Abstract
Genes of the major histocompatibility complex (MHC) play a central role in immune recognition, yet they also influence the odor of individuals. Mice can be trained to distinguish odors mediated by classical MHC loci; however, training can introduce confounding behavioral artifacts. This study demonstrates that mice can distinguish some, but not all, naturally occurring allelic variants at classical MHC loci without prior training. This result suggests that MHC-disassortative mating preferences might operate by means of small MHC-based odor differences, and could therefore contribute to diversifying selection acting on MHC loci. Here we show that odors of two MHC mutant mouse strains (bm1 and bm3) can be distinguished, even after genetic background is controlled by intercrossing strains. These two strains differ by five amino acids, three of which are predicted to chemically contact peptides bound to the peptide-binding region (PBR), the site of antigen presentation for T cell recognition. However, the odors of neither bm1 nor bm3 were distinguished from their parental B6 haplotype after randomizing genomic background, despite discrimination of pure-bred B6 and bm1 strain odors. These combined results suggest that (i) there may be an MHC odor discrimination threshold based on divergence in PBR residues, providing a more logical pattern of MHC-based odor discrimination than found in previous training studies, where discrimination ability was not correlated with PBR divergence; and (ii) additional (non-MHC) mutations that influence odor have accumulated in these strains during the 100 generations of divergence between pure B6 and bm1 strains.
The highly polymorphic genes of the major histocompatibility complex (MHC) encode cell surface glycoproteins (class I and II molecules) that bind peptides for T lymphocyte-mediated immune recognition of pathogens (1). The genetic diversity of MHC genes is found largely in the peptide-binding region (PBR) of MHC molecules, which evolves under positive Darwinian selection (2, 3). Although the selection on MHC alleles is generally presumed to be from pathogens, evidence in mice, humans (4, 5), and salmon (6) indicates that mating preferences, mediated through MHC-based odor recognition, may also provide a force driving the evolution and maintenance of MHC diversity. The generality of MHC-based mating preferences is in doubt, as some natural population studies have failed to find an effect (7–9). However, when the opportunities for mate choice exist, MHC-disassortative mating preferences are expected to evolve under the two major pathogen-driven models of MHC evolution: heterozygote advantage and antagonistic host–pathogen coevolution. Disassortative mating preferences might function to produce progeny with greater disease resistance to pathogens and parasites (4), and in addition, could function to reduce inbreeding (10, 11).
Numerous studies indicate that mice can recognize MHC-identity through the use of olfactory cues (12, 13). Male and female house mice prefer to mate with individuals that carry dissimilar MHC haplotypes from their parents (14, 15), even when MHC haplotypes are bred onto an outcrossed wild genomic background (16). However, although untrained mice can discriminate whole MHC-haplotype differences and single gene knockouts (17), studies have not clearly shown that they can distinguish allelic variants resulting from single-locus MHC mutations, the evolutionary ingredients from which the modern MHC pool was derived. There is clearly some form of selection favoring new mutations as evidenced by the high rates of nonsynonymous vs. synonymous substitutions in the PBR codons (18, 19). If such allelic variants mediate novel odors, then disassortative mating preferences driven by MHC-based odors could contribute to the diversifying selection on PBR codons. This model of MHC evolution mediated by sexual selection requires that untrained mice distinguish odors mediated by naturally occurring variants of MHC alleles.
The physiochemical origin and nature of MHC-based odors remains unclear. However, classical MHC genes are the most promising candidate loci for MHC-based odor mediation, and a number of plausible mechanisms have been postulated. Some evidence indicates that these genes influence ratios of volatile carboxylic acids, which may arise as metabolic by-products of peptides (20). Alternatively, volatile odorants might bind to the PBS of degrading MHC molecules, implying a dual role for the PBS in both peptide and odorant binding (the “carrier hypothesis,” ref. 21). Volatile odorants, as well as degraded MHC molecules and peptides, appear in the urine and bodily fluids of vertebrates and may thus facilitate odor detection (22–24). If genetic variation at these candidate loci imparts relevant olfactory information, this would provide the mechanistic basis for MHC-disassortative mating preferences.
Naturally occurring H2 (mouse MHC) mutant congenic mice provide the opportunity to study the role of odor variation and sexual selection at the level of individual gene differences. These spontaneous mutants were identified by skin graft rejection experiments (1, 25), and all possess potential for novel immunological functionality conferred by a change in their peptide-binding properties. Sexual selection is a powerful evolutionary force, equal to or greater than viability selection over both short and long time scales (26). If spontaneous H2 mutants can be distinguished through odor recognition, then mating preferences, in addition to novel antigen epitope presentation, could be a rapid and powerful selective force determining which new alleles survive and which ones die.
Three research groups have provided support for the hypothesis that classical MHC genes influence individual odors. Penn and Potts (17) showed that untrained mice can discriminate between the odors of individuals that differ by either possessing or entirely lacking the H2–Kd class I locus (dm2 mutants). Recently, Montag et al. (27) used an “electronic nose” to detect odor differences among various strains, including parental and H2 mutant odor combinations. However, neither of these studies used crossed strains to randomize background gene differences, and it is possible that discrimination was caused by accumulated background mutations that affected strain-specific odor types. Many MHC congenic strains (including the dm and bm mutant series) were established decades ago and are likely to differ from one another by dozens of fixed mutational differences at background loci (28). This growing problem can be largely solved by crossing congenic strains and intercrossing heterozygotes to produce F2 segregants (F2 generation animals that are homozygous for relevant genes but carry randomly segregating background genes). Yamazaki and colleagues (29, 30) successfully trained mice to distinguish a number of parental vs. H2 mutant odor combinations, including odors of F2 segregants. However, when a modified training approach was used, mice were unable to distinguish odors among H2 mutants. This result is rather puzzling, because in most cases, mice were able to distinguish few, but not larger differences in PBS amino acids. Training can introduce behavioral artifacts (13), and sexual selection will favor only novel odors that are detectable without training. Therefore, we tested the discrimination of odor differences by using a habituation/dishabituation approach that employs untrained mice.
The results we present here are largely opposite to those of prior training assays. After crossing strains to randomize background variation, the odor effects of two H2 mutations (bm3 and bm1) were each tested against the parental genotype (B6), and also against each other. We used this incremental approach and found that only odors from the bm1 vs. bm3 strain pair having the greatest PBR divergence were distinguished (five amino acids, at least three of which are predicted to directly influence antigen presentation; refs. 31–33). These data are biologically consistent with an odor detection threshold based on PBR divergence (although other effects are also plausible), and are prerequisite for a model of MHC diversifying selection based on odor detection and sexual selection driving both incorporation and further diversification of newly arisen alleles. Our results also suggest that accumulated background mutations influence odor variation, as pure-strain (but not F2 segregant) odors of the B6 vs. bm1 combination were readily distinguished.
Materials and Methods
We conducted these experiments from December 1998 through June 2000 at the Biology Department of the University of Utah in Salt Lake City.
Animals.
Six-week-old male and female C57BL/6J (B6, parental strain), B6.C-H2bm1/ByJ (bm1, mutant strain), and C57BL/6J-H2bm3/Eg (bm3, mutant strain) were purchased from The Jackson Laboratories. Additional bm3 mice were kindly donated by Larry Pease at Mayo Clinic (Rochester, MN). All mice were placed in individual maintenance cages and provided with bedding, Purina Rodent Chow, and water ad libitum.
Pure-Bred B6 and bm1 Odors.
After 1 week of acclimation to our animal room, ≈2 ml of urine was collected from each of the B6 and bm1 males (nine individuals of each strain) over the following 2 weeks. Urine was obtained daily by gently palpating bladder contents into autoclaved Eppendorf tubes. Fresh urine was immediately stored at −20°C. At the end of collections, urine was pooled for each male, and males (within an MHC type) were randomly assigned to groups of three odor donors for the urine pools in each control and experimental assay. Penn and Potts (17) previously found that pooling urine reduces nongenetic sources of individual variation in odor. Pooled urine (10 μl per male per trial, 30 μl per pool) was coded and stored at −20°C until testing. No male was ever represented in more than one pool sniffed by a given subject to prevent possible biases caused by inadvertent familiarization of subjects to any particular male odor.
Rederived Strains (F2 Segregants).
To rule out the possibility that odor differences among strains are caused by variation in background genes, we randomized genomic background by crossing strains. Parental generation breedings were performed across all three possible strain pairs (B6 × bm1; B6 × bm3, and bm1 × bm3) to mix inbred genomes (Table 1). All parents and progeny from B6 × bm1 and B6 × bm3 crosses were placed on adjacent racks in the same room. Parents and progeny from the bm1 × bm3 cross were placed in a separate room in our rodent facility because of space limitations, although maintenance conditions and air supply were essentially equivalent among rooms. F1 (first) generation heterozygous males and females from the parental crosses were intercrossed to produce F2 segregants. At 21 days, F2 pups were removed from their parental cages and housed in same-sex cages of littermates. Three millimeters of tail tip was clipped for DNA extraction and subsequent F2 segregant genotyping (in accordance with regulations issued by the state of Utah Animal Care and Use Committee).
Table 1.
Breeding scheme for producing F2 segregants
| Generation | Function | B6 × bm1 | B6 × bm3 | bm1 × bm3 |
|---|---|---|---|---|
| P | Mix inbred genomes | B6/B6 × bm1/bm1 | B6/B6 × bm3/bm3 | bm1/bm1 × bm3/bm3 |
| F1 | Create F2 segregants | B6/bm1 × B6/bm1 | B6/bm3 × B6/bm3 | bm1/bm3 × bm1/bm3 |
| F2 | Collect urine from homozygous F2 males | B6/B6, B6/bm1, and bm1/bm1 | B6/B6, B6/bm3, and bm3/bm3 | bm1/bm1, bm1/bm3, and bm3/bm3 |
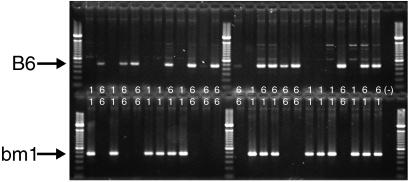
Genotyping was performed by using a PCR amplification-based assay (see Fig. 1 legend). We used primers designed to anneal to the actual mutation sites in an allele-specific presence/absence approach. Fig. 1 shows an agarose gel containing amplified products from 25 known B6, B6/bm1, and bm1 individuals. All individuals were correctly scored in this test trial, and all F2 progeny for each of the three inbred crosses were subsequently determined by using this PCR-based approach. When F2 litters reached 7 weeks of age, urine was collected from all F2 segregant males for 2 weeks (as for the pure-bred strains), pooled, coded, and stored at −20°C for discrimination trials. Thus, all urine came from age-matched donors. No more than one male per litter contributed to any given urine pool to prevent potential litter effects.
Figure 1.
Agarose gel showing genotypes of B6 (6), bm1 (1) and B6/bm1 (6/1) individuals. All 25 individuals were correctly scored in this initial test of amplification efficiency. The upper row reveals presence or absence of the B6 haplotype, and the lower row reveals presence or absence of the bm1 haplotype. Individual genotypes are written in the center of each lane. We used sequence-specific primers in conjunction with PCR (SSP-PCR) to discriminate F2 segregants. For each genomic DNA, a pair of SSP-PCR reactions was performed; one using specific primer pairs that recognize one allele and the other reaction using a primer pair specific for the other allele. Allele-specific primers were B6 (WP335, 5′-TGCCCTCCAGGTAGGCCCTGA-3′; WP342, 5′-AATGAGCAGAGTTTCCGAGTGGA-3′); bm1 (WP334, 5′- CAATGAGCAGAGTTTCCGAGTGG-3′; WP337, 5′-CCCTCCAGGTAGGCCCTGTAA-3′); and bm3 [WP335 (also used for B6), and WP338, 5′-AATGAGCAGAGTTTCCGAGTGAG-3′]. Amplification reactions contained 100–200 ng of DNA template, 0.2 μM of each forward and reverse oligonucleotide primer, 0.125 mM of each dNTP, 10 mM Tris⋅HCl (pH 8.3), 2.5 mM magnesium chloride, 50 mM potassium chloride, 4% DMSO, and 1.5 units of Taq polymerase in a total reaction volume of 25 μl. PCR conditions were 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. A final extension step at 72°C for 7 min followed the last cycle. The paired products from each genomic DNA are run in parallel on 1.5% agarose at 125 V for 25 min, stained with 1 μg/μl ethidium bromide and visualized with ultraviolet light.
Habituation/Dishabituation Assay.
A detailed description of the habituation/dishabituation assay is given in Penn and Potts (17). The basis of this assay is the observation that subjects decrease investigatory behavior toward a repeatedly presented odor, but increase investigatory behavior when a novel odor is presented (34). Investigatory behavior was measured as both the number and the duration of sniffs directed toward a presented odor. After a 5-min acclimation period (blank trial), a female subject was “habituated” to a pool of urine (odor 1) from three unrelated MHC-identical males for three successive 2-min presentations. During a final 2-min presentation, a new MHC urine pool (odor 2) was presented to the female and the degree of “dishabituation” was measured. In control trials, this new pool was from unrelated males that matched the MHC type of the donors in the three previous presentations. In test trials, the new pool was from unrelated males of an MHC-type dissimilar to donors in the three previous presentations. Each female subject was tested twice, 1 week apart, in paired control and test trials, and dishabituation responses of these paired control and test trials were compared. If urine samples from different MHC strains (test trials) elicit an increase in investigatory behavior above the control trials, then this indicates that mice can distinguish odors based on genetic differences between the strains.
The order of test and control sessions was balanced to control for bias caused by long-term habituation to the experimental setup. All urine samples were previously coded so that observers were unaware whether trials were controls or tests. MHC-donor types (i.e., B6 or bm1) were represented equally within and among control and test trials for all groups to control for any preference to a particular MHC type.
The habituation/dishabituation assay was designed to test odor discrimination ability, not to determine odor preferences. However, to prevent inadvertent MHC-based odor preferences, only mice that had MHC haplotypes dissimilar from all odor donors were selected as subjects (sniffers). Sniffer mice (tested between 6 and 10 weeks old) were F1 female B10.D2-H2dTlacHC1/nSaJ (H2-D), B10.Q-H2q/SgJ (H2-Q), B10.BR-H2kH2-Tl8 a/SgSnJ(H2-K), and F1 crosses of these three congenic strains. No genotype effect was found for discrimination ability (ANOVA, sniff bouts: F = 1.8, P < 0.18; sniff duration: F = 1.11, P < 0.36). Sniffer genotypes were therefore pooled within each experiment for analysis.
Data Analysis.
Sniffers that failed to sniff the odor during the first two successive 2-min trials of an assay were excluded from analyses. In both control and test trials, dishabituation scores were recorded as the difference between sniff responses to the second odor (fourth trial) and sniff responses to the last trial of the first odor (third trial). All statistical tests were conducted with jmp software (SAS Institute, Cary, NC). The habituation/dishabituation assay tests a directional hypothesis, specifically, whether mean dishabituation responses are increased above the baseline controls. We therefore compared dishabituation scores for each experiment by using one-tailed Student's t tests when statistical assumptions were justified (35). Non-normally distributed data were tested with one-tailed Wilcoxon signed-rank tests (W). Based on Penn and Potts' experimental design (17), 20 sniffer subjects were used for pure-bred B6 vs. bm1 assays, and for bm1 vs. bm3 F2 segregant assays. After testing these two groups, the number of subjects in F2 segregant B6 vs. bm1 and B6 vs. bm3 assays (the last two groups tested) was a priori increased to 28 to increase the potential power (β > 0.7) of detecting an effect of similar magnitude.
Results
Pure-Bred B6 and bm1 Odors.
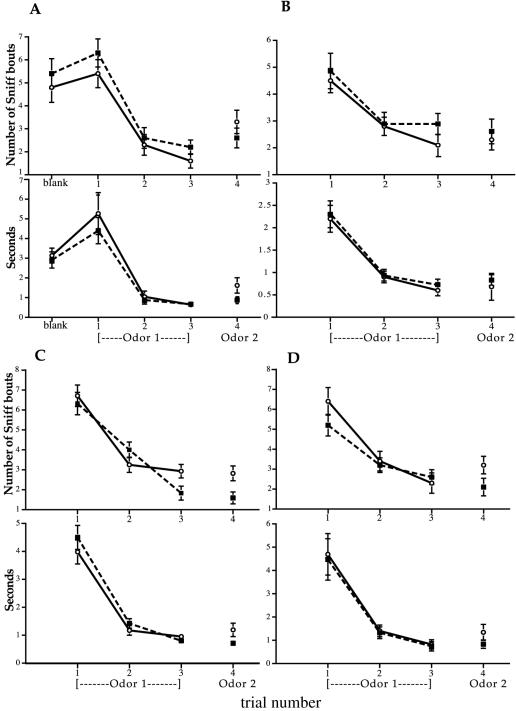
Sniffers readily distinguished the urine odors of pure-bred B6 versus bm1 strains in test sessions [test trials: sniff bouts (SB), P = 0.0001; sniff duration (SD), P = 0.001; Fig. 2A)]. Subjects in these trials also distinguished control urine pools as shown by a significant increase in SB (control trials: P = 0.042), although not in SD (P = 0.11). Test sessions showed significantly greater mean dishabituation scores than control sessions for both parameters measured (SB, P = 0.042; SD, P = 0.036). Table 2 summarizes the statistical analyses of dishabituation responses within each test assay and control assay, as well as test vs. control comparisons.
Figure 2.
Responses of female sniffers (mean ± SE) during 2-min odor presentations (trials 1, 2, 3, and 4). Test sessions (○, solid lines) show number of sniff bouts (SB) or sniff duration (SD) (in seconds) when females are presented with odors from two different strains (i.e., B6 vs. bm1). Control sessions (■, broken lines) show number of SB or SD (in seconds) when females are presented with odors from the same strain (i.e., B6 vs. B6). Dishabituation scores were measured as the difference between the second odor response (fourth trial) and the final first odor response (third trial). Significance values for all four strain comparisons are presented in Table 2. (A) Pure-bred B6 vs. bm1 congenic odors. During this experiment, SB and SD were also recorded during the initial blank trial (water was used instead of pooled urine), but because the water blank did not add substantial information, we ceased to record behavior during blank trials in subsequent experiments. (B) B6 vs. bm1 F2 segregant odors. (C) B6 vs. bm3 F2 segregant odors. (D) bm1 vs. bm3 F2 segregant odors.
Table 2.
Results of statistical tests performed on dishabituation scores [using number of sniff bouts (SB) and sniff bout duration (SD)]
| Experiment | Measured behavioral response | Control trials | Test trials | Control vs. test trials |
|---|---|---|---|---|
| Pure-bred B6 vs. bm1 | SB | T = 1.8, P = 0.042* | T = 4.6, P = 0.0001** | T = 1.8, P = 0.042* |
| (n = 19) | SD | T = 1.3, P = 0.11 | W = 70.5, P = 0.001** | W = 45, P = 0.036* |
| F2:B6 vs. bm1 | SB | T = −0.98, P = 0.83 | T = 0.5, P = 0.30 | T = 0.98, P = 0.17 |
| (n = 20) | SD | W = 12.5, P = 0.38 | T = 0.83, P = 0.20 | T = −0.07, P = 0.53 |
| F2:B6 vs. bm3 | SB | T = −1.05, P = 0.84 | T = −0.29, P = 0.61 | T = 0.52, P = 0.30 |
| (n = 28) | SD | T = −0.63, P = 0.73 | W = 15.5, P = 0.36 | W = 39.5, P = 0.19 |
| F2:bm1 vs. bm3 | SB | T = −1.02, P = 0.84 | T = 1.81, P = 0.043* | T = 1.9, P = 0.039* |
| (n = 28) | SD | W = −4, P = 0.55 | W = 71.5, P = 0.001** | T = 1.5, P = 0.070 |
The Control trials column shows whether sniffers could discriminate between the odor of different individuals with the same MHC. The Test trials column shows whether mice could discriminate between the odor of individuals with different MHC. The final column indicates whether dishabituation scores for test trials were significantly greater than control trials. Paired, one-way Student's t tests (T) or Wilcoxon tests (W) were performed.
, P < 0.05 significance level;
, P < 0.001 significance level;
, P < 0.0001 significance level.
F2 Segregant Odors.
In contrast to odors from the pure-bred strains, neither test nor control trials of F2 segregant odors from the B6 × bm1 group were distinguished (test trials: SB, P = 0.30; SD, P = 0.20; control trials: SB, P = 0.83; SD, P = 0.38) (Fig. 2B; Table 2). Although mean dishabituation score was greater in test trials, this increase was not statistically significant (P = 0.17). A post hoc power analysis indicated that this nonsignificant trend, if it held, would require over 140 subjects to achieve significance, 8-fold more subjects than was sufficient to detect a significant difference between test and control assays in the pure-bred B6 vs. bm1 strain. Similarly, in discrimination trials of F2 segregants from the B6 × bm3 group, neither control nor test trials were distinguished, nor were test dishabituation scores significantly greater than control scores (Fig. 2C; Table 2). Again, post hoc power analyses suggests that an enormous sample size (>600) would be necessary to achieve significance.
In contrast to the other two F2 segregant groups, sniffers for the bm1 vs. bm3 group significantly discriminated test session odors (SB, P = 0.043; SD, P = 0.001), but not control odors (SB, P = 0.84, SD, P = 0.55) (Fig. 2D; Table 2). This group also showed a significantly greater mean dishabituation score for test trials (compared with control trials) in SB (P = 0.039). SD score was greater in the test trials than the control trials, although only marginally significant (P = 0.070).
Discussion
Our study demonstrates that mice can detect odor differences among natural variants of a single class I classical H2 locus without prior training. However, among the F2 segregant pairs, only the comparison with the largest difference between antigen-binding codons (bm1 vs. bm3) was distinguished. Neither of these mutants was distinguished from the parental B6 strain. These results are consistent with a threshold requirement for mutation-based odor detection, whereby multiple PBS codons must differ to yield a detectable new odor. Only contact amino acids directed into the antigen-binding groove are predicted to have an effect on the binding properties that determine which ligands and potential odorants are presented to T cells (33). MHC mutants bm1 and bm3 differ from their parental strain B6, respectively, by two and one contact amino acids directed into and on opposite sides of the peptide-binding groove. All three amino acid sites have been experimentally demonstrated to influence binding properties in studies of mouse and human MHC (31, 36, 37). Although our three comparisons are consistent with a threshold effect, more comparisons will be required to determine whether a general pattern exists.
Alternatively, overall PBR divergence may be less important in determining odors than are complex, context-specific interactions among PBR residues. Each of the 57 class I Mus PBR residues may influence antigen binding to a different degree, and amino acid substitutions at a given site are not equivalent (33). Therefore, odor properties may be highly synergistic and generally more difficult to predict than the threshold effect we observed.
Sniffers in our study were able to distinguish between pure-bred B6 and bm1 odors. Because this ability was not found when we tested odors from F2 segregants, discrimination of these pure-bred lines may be entirely due to effects from background genes. There is mounting evidence that genetic background variation mediates manifold phenotypic differences among congenic strains or substrains (38, 39) in traits such as disease susceptibility, physiology, and behavior (28). Documented differences among H2 congenics, such as variation in activity level, urination, and ultrasonic vocalizations (40–42), may likewise be caused by genetic background variation and should be reevaluated after controlling for such effects. However, it is also possible that using F2s removed MHC correlated environmental effects in the pure strains (i.e., litter effects) that were present before the arrival of original B6 and bm1 odor donors at our animal facility. Our results should at least instill caution in drawing conclusions about any observed differences between congenic lines, especially those lines that have been separated for an appreciable time.
Estimates of mutational background differences among congenic strains can be calculated from the equation n = (G1 + G2)μγ, where n is the expected number of differentially fixed alleles arising from new mutations, G1 + G2 is the number of combined generations since the branching between strains, μ is the mutation rate per locus per generation, and γ is the number of coding loci affected by the mutation rate, μ (38, 43). Using an averaged male and female mouse mutation rate of 8.7 × 10−6 per locus × generation (44, 45), two congenic mouse strains separated for 20 years with two generations per year (G1 + G2 = 80) are predicted to differ by 35 unique new mutations, assuming 50,000 ( = γ) coding and regulatory genes in mice. Background mutations might therefore constitute an important source of phenotypic variation among laboratory strains. Although the possibility is controversial, histocompatibility loci may mutate faster than background genes (25), at ≈1.5 × 10−5 per locus × generation. Mutations arising within the tightly linked H2 region do not segregate independently of classical H2 genes, and therefore breeding F2 segregants will not eliminate this potential source of genetic variation. However, the number of new mutations arising within the H2 complex over 40 generations of strain separation is only 0.12 based on a similar calculation. Therefore, it is 300 times less likely that phenotypic variation will arise because of mutations in H2 genes. The H2-K locus has an exceptionally rapid mutation rate—one to two orders of magnitude higher than empirical estimates of other genes (25, 46). Using a μ of 1.7 × 10−4 at the H2-K locus, two congenic strains separated for 20 years should diverge by only 0.0136 new mutations. These calculations suggest that most new mutations in MHC congenic strains are in background genes and not MHC-linked genes. This finding could explain why the mice could easily discriminate B6 vs. bm1 pure-bred strains (which have been separated for over 100 generations, but not the B6 and bm1 F2 segregants.
There are two apparent inconsistencies between our results and those of training assays performed by Yamazaki and coworkers. First, trained mice in Yamazaki's group were able to distinguish differences between odors of B6 vs. bm1 F2 segregant mice, in contrast to our untrained mice (29). Because the goal of our research is to determine the role of odor recognition in mate choice, it is crucial to test the detection of MHC-mediated odors without using trained animals or mechanical sensors (27). Habituation-dishabituation assays may not be as sensitive as training assays, but they better reflect the sensory capabilities of animals in a natural context. The second inconsistency between our results and those of Yamazaki's group is that their mice could not distinguish among the odors of various bm mutants when an indirect training approach was used (mice were tested for their ability to discriminate odor combinations slightly different from those they were trained on) (30, 47). It is not known whether mice can be directly trained to make such among-mutant distinctions; however, based on the results of our bm1 vs. bm3 comparison, we reject the conclusion that H2 mutants are not mutually discriminable.
MHC supermotif patterns in humans suggest that within ethnic populations, selection favors sets of alleles with divergent antigen-binding properties and immune functionality (48). If odortype expression coincides with differences in immune functionality, mating preferences based on MHC chemosensory information will additionally favor allele sets with greater divergence in PBR residues, as recently demonstrated by Landry et al. (6) in a natural population of Atlantic salmon. The salmon study identified a significant positive correlation between mate choice and PBR divergence at the MHC class II β locus, but failed to identify MHC allele sharing as a significant factor in mate choice. Accordingly, ignoring PBR divergence among alleles could confound attempts to identify MHC-based mate choice in general.
Unlike the salmon study, our experimental approach and use of inbred H2 mutant mice allows the exclusion of effects mediated by all but a single class I locus. Thus, we determined minimum detectable odor changes arising from single, naturally occurring mutations. Sequence analyses indicate that the vast majority of spontaneous H2 mutations detected through transplant rejection carry multiple amino acid substitutions resulting from microrecombinational events involving nonreciprocal exchange of DNA sequence from related MHC coding and noncoding loci (49–55). Such events are more likely to surpass a physiochemical threshold for odor detection than are single point mutations. The opportunity for sexual selection to act on novel alleles may therefore depend on the special molecular properties of the MHC, whose multiple duplications and GpC-rich regions seem to facilitate the process of microrecombination (56).
This study provides the requisite olfactory basis for the role of MHC-mediated odors and chemosensory-based mate choice in the evolutionary origin and maintenance of MHC diversity. We have demonstrated that untrained mice detect odor differences mediated by naturally occurring and mutually expressed alleles at a single MHC locus. It remains to be determined whether sexual selection (mating preferences) can operate on these small differences to drive the spread of novel alleles.
Acknowledgments
We thank Chris Bodily, Kristy Damjanovich, David Francyk, and Tish Oberly for assistance, and Linda Morrison for technical support. Female mice used as odor sniffers were kindly bred by Erin McClelland. Members of our animal maintenance staff cared for the animals. Comments on an earlier draft by Bradley Demarest and two anonymous reviewers greatly improved this manuscript. This work was supported by National Science Foundation Grant IBN-9904609 (to W.K.P. and D.J.P.).
Abbreviations
- MHC
major histocompatibility complex
- PBR
peptide-binding region
- SB
sniff bouts
- SD
sniff duration
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Klein J. Natural History of the Histocompatibility Complex. New York: Wiley; 1986. [Google Scholar]
- 2.Apanius V, Penn D, Slev P, Ruff L R, Potts W K. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 3.Edwards S V, Hedrick P W. Trends Ecol Evol. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- 4.Penn D, Potts W. Am Nat. 1999;153:145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- 5.Jordan W C, Bruford M W. Heredity. 1998;81:127–133. doi: 10.1046/j.1365-2540.1998.00428.x. [DOI] [PubMed] [Google Scholar]
- 6.Landry C, Garant D, Duchesne P, Bernatchez L. Proc R Soc London B. 2001;268:1279–1285. doi: 10.1098/rspb.2001.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedrick P W, Black F L. Am J Hum Genet. 1997;61:505–511. doi: 10.1086/515519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson S, Pemberton J M. Proc R Soc London B. 1997;264:1813–1819. doi: 10.1098/rspb.1997.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenink P W, Groen A F, Roelke-Parker M E, Prins H H. Mol Ecol. 1998;7:1315–1322. doi: 10.1046/j.1365-294x.1998.00463.x. [DOI] [PubMed] [Google Scholar]
- 10.Brown J L, Eklund A. Am Nat. 1994;143:435–461. [Google Scholar]
- 11.Tregenza T, Wedell N. Mol Ecol. 2000;9:1013–1027. doi: 10.1046/j.1365-294x.2000.00964.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi M, Yamazaki K, Beauchamp G K, Bard J, Thomas L, Boyse E A. Proc Natl Acad Sci USA. 1981;78:5817–5820. doi: 10.1073/pnas.78.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penn D, Potts W. Adv Immunol. 1998;69:411–435. doi: 10.1016/s0065-2776(08)60612-4. [DOI] [PubMed] [Google Scholar]
- 14.Yamazaki K, Boyse E A, Mike V, Thaler H T, Mathieson B J, Abbott J, Boyse J, Zayas Z A. J Exp Med. 1976;144:1324–1335. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penn D, Potts W. Proc R Soc London B. 1998;265:1299–1306. doi: 10.1098/rspb.1998.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potts W K, Manning C J, Wakeland E K. Nature (London) 1991;352:619–621. doi: 10.1038/352619a0. [DOI] [PubMed] [Google Scholar]
- 17.Penn D, Potts W. Physiol Behav. 1998;64:235–243. doi: 10.1016/s0031-9384(98)00052-3. [DOI] [PubMed] [Google Scholar]
- 18.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 19.Hughes A L, Nei M. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer A G, Beauchamp G K, Yamazaki K. Proc Natl Acad Sci USA. 1997;94:2210–2214. doi: 10.1073/pnas.94.6.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearse-Pratt R, Schellinck H, Brown R, Singh P B, Roser B. Genetica. 1998;104:223–230. doi: 10.1023/a:1026489524199. [DOI] [PubMed] [Google Scholar]
- 22.Schwende F J, Jorgenson J W, Novotny M. J Chem Ecol. 1984;10:1603–1615. doi: 10.1007/BF00988428. [DOI] [PubMed] [Google Scholar]
- 23.Singh P B, Brown R E, Roser B. Nature (London) 1987;327:161–164. doi: 10.1038/327161a0. [DOI] [PubMed] [Google Scholar]
- 24.Eggert F, Höller C, Luszyk D, Müller-Ruchholtz W, Ferstl R. Physiol Behav. 1996;59:57–62. doi: 10.1016/0031-9384(95)02029-2. [DOI] [PubMed] [Google Scholar]
- 25.Melvold R W, Wang K, Kohn H I. Immunogenetics. 1997;47:44–54. doi: 10.1007/s002510050325. [DOI] [PubMed] [Google Scholar]
- 26.Hoekstra H E, Hoekstra J M, Berrigan D, Vignieri S N, Hoang A, Hill C E, Beerli P, Kingsolver J G. Proc Natl Acad Sci USA. 2001;98:9157–9160. doi: 10.1073/pnas.161281098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montag S, Frank M, Ulmer H, Wernet D, Gopel W, Rammensee H. Proc Natl Acad Sci USA. 2001;98:9249–9254. doi: 10.1073/pnas.161266398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll L, Potts W. J Immunol Methods. 2001;257:137–143. doi: 10.1016/s0022-1759(01)00456-2. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki K, Beauchamp G K, Egorov I K, Bard J, Thomas L, Boyse E A. Proc Natl Acad Sci USA. 1983;80:5685–5688. doi: 10.1073/pnas.80.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki K, Beauchamp G K, Thomas L, Bard J, Boyse E. In: Transgenic Mice and Mutants in MHC Research. Egorov I R, David C S, editors. Berlin: Springer; 1990. pp. 62–64. [Google Scholar]
- 31.Pullen J K, Hunt H D, Horton R M, Pease L R. J Immunol. 1989;143:1674–1679. [PubMed] [Google Scholar]
- 32.van Bleek G M, Nathenson S G. Proc Natl Acad Sci USA. 1991;88:11032–11036. doi: 10.1073/pnas.88.24.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumura M, Fremont D H, Peterson P A, Wilson I A. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 34.Gregg B, Thiessen D D. Physiol Behav. 1981;26:1133–1136. doi: 10.1016/0031-9384(81)90221-3. [DOI] [PubMed] [Google Scholar]
- 35.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice-Hall; 1999. [Google Scholar]
- 36.Rojo S, Calvo V, Lopez D, Galocha B, Lopez De Castro J A. In: Transgenic Mice and Mutants in MHC Research. Egorov I K, David C S, editors. Berlin: Springer; 1990. pp. 67–76. [Google Scholar]
- 37.Zhang W, Young A C, Imarai M, Nathenson S G, Sacchettini J C. Proc Natl Acad Sci USA. 1992;89:8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey D W. Immunol Today. 1982;3:210–214. doi: 10.1016/0167-5699(82)90093-7. [DOI] [PubMed] [Google Scholar]
- 39.Simpson E M, Linder C C, Sargent E E, Davisson M T, Mobraaten L E, Sharp J J. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 40.Klein J. Adv Immunol. 1978;26:55–146. doi: 10.1016/s0065-2776(08)60229-1. [DOI] [PubMed] [Google Scholar]
- 41.Ivanyi P. Proc R Soc London B. 1978;202:117–158. doi: 10.1098/rspb.1978.0060. [DOI] [PubMed] [Google Scholar]
- 42.Brown R E, Schellinck H M, Jagosh J. Genetica. 1998;104:249–257. doi: 10.1023/a:1026438010723. [DOI] [PubMed] [Google Scholar]
- 43.Bailey D W. In: Origins of Inbred Mice. Morse H C I, editor. New York: Academic; 1978. pp. 197–215. [Google Scholar]
- 44.Russell L B, Russell W L. Proc Natl Acad Sci USA. 1996;93:13072–13077. doi: 10.1073/pnas.93.23.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeom Y I, Abe K, Artzt K. Genetics. 1992;130:629–638. doi: 10.1093/genetics/130.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamazaki K, Beauchamp G K, Bard J, Boyse E A. In: Chemical Signals in Vertebrates. MacDonald D W, Möller-Schwarze D, Natynczuk S E, editors. Vol. 5. Oxford: Oxford Univ. Press; 1990. pp. 255–259. [Google Scholar]
- 48.Sidney J, Grey H M, Kubo R T, Sette A. Immunol Today. 1996;17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 49.Mellor A L, Weiss E H, Ramachandran K, Flavell R A. Nature (London) 1983;306:792–795. doi: 10.1038/306792a0. [DOI] [PubMed] [Google Scholar]
- 50.Pease L R, Schulze D H, Pfaffenbach G M, Nathenson S G. Proc Natl Acad Sci USA. 1983;80:242–246. doi: 10.1073/pnas.80.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulze D H, Pease L R, Geier S S, Reyes A A, Sarmiento L A, Wallace R B, Nathenson S G. Proc Natl Acad Sci USA. 1983;80:2007–2011. doi: 10.1073/pnas.80.7.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss E H, Mellor A, Golden L, Fahrner K, Simpson E, Hurst J, Flavell R A. Nature (London) 1983;301:671–674. doi: 10.1038/301671a0. [DOI] [PubMed] [Google Scholar]
- 53.Geliebter J, Nathenson S G. Mol Cell Biol. 1988;8:4342–4352. doi: 10.1128/mcb.8.10.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pease L R, Horton R M, Pullen J K, Yun T J. Mol Cell Biol. 1993;13:4374–4381. doi: 10.1128/mcb.13.7.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun T J, Melvold R W, Pease L R. Proc Natl Acad Sci USA. 1997;94:1384–1389. doi: 10.1073/pnas.94.4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hogstrand K, Bohme J. Immunogenetics. 1999;49:446–455. doi: 10.1007/s002510050518. [DOI] [PubMed] [Google Scholar]