas. (C) H&E staining of pheochromocytoma (original magnification ×60), demonstrating mitotic figures as indicated by arrowheads. (D) H&E staining of lung metastasis of pheochromocytoma (original magnification ×40). An arrow indicates a single metastasis exhibiting a histological appearance similar to the primary tumor. (E) Immunohistochemical staining of pheochromocytoma with anti-chromogranin A. Positive staining is in a dark brown color. (F) Immunohistochemistry staining of pheochromocytoma with anti-neuron-specific enolase. Positive staining is in a brown color. (G) Gross view of a cutaneous melanoma. Each unit of the ruler represents 1 mm. (H) H&E staining of cutaneous melanoma (original magnification ×40). Focal pigmentation is indicated with arrowheads. (I) H&E staining of cutaneous melanoma (original magnification ×40). Focal necrosis and invasion of adipose tissue is shown. (J) Fontana–Masson staining of cutaneous melanoma (original magnification ×40). Arrowheads indicate positive staining of focal brown–black melanin pigment. (K) Positive control for iron staining (human liver, original magnification ×40). (L) Negative iron staining of cutaneous melanoma (original magnification ×40).
Pten+/− mice are highly prone to the development of prostatic intraepithelial neoplasia (4). In our studies, prostatic intraepithelial neoplasia typically presented in Ink4a/Arf+/+ Pten+/− mice at an average age of 49 weeks or older and was not evident in mice younger than 36 weeks (Table 1), observations that are consistent with previous reports (4, 6). In contrast, prostatic intraepithelial neoplasia was diagnosed in Ink4a/Arf−/−Pten+/− and Ink4a/Arf+/−Pten+/− mice at an average age of 27 and 40 weeks (Table 1), respectively, and was not detected in Ink4a/Arf−/− or wt littermates by 59 weeks (n = 18). A similar pattern of latencies was observed in the endometrium. Compared with Pten+/− mice, Ink4a/Arf−/−Pten+/− and Ink4a/Arf+/−Pten+/− mice presented with a much earlier onset of endometrial atypical hyperplasia (Table 1, P < 0.05), a condition considered to be a precursor of endometrial carcinoma. Two of nine Ink4a/Arf−/−Pten+/− mice developed endometrial carcinoma at 38 and 48 weeks of age, with both displaying focal squamous differentiation as observed in a high proportion of human endometrial carcinomas (19). Thus, these observations demonstrate significant cooperativity between Ink4a/Arf and Pten in the development of cancer precursor lesions in both the endometrium and prostate, sites where loss of PTEN has a clear etiological significance and where inactivation of INK4a/ARF has been detected in derivative human malignancies (10, 11).
Several tumor types not reported previously in Pten+/− or in Ink4a/Arf mutant mice were observed in the compound mutant mice and included cutaneous melanoma and squamous cell carcinoma. Melanomas were observed in 4 of 37 and 3 of 46 Ink4a/Arf−/−Pten+/− and Ink4a/Arf+/−Pten+/− mice, respectively, the latter at an older age (Table 1). Melanomas presented as pigmented, ulcerating, and invasive cutaneous lesions composed of large epithelioid cells (Fig. 3 G–I). Histochemical analysis revealed the presence of melanin and an absence of iron, thus confirming the melanocytic origin of the tumor (Fig. 3 J and L). The occurrence of melanoma in this model is consistent with mutational or epigenetic inactivation of PTEN and INK4a/ARF found in a significant proportion of human melanomas (10, 20) as well as with the capacity of Ink4a/Arf deficiency and activated H-Ras to generate melanoma in mice (13). Squamous cell carcinoma s detected in Ink4a/Arf−/−Pten+/− and Ink4a/Arf+/−Pten+/− mice presented as widespread peritoneal involvement (Table 1 and data not shown). The extensive nature of the disease at the time of diagnosis precluded identification of the primary disease site. Whereas squamous cell carcinomas have not been reported in Pten+/− or Ink4a/Arf−/− mice, it is noteworthy that PTEN or INK4a mutations have been detected in human squamous cell carcinoma (10, 11), and p16Ink4a loss has been implicated in the transition from benign to malignant growth in mouse skin carcinogenesis model systems (15).
To provide molecular evidence for the cooperative nature of Pten and Ink4a/Arf mutations in tumorigenesis, Southern blot analysis was performed to determine the status of their respective wt alleles in a panel of pheochromocytomas. As shown in Fig. 2B, pheochromocytomas from Ink4a/Arf+/+Pten+/−, Ink4a/Arf−/−Pten+/−, and Ink4a/Arf+/−Pten+/− mice exhibited loss of the wt Pten allele and retention of the null allele (Fig. 2B). Similarly, reduction to homozygosity for Ink4a/Arf was demonstrated in pheochromocytomas arising in Ink4a/Arf+/−Pten+/− mice (Fig. 2C). The cancer relevance of loss of Pten and Ink4a/Arf function was supported further by impaired growth of Pten and Ink4a/Arf melanoma cell lines upon reconstitution of p16INK4a, p19ARF, or Pten by retroviral transduction (data not shown).
Molecular Analysis of Genes and Loci Implicated in the Genesis of Pheochromocytoma.
The strong cooperative interactions between Pten and Ink4a/Arf mutations in the formation of mouse pheochromocytoma prompted a survey of genes and loci associated with this rare human neoplasm. We reasoned that cross-species comparisons of similar and contrasting mutational patterns in mouse versus human pheochromocytomas may be informative with respect to whether a given pheochromocytoma gene/locus intersects with or remains functionally distinct from pathways engaged by p16INK4, p19ARF, and/or Pten.
Candidate gene analysis.
Most human pheochromocytomas are sporadic, and ≈10% exhibit hereditary patterns with causality linked to activation of the RET receptor tyrosine kinase or to inactivation of the von Hippel–Lindau (VHL) or neurofibromatosis type 1 (NF1) genes (21, 22). Direct sequence analysis of the ret coding region in the mouse pheochromocytomas did not identify classical activating point mutations (n = 10, data not shown). Nf-1 protein levels and molecular mass in the pheochromocytomas were comparable to that of normal adrenal control tissue (n = 10, data not shown). Because loss of VHL function results in HIF stabilization and overexpression (23), we documented the functional and structural integrity of Vhl by Western blot analysis showing detectable Vhl protein and, correspondingly, barely detectable levels of the hypoxia-inducible factors, HIF1α and HIF2α (n = 10, data not shown). In addition, because pheochromocytoma in the setting of VHL disease is generally associated with subtle VHL mutations, including missense mutations that retain their ability to regulate HIFs (23), direct sequence analysis of the vhl ORF documented the lack of such mutations (n = 10, data not shown). In summary, this limited survey failed to uncover alterations in known classical pheochromocytoma susceptibility genes.
Genome-wide analysis.
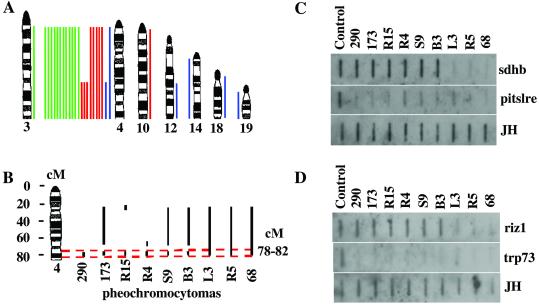
The above candidate gene analysis was complemented by conventional and array-CGH assays designed to detect the presence of recurrent chromosomal copy number aberrations in tumors (16). The pheochromocytomas analyzed here by conventional CGH were derived from 12 Ink4a/ Arf−/−Pten+/−, 8 Ink4a/Arf+/−Pten+/−, and 4 Ink4a/Arf+/+Pten+/− mice (Fig. 4 and Table 2). The most frequent chromosomal alteration detected by the CGH assays in this panel of pheochromocytomas is loss of chromosome 4. One of four pheochromocytomas from the Ink4a/Arf+/+Pten+/− and three of eight pheochromocytomas from Ink4a/Arf+/−Pten+/− mice, respectively, sustained loss of distal chromosome 4 only, whereas 17 of the remaining 20 pheochromocytomas from Ink4a/Arf−/−Pten+/−, Ink4a/Arf+/−Pten+/−, or Ink4a/Arf+/+Pten+/− mice lost the entire chromosome 4. Two of four tumors derived from Ink4a/Arf+/+Pten+/− mice did not exhibit apparent loss in chromosome 4. As the CGH assays implicated the loss of the distal portion of chromosome 4 in pheochromocytomas, we attempted to delimit further the minimal region of involvement employing array-CGH (24). Array-CGH analysis resolved regional chromosomal aberrations at ≈3 Megabase intervals (see Materials and Methods). These assays confirmed and refined a common regional loss involving 78–82 cM in 9 of the12 tumors exhibiting loss of distal chromosome 4 (Fig. 4B and Table 2). Data obtained by conventional and array-based CGH were highly concordant. However, 2 of 11 cases showed discordance between the two methods. In one of the nine pheochromocytomas (case B1 of Table 1), a decreased dosage of chromosome 4 was detected by conventional CGH, but array-CGH and Slot blot analysis failed to confirm this change (data not shown). In the second case (number 68 of Table 1), a large portion of chromosome 4 was lost as determined by array-CGH, whereas the loss was not detected by conventional CGH. The loss of the distal chromosome 4 was confirmed by Slot blot analysis (Fig. 4 C and D). DNA was extracted from paraffin blocks for conventional CGH and from snap-frozen tissue for array-CGH and Slot blot. Therefore, differences between the two methods could be because of differences in the DNA sources or tumor heterogeneity, or differences in sensitivity between the two methods.
Figure 4.
Chromosomal alterations detected by conventional CGH and array-CGH in mouse pheochromocytomas. (A) Lines to the right of the ideograms indicate gains; lines to the left indicate losses. Pheochromocytomas from 4 Pten+/− mutant mice (blue), 8 Ink4a/Arf+/− Pten+/− (red), and 12 Ink4a/Arf−/− Pten+/− (green) mice were analyzed by conventional CGH. (B) Schematic illustration of deleted regions in chromosome 4 by array-CGH. Vertical bars indicate regions deleted. Horizontal dashed lines define the most commonly deleted region. (C and D) Slot blot analysis of mouse sdhd and pitslre, riz1 and trp73 genes. One microgram of normal or tumor genomic DNA was loaded in each well. Corresponding DNA fragments were radiolabeled as probes. A DNA fragment of mouse Ig JH gene was used to confirm the DNA quantity and quality.
Table 2.
Aberration of chromosomes in mouse pheochromocytomas
| Case | CGH | array-CGH | Genotype
|
|
|---|---|---|---|---|
| Ink4a/Arf | Pten | |||
| L3 | D4 | D4:20–82, E5:40–42 | +/− | +/− |
| R6 | D4 | NA | +/− | +/− |
| R4 | D4:C–E, E10 | D4:65–67 & 78–82, D12:40 | +/− | +/− |
| L7 | D4 | NA | +/− | +/− |
| L1 | D4 | NA | +/− | +/− |
| R5 | D4:C–E | D4:20–82 | +/− | +/− |
| R15 | D4:C–E | D4:18–23 & 78–82, D6:19, E5:40–42 | +/− | +/− |
| R1 | D4 | NA | +/− | +/− |
| B3 | D4 | D4:20–67 & 78–82, D12:40 | −/− | +/− |
| B1 | D4, E3 | ND4 | −/− | +/− |
| S9 | D4 | D4:20–67 & 78–82, D12:40 | −/− | +/− |
| S29 | D4 | NA | −/− | +/− |
| S25 | D4 | NA | −/− | +/− |
| S13 | D4 | NA | −/− | +/− |
| S5 | NC | ND4, D19:40–44 | −/− | +/− |
| S3 | D4 | NA | −/− | +/− |
| S1 | D4 | NA | −/− | +/− |
| S2 | D4 | NA | −/− | +/− |
| S16 | D4 | NA | −/− | +/− |
| S19 | D4 | NA | −/− | +/− |
| 63 | NA | ND4, D6:19 | +/+ | +/− |
| 68 | E12:D–F | D4:24–82 | +/+ | +/− |
| 173 | D4 | D4:20–67 & 78–82, D19:42–44 E5:40–44 | +/+ | +/− |
| 290 | D4:C–E | D4:78–82, E5:40–44 | +/+ | +/− |
| 300 | D14, D19, E18 | NA | +/+ | +/− |
Chromosomal losses (D) and gains (E) are indicated. C–E is the distal half of chromosome 4. Regional losses and gains are indicated in the unit of cM. ND4, no change of chromosome 4; NA, no analysis was performed; NC, no chromosomal changes.
Conventional and array-CGH failed to detect copy number change in the pten (located at 24.5 cM of mouse chromosome 19) locus, despite reduction to homozygosity in the Southern blot analysis (Fig. 3B). This may reflect the common scenario of chromosomal loss followed by duplication of the chromosome bearing the engineered Pten null allele or the inability of either assay to detect very small deletions. Although the Ink4a/Arf locus maps to 42.7 cM of chromosome 4 in the mouse, localization of the minimal recurrent region of loss to distal 4 in these tumors points to the existence of additional tumor suppressor(s) in driving pheochromocytomas in this model. This possibility is strengthened by the fact that this region bears synteny to human chromosome 1p31-pter, a hotspot for deletion in both sporadic and hereditary forms of human pheochromocytoma and several other tumor types (25–29). The utility and specificity of cross-species comparisons are evident with absence of aberrations on the distal part of mouse chromosome 3, a region syntenic to human chromosome 1cen-1p31 (adjacent to the 1p hotspot) (Fig. 4 and Table 2). Consistent with the candidate gene analysis, conventional CGH and array-CGH did not detect copy number changes in regions harboring classical pheochromocytoma-related genes (Table 2 and Fig. 4A), including ret (located at 53.2 cM of mouse chromosome 6), vhl (located at 49.45 cM of mouse chromosome 6), and nf1 (located at 46.06 cM of mouse chromosome 11). Similarly, Ink4c (located at 24.7 cM of chromosome 4) inactivation, which predisposes mice to pheochromocytoma (30, 31), does not seem to be a key tumor suppressor in this model as inferred by the lack of deletions that specifically target the ink4c locus (n = 4; Fig. 4, Table 2, and Southern blot, data not shown). That is, the only tumors showing a reduction in ink4c gene dosage were those that had sustained loss of most or all of chromosome 4 by CGH (data not shown), although we have not ruled out epigenetic mechanisms. The lack of a consistent inactivation of Ink4c may relate in part to functional relatedness among members of the Ink4a family.
Possible candidate tumor suppressors encoded in the distal region of mouse chromosome 4 include Riz1 (located at 72.0 cM), Pitslre kinase isoforms (located at 79.4 cM), and Trp73 (located at 82.0 cM). RIZ1 was identified as an Rb-binding zinc finger protein that is commonly lost in cancer cells. Enforced expressions of RIZ1 lead to cell-cycle arrest at G2/M and/or apoptosis and suppress tumorigenicity (32). Mice mutant for the riz1 locus develop a broad spectrum of tumors (33). The PITSLRE kinases are related to the master mitotic protein kinase p34cdc2, and these PITSLRE kinases inhibit cell growth, cause apoptosis, and may regulate RNA splicing and transcription. In addition, the expression of PITSLRE kinases is reduced in a variety of tumors (34, 35). Trp73 is a homolog of the tumor suppressor p53 and induces apoptosis independent of p53 (36). Recently, mutations of human succinate dehydrogenase subunit B (SDHB) and subunit D (SDHD) have been implicated in the development of pheochromocytoma (37, 38). Both SDHB and SDHD proteins localize to the mitochondria, and their loss of function may impair apoptotic processes linked to tumor suppression. Germ-line SDHD mutations have been detected in ≈11% of sporadic pheochromocytomas (38); however, the mouse ortholog has not been identified. Notably, SDHB is located in the human chromosome 1p36 and mutated in familial (4 of 8) and sporadic tumor (1 of 24) pheochromocytomas (37). The mouse sdhb is located at ≈73 cM of chromosome 4, although a precise location is not yet available. In the pheochromocytomas analyzed in this study, Slot-blotting analyses confirmed the loss of trp73 and pitslre in all of tumors analyzed, whereas sdhb and riz1 were lost in only three of nine tumors analyzed (Fig. 4 C and D). Thus, establishment of a role for Sdhb in the development of mouse pheochromocytomas will require more direct genetic validation efforts.
The above gene survey results are in line with the functional overlap between these classical pheochromocytoma genes and p16INK4a, p19ARF, and/or Pten. In the case of RET, activation of RET leads to activation of RAS/MAPK and PI3K/AKT pathways (21, 22). Similarly, Neurofibromin, the protein product of the NF1 tumor suppressor gene, acts as a RAS-GAP by catalyzing the hydrolysis of RAS-GTP to RAS-GDP, thus negatively regulating the Ras protooncogene and reducing cell growth (21). Ras activates multiple effector pathways including PI3K and RAF (14). Finally, VHL down-regulates HIF activity under normoxic conditions; intriguingly, PTEN attenuates hypoxia-mediated HIF1α stabilization (39). A role of PI3K pathway in the induction of HIF1α has emerged from recent studies (40).
Similarly, a genetic and biochemical link between p19ARF and p53 has been well substantiated (8), hence a role for p19ARF in mouse pheochromocytomas gains support from the occurrence of p53 mutations in human pheochromocytomas. However, the actions of p19ARF may also extend to the VHL pathway as implied by a role for p53 in modulating Mdm2-mediated degradation of HIF1 (41). This link is strengthened further by the observation of another rare VHL-linked tumor, pancreatic serous cystadenoma, in mice deficient for Ink4a/Arf (42). A similar body of evidence supports a role for p16INK4a in the genesis of pheochromocytoma and includes reduced expression of Rb in human pheochromocytomas (43) and occurrence of pheochromocytoma in mice mutant for RB (44) or Ink4c (30, 31).
That pheochromocytomas have not been detected in mice deficient for both p16INK4a and p19ARF (this study and others) points to the critical roles exerted by the loss of Pten and/or the presumed tumor suppressor located on distal chromosome 4. On the basis of the high penetrance, earlier onset, and bilateral presentation of this rare neoplasm in our compound mutant mice, it seems that at least four pathways play significant roles in the formation of pheochromocytoma: inactivation of p16INK4a-RB and p19ARF-p53-VHL, activation of the RET receptor tyrosine kinase or disruption of its signaling surrogates (e.g., PTEN/NF-1/PI3K), and deletion of a tumor suppressor(s) residing on mouse distal 4. With regard to the last pathway mentioned, we are mindful that this region could encode multiple tumor suppressors, a possibility suggested by the frequent occurrence of large deletions encompassing the locus in mice and humans (25–28). Indeed, recent data in humans are consistent with the presence of up to three tumor suppressor loci residing on the distal region of human chromosome 1p (25). The mouse model here, together with emerging genomic technologies, provides a genetic system with which to refine the region of deletion and to validate the roles of candidate tumor suppressor genes in the genesis and progression of pheochromocytoma and possibly a number of additional tumor types.
Acknowledgments
We thank Jun Liu, Susan Charzan, Jessica M. DeFrances, and Christine Lam for their excellent technical assistance. We also thank Drs. Sandy Chang, Ned Sharpless, Haifeng Yang, Nabeel Bardeesy, and Richard Maser for their assistance and helpful discussions. We acknowledge support by the National Institutes of Health National Research Service Award (to M.J.Y.), the Brain Tumor Association (to M.J.Y.), the Damon Runyon-Walter Winchell Foundation (to D.H.C.), the Marvin and Roma Auerback Melanoma Research Fund (to B.C.B.), and Howard Hughes Medical Institute fellowship (to M.W.B.). L.C. is a V Foundation Scholar, and R.A.D. is an American Cancer Society Professor and a recipient of the Steven and Michele Kirsch Foundation Investigator Award. This work was supported by National Institutes of Health and American Cancer Society grants and by the Arthur and Rochelle Belfer Cancer Genomics Center.
Abbreviations
- MEF
mouse embryonic fibroblast
- CGH
comparative genomic hybridization
- PI3K
phosphatidylinositol 3-kinase
- wt
wild type
- H&E
hematoxylin and eosin
References
- 1.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson L, Parsons R. Exp Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 3.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 4.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi P P. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 6.Stambolic V, Tsao M S, Macpherson D, Suzuki A, Chapman W B, Mak T W. Cancer Res. 2000;60:3605–3611. [PubMed] [Google Scholar]
- 7.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Catoretti G, Fisher P E, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharpless N E, DePinho R A. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 9.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 10.Ali I U, Schriml L M, Dean M. J Natl Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 11.Ruas M, Peters G. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 12.Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, Schorderet D, Bosman F T, Chaubert P. Lab Invest. 2001;81:217–229. doi: 10.1038/labinvest.3780230. [DOI] [PubMed] [Google Scholar]
- 13.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W, II, DePinho R A. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frame S, Balmain A. Curr Opin Genet Dev. 2000;10:106–113. doi: 10.1016/s0959-437x(99)00052-0. [DOI] [PubMed] [Google Scholar]
- 15.Sharpless N E, Bardeesy N, Lee K H, Carrasco D, Castrillon D H, Aguirre A J, Wu E A, Horner J W, DePinho R A. Nature (London) 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 16.Bardeesy N, Bastian B C, Hezel A, Pinkel D, DePinho R A, Chin L. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng L, Brown J, Eng C. Hum Mol Genet. 2001;10:237–242. doi: 10.1093/hmg/10.3.237. [DOI] [PubMed] [Google Scholar]
- 18.Frank K M, Sharpless N E, Gao Y, Sekiguchi J M, Ferguson D O, Zhu C, Manis J P, Horner J, DePinho R A, Alt F W. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 19.Abeler V M, Kjorstad K E. Cancer. 1992;69:488–495. doi: 10.1002/1097-0142(19920115)69:2<488::aid-cncr2820690236>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X P, Gimm O, Hampel H, Niemann T, Walker M J, Eng C. Am J Pathol. 2000;157:1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch C A, Vortmeyer A O, Huang S C, Alesci S, Zhuang Z, Pacak K. Endocr Regul. 2001;35:43–52. [PubMed] [Google Scholar]
- 22.Eng C. J Clin Oncol. 1999;17:380–393. doi: 10.1200/JCO.1999.17.1.380. [DOI] [PubMed] [Google Scholar]
- 23.Kondo K, Kaelin W G., Jr Exp Cell Res. 2001;264:117–125. doi: 10.1006/excr.2000.5139. [DOI] [PubMed] [Google Scholar]
- 24.Albertson D G, Ylstra B, Segraves R, Collins C, Dairkee S H, Kowbel D, Kuo W L, Gray J W, Pinkel D. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 25.Benn D E, Dwight T, Richardson A L, Delbridge L, Bambach C P, Stowasser M, Gordon R D, Marsh D J, Robinson B G. Cancer Res. 2000;60:7048–7051. [PubMed] [Google Scholar]
- 26.Edstrom E, Mahlamaki E, Nord B, Kjellman M, Karhu R, Hoog A, Goncharov N, Teh B T, Backdahl M, Larsson C. Am J Pathol. 2000;156:651–659. doi: 10.1016/S0002-9440(10)64769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dannenberg H, Speel E J, Zhao J, Saremaslani P, van Der Harst E, Roth J, Heitz P U, Bonjer H J, Dinjens W N, Mooi W J, et al. Am J Pathol. 2000;157:353–359. doi: 10.1016/S0002-9440(10)64547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vargas M P, Zhuang Z, Wang C, Vortmeyer A, Linehan W M, Merino M J. Hum Pathol. 1997;28:411–415. doi: 10.1016/s0046-8177(97)90028-9. [DOI] [PubMed] [Google Scholar]
- 29.Knuutila S, Aalto Y, Autio K, Bjorkqvist A M, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy M L, et al. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latres E, Malumbres M, Sotillo R, Martin J, Ortega S, Martin-Caballero J, Flores J M, Cordon-Cardo C, Barbacid M. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franklin D S, Godfrey V L, O'Brien D A, Deng C, Xiong Y. Mol Cell Biol. 2000;20:6147–6158. doi: 10.1128/mcb.20.16.6147-6158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chadwick R B, Jiang G L, Bennington G A, Yuan B, Johnson C K, Stevens M W, Niemann T H, Peltomaki P, Huang S, de la Chapelle A. Proc Natl Acad Sci USA. 2000;97:2662–2667. doi: 10.1073/pnas.040579497. . (First Published February 25, 2000; 10.1073/pnas.040579497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steele-Perkins G, Fang W, Yang X H, Van Gele M, Carling T, Gu J, Buyse I M, Fletcher J A, Liu J, Bronson R, et al. Genes Dev. 2001;15:2250–2262. doi: 10.1101/gad.870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loyer P, Trembley J H, Lahti J M, Kidd V J. J Cell Sci. 1998;111:1495–1506. doi: 10.1242/jcs.111.11.1495. [DOI] [PubMed] [Google Scholar]
- 35.Lahti J M, Xiang J, Heath L S, Campana D, Kidd V J. Mol Cell Biol. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irwin M, Marin M C, Phillips A C, Seelan R S, Smith D I, Liu W, Flores E R, Tsai K Y, Jacks T, Vousden K H, Kaelin W G., Jr Nature (London) 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 37.Astuti D, Latif F, Dallol A, Dahia P L, Douglas F, George E, Skoldberg F, Husebye E S, Eng C, Maher E R. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimm O, Armanios M, Dziema H, Neumann H P, Eng C. Cancer Res. 2000;60:6822–6825. [PubMed] [Google Scholar]
- 39.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk A R, Ryan H E, Johnson R S, Jefferson A B, et al. Genes Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]
- 40.Laughner E, Taghavi P, Chiles K, Mahon P C, Semenza G L. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravi R, Mookerjee B, Bhujwalla Z M, Sutter C H, Artemov D, Zeng Q, Dillehay L E, Madan A, Semenza G L, Bedi A. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 42.Bardeesy N, Morgan J, Sinha M, Signoretti S, Srivastava S, Loda M, Merlino G, DePinho R A. Mol Cell Biol. 2002;22:635–643. doi: 10.1128/MCB.22.2.635-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam K Y, Lo C Y, Wat N M, Luk J M, Lam K S. J Clin Pathol. 2001;54:443–448. doi: 10.1136/jcp.54.6.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikitin A Y, Juarez-Perez M I, Li S, Huang L, Lee W H. Proc Natl Acad Sci USA. 1999;96:3916–3921. doi: 10.1073/pnas.96.7.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]