Abstract
Linkage disequilibrium (LD) provides information about positional cloning, linkage, and evolution that cannot be inferred from other evidence, even when a correct sequence and a linkage map based on more than a handful of families become available. We present theory to construct an LD map for which distances are additive and population-specific maps are expected to be approximately proportional. For this purpose, there is only a modest difference in relative efficiency of haplotypes and diplotypes: resolving the latter into 2-locus haplotypes has significant cost or error and increases information by about 50%. LD maps for a cold spot in 19p13.3 and a more typical region in 3q21 are optimized by interval estimates. For a random sample and trustworthy map the value of LD at large distance can be predicted reliably from information over a small distance and does not depend on the evolutionary variance unless the sample size approaches the population size. Values of the association probability that can be distinguished from the value at large distance are determined not by population size but by time since a critical bottleneck. In these examples, omission of markers with significant Hardy–Weinberg disequilibrium does not improve the map, and widely discrepant draft sequences have similar estimates of the genetic parameters. The LD cold spot in 19p13.3 gives an unusually high estimate of time, supporting an argument that this relationship is general. As predicted for a region with ancient haplotypes or uniformly high recombination, there is no clear evidence of LD clustering. On the contrary, the 3q21 region is resolved into alternating blocks of stable and decreasing LD, as expected from crossover clustering. Construction of a genomewide LD map requires data not yet available, which may be complemented but not replaced by a catalog of haplotypes.
Positional cloning of genes for disease susceptibility depends on linkage and “allelic association” (also called “linkage disequilibrium” or LD). A cold spot for LD is an interval in which LD declines rapidly with distance: neither linkage nor LD is proportional to the sequence-based map. To the extent that LD mirrors recombination it can extend the low resolution of linkage: a cold spot for LD is a hot spot for recombination and vice versa. However, this correspondence is disturbed by other factors that cannot be reliably predicted. To the extent that these phenomena are important, both the physical and linkage maps are unreliable guides to LD. We need an LD map to facilitate positional cloning, extend the resolution of the linkage map, compare populations, infer their paleodemography, and detect selective sweeps and other events of evolutionary interest. LD mapping is at the stage of linkage maps nearly a century ago, with the same promise.
The definitive property of a chromosome map, whether physical or genetic, is that its distances are additive. With this constraint, we require a standard LD map to which population-specific maps are approximately proportional. Here we develop LD mapping, examine the relative efficiency of haplotypes and diplotypes, and optimize LD maps for a cold spot in 19p13.3 and a more typical interval in 3q21.
LD Mapping Theory
A map interval is completely specified by a pair of DNA sites, which we shall call “markers.” Theory to estimate the covariance D for a random sample of haplotypes or disomic genotypes (diplotypes) may be extended to the association probability ρ = D/Q(1 − R), where Q is the frequency of the rarest and therefore putatively youngest allele, R is the frequency of the associated marker allele, and D is the absolute value of the difference between a haplotype frequency and its equilibrium value as the product of allele frequencies (1, 2). The optimality of ρ and its basis in evolutionary theory derives from its uniqueness as a probability conditional on R and Q, giving the frequency of the rarest haplotype as Q(1 − R)(1 − ρ). The information Kρ under the null hypothesis that D = 0 is N Q(1 − R)/R(1 − Q) for N random haplotypes or diplotypes. Under the alternative hypothesis the information from haplotypes is a closed form in D (3), but the information from diplotypes must be evaluated by inversion of the 3 × 3 information matrix for Q, R, and D (4). To validate our analysis we randomly paired X chromosomes from males with replacement to create diplotypes and fitted the Malecot model: the estimates from haplotypes and diplotypes were virtually identical. Diplotype analysis has been incorporated into the allass program together with the LD mapping procedure used here (http://cedar.genetics.soton.ac.uk/public_html/).
There are three sources of variation in Kρ: the gene frequencies Q and R, the association ρ, and the inference of haplotype (4). These factors are summarized in Table 1, where Kρ is conditional on ρ, and efficiency E is defined as the ratio of Kρ for haplotypes and diplotypes. Kρ increases with gene frequency when Q = R but decreases as R increases for given Q. There are only 2 haplotypes and therefore 3 diplotypes when Q = R, ρ = 1, and only 3 haplotypes with 6 diplotypes when ρ = 1 but Q < R. This explains why Kρ increases so steeply near the lower right-hand corner of Table 1. All other cases have 4 haplotypes and 10 diplotypes, 2 of which are double heterozygotes differing in phase. Diplotypes and haplotypes contribute the same information when ρ = 0, but haplotype efficiency is half as great when ρ = 1 and the haplotypes of the double heterozygote are certain. In the intervening range the efficiency of a haplotype can slightly exceed a diplotype when Q is moderate and R is large, but then the information is small. In the most favorable case, haplotyping doubles the amount of information, but typically E is roughly 0.75 and thus the gain is about 50%, which must be balanced against the added expense of determining haplotypes by family studies or somatic cell hybrids (5). An earlier comparison of haplotypes and diplotypes used operating characteristics for a very different metric than ρ, with no probabilistic interpretation (6), but the conclusions were similar.
Table 1.
Information K for allelic association ρ in N = 10,000 haplotypes, and efficiency E relative to N diplotypes
| Q | R | ρ = 0
|
ρ = 0.1
|
ρ = 0.5
|
ρ = 0.9
|
ρ = 1
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K | E | K | E | K | E | K | E | K | E | ||
| 0.01 | 0.01 | 10,000 | 1 | 935 | 0.5385 | 203 | 0.5026 | 114 | 0.5001 | 103 | 0.5 |
| 0.10 | 909 | 1 | 508 | 0.7639 | 184 | 0.5531 | 113 | 0.5063 | 103 | 0.5 | |
| 0.10 | 0.10 | 10,000 | 1 | 5,878 | 0.7627 | 2,323 | 0.5318 | 1,518 | 0.5008 | 1,407 | 0.5 |
| 0.50 | 1,111 | 1 | 1,112 | 0.9938 | 1,143 | 0.8549 | 1,221 | 0.5814 | 1,250 | 0.5 | |
| 0.30 | 0.30 | 10,000 | 1 | 9,379 | 0.9417 | 8,842 | 0.6365 | 11,419 | 0.5060 | 13,104 | 0.5 |
| 0.70 | 1,837 | 1 | 1,902 | 1.0115 | 2,323 | 0.9344 | 3,296 | 0.6248 | 3,745 | 0.5 | |
| 0.50 | 0.50 | 10,000 | 1 | 10,101 | 0.9806 | 13,333 | 0.7143 | 52,632 | 0.5277 | 5,002,501 | 0.5 |
LD can be mapped efficiently in diplotype samples. Rare genes of major effect are assigned to haplotypes by family study. Oligogenes by definition have effects so small that they cannot be confidently attributed to an individual, let alone to one or the other or both haplotypes. This uncertainty greatly diminishes the value of haplotyping normal and affected unless the oligogene is unambiguously defined by DNA typing rather than by its phenotypic effect. Therefore, exceptional effort to haplotype valuable samples is seldom justified. The most favorable condition for haplotyping is when an oligogene is predicted to be present on a particular haplotype that has not been verified by family study: selection of a donor for tissue transplantation is a practical example. When expression in cell culture is relevant to a disease, aneuploid cell lines provide information about candidate gene dosage, and monosomic haplotypes are informative for allelic association.
Whether pairwise association is inferred from haplotypes or diplotypes, the Malecot prediction of association is ρ = (1 − L)Me−ɛd + L, where the asymptote L is the bias at large distance, M is the proportion of the youngest haplotype that is monophyletic, and ɛ is the exponential decline of ρ with physical distance d (1). A natural measure of LD is ɛd = θt, where θ is a small frequency of recombination, and t is the number of generations since the population frequency of the rarest two-marker haplotype was minimal (3). In general t exceeds 100 generations, and therefore e−θt is negligible unless θ is so small that θt is proportional to the genetic distance in centimorgans (cM). Because ɛd is not biased in favor of the linkage map and is much more accurately known than θt, it is a more useful metric for LD. To compare the LD map with genetic and physical maps we fit the Malecot model with distance expressed in cM or kb and calculate the residual variances (1, 2).
Over small distances the L parameter is poorly determined when ɛ is estimated simultaneously, and thus ɛ has a large SE. It is therefore useful to have an independent estimate of L, which is the mean value of ρ as e−ɛd = e−θt approaches zero. This condition is clearly satisfied for unlinked genes (θ = 1/2, t >10). Recall that e−θt is an approximation to (1 − θ)t, and that unlinked genes go halfway to linkage equilibrium in one generation. To formalize the argument, let L = LE + (1 − LE)LS, where LE is the contribution of past generations and LS is the bias because of sample size. We assume that LE = 1/(1 + 2Ne) for θ = 1/2 (3), where Ne is the recent effective size. Therefore, LE is far too small to be measurable except in an extreme isolate, where it would not approach significance. On the contrary, LS may be large. For simplicity, assume a random sample, a trustworthy map, and an estimate of ρ that is the average of the absolute deviations in a normal distribution with mean 0 and information K for a particular pair of alleles. If K were constant, LS would be 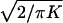 (1). If K varies randomly with respect to distance, LS =
(1). If K varies randomly with respect to distance, LS = 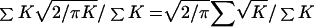 , where the summation is over all pairs of alleles. Because 1/K is the variance in drawing a single sample, LS includes no evolutionary variance. These results may be extended from θ = 1/2 to much smaller values, because the mean value of ρ is effectively L at distances much greater than the swept radius, which is 1/ɛ kb or 100/t cM. Table 2 shows the adequacy of our simple model for L, neglecting LE. We may be confident that the asymptote for LD cannot support a bottleneck 40,000 years ago, as recently proposed (12). L is a nuisance parameter of no evolutionary interest, a source of error for positional cloning, and should be minimized by taking large samples.
, where the summation is over all pairs of alleles. Because 1/K is the variance in drawing a single sample, LS includes no evolutionary variance. These results may be extended from θ = 1/2 to much smaller values, because the mean value of ρ is effectively L at distances much greater than the swept radius, which is 1/ɛ kb or 100/t cM. Table 2 shows the adequacy of our simple model for L, neglecting LE. We may be confident that the asymptote for LD cannot support a bottleneck 40,000 years ago, as recently proposed (12). L is a nuisance parameter of no evolutionary interest, a source of error for positional cloning, and should be minimized by taking large samples.
Table 2.
Prediction and estimation of L
| Ch | Type | Population | Predicted | Estimated | Ref. |
|---|---|---|---|---|---|
| X | H | Wessex | 0.0207 | 0 | 7 |
| 18 | H | U.K. | 0.0387 | 0.0342 | 8 |
| 18 | H | U.S. | 0.0394 | 0.0320 | 8 |
| 18 | H | Finland | 0.0403 | 0.0515 | 8 |
| 18 | H | Sardinia | 0.0393 | 0.0331 | 8 |
| X | H | CEPH | 0.1215 | 0.1276 | 9 |
| X | H | Sardinia | 0.1113 | 0.1300 | 9 |
| X | H | Finland | 0.1144 | 0.1215 | 9 |
| 19 | D | U.S. | 0.0501 | 0.0622 | 10 |
| 3 | D | Sweden | 0.0706 | 0.0453 | 11 |
| Mean | — | — | 0.0646 | 0.0637 | — |
Ch, chromosome; H, haplotype; D, diplotype.
Given n markers on the LD map, let the length of the ith interval be ɛidi LD units (LDU), where ɛi estimates the Malecot parameter, and di is the length of the interval on the physical map in kb. A region has Σɛidi LDU and Σdi kb, with their ratio as a rough estimate of regional ɛ. Here we consider two estimates of ɛi: the estimate when all pairs that include flanking markers i and i + 1 are considered simultaneously, with the adjacent pair entered only once; and the estimate when all pairs that include the interval between markers i and i + 1 are efficiently weighted and pooled. The logic of LD mapping may be inverted to compare physical or genetic maps that differ in assumptions about interference, error rate, sequence length, order, or mapping algorithm; however constructed, a map is optimal if its distances consistently maximize the fit of the Malecot model.
No significant variation in linkage has been detected among human populations. On the contrary, LD varies with population history. A standard map can be created by scaling each partial map by T/tj, where tj is the mean duration estimated for population j and T is the value in a representative population. If z is the ratio of physical distance in kb to the genetic distance in cM over an interval that includes the partial map and has the same value of ɛ, then t = 100zɛ (1). Although LD mapping is too young to solve all of the problems associated with estimation of T, rapid progress may be anticipated as the draft sequence is improved and larger intervals are densely mapped.
Materials and Methods
The data consist of 22 single-nucleotide polymorphisms (SNPs) mapped to a small interval on 19p13.3 (10) and 28 SNPs similarly mapped to 3q21 (11), typed in unrelated individuals of Caucasian ancestry. Maps in these references are termed local. Before diplotype analysis the samples were subjected to Hardy–Weinberg quality control (13) and three significant deviations were identified. The Malecot model was fitted with and without these SNPs, with the error variance estimated by V = −2ln lk/(q − m), where q is the number of SNP pairs, m is the number of parameters estimated, and ln lk = −ΣKρ(ρ̂ − ρ)2/2 is the logarithm of the composite likelihood. A subhypothesis specifying r of these m parameters is tested by χ = Δ/V, where Δ is the difference in −2 ln lk, and the SE of ɛ̂ is taken as σɛ =
= Δ/V, where Δ is the difference in −2 ln lk, and the SE of ɛ̂ is taken as σɛ = 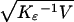 . Estimates of V are inflated by the evolutionary variance, which is unpredictably greater for large estimates of ρ. V is a valid basis for comparison of two analyses of the same data with the same estimates of Kρ and ρ. Comparisons with different estimates of Kρ and ρ are made with σɛ.
. Estimates of V are inflated by the evolutionary variance, which is unpredictably greater for large estimates of ρ. V is a valid basis for comparison of two analyses of the same data with the same estimates of Kρ and ρ. Comparisons with different estimates of Kρ and ρ are made with σɛ.
Because both chromosomes are currently without finished sequences, we performed these analyses for all relevant databases. In this way, the robustness of our LD maps was tested. The flanking method to estimate ɛi depends on a local fit to the Malecot model. It is appropriate if some intervals are large relative to the swept radius, but may smooth the LD map too much. The interval method, which is formally the same as for locus-oriented linkage analysis (14), should give detail at the high resolution of a haplotype catalog (15). Let Shk = Σɛjdj for all disjoint intervals with j between SNPs h and k and ρhk = (1 − L) M exp(−Shk) + L, where M, L, and the trial value of ɛi are taken from the Malecot model for the physical map. Let i be a particular value of j. Then an iterative estimate of ɛi is given by ɛi(t) = ɛi(t−1) + (Ui/Ki)(t−1), where
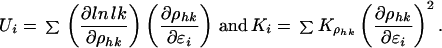 |
This gives a tolerably good estimate of ɛi unless the information Ki is small (say <100), in which case the corresponding flanking estimate or mean adjacent value of ɛi is preferable. The latter is easier to implement and is taken as the default because there is little difference in the few examples here.
The number m of parameters estimated is typically n for n − 1 intervals and M, whether only ɛi or ɛi and Mi are estimated. The interval method does not allow Mi to be estimated, although the value from the flanking method could be used if regions with high and low estimates of Mi are interspersed. When the number of SNPs is small, the correlation between ɛi and di is expected to vary symmetrically around 0, making Σɛidi unequal to ɛΣdi, where ɛ is the regional value. To correct for this, interpolation of small LD maps into a standard map should scale LDU by ɛΣdi/Σɛidi. We omit this refinement pending a standard LD map based on dense markers and a trustworthy sequence.
The two methods to estimate ɛi were applied to maps with minimal deviation from the Malecot model. Finally, the duration t was estimated from sequence-based integrated maps in the LDB2000 database (http://cedar.genetics.soton.ac.uk/public_html/LDB2000.html).
Chromosome 19
The 795 individuals are controls for a migraine study of the insulin receptor (INSR) region (10). Markers have been entered in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/), which creates nine-character accession numbers that are far too long for human communication. Fortunately, the higher-order dbSNP characters have redundancy in this study and are unambiguously reduced to 3 characters (A61, B41) by assigning A = ss40492, B = ss43188.
Primers for all SNPs were located at high stringency in the Celera map (16) but the same blast algorithm located only eight of them in Golden Path (http://genome.ucsc.edu). All these draft sequences have many gaps and errors in contig assembly, with ambiguous orders resolved by fallible radiation hybrid and genetic maps (16–18). It has been reported that errors in order are as frequent in draft sequences as in those maps (19), and therefore “all assemblies of draft sequences should be treated with suspicion” (20). Pending a definitive sequence of chromosome 19, we examined both the Celera and local maps (Tables 3 and 4). The former (model 1) has a much smaller value of V than the corresponding local model 5, as well as a smaller value of σɛ. These differences are maintained if the two SNPs with significant Hardy–Weinberg disequilibrium are omitted or Kρ is evaluated under the alternative hypothesis that ρ is given by the Malecot model when estimates are iteratively reweighted; limiting analysis to the eight markers in all three databases, the Golden Path map fits least well (data not shown).
Table 3.
Application of the Malecot model to 19p13.3 under H0 (parameters specified by hypothesis in parentheses)
| Model | Sequence | Unit | Estimate of ɛi | No. of pairs | df† | ɛ | σɛ | L | M | V |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Celera | kb | — | 231 | 229 | 0.0583 | 0.0052 | (0.0501) | 0.9541 | 6.32 |
| 2 | Celera | kb | — | 210* | 208 | 0.0584 | 0.0054 | (0.0505) | 0.9542 | 6.88 |
| 3 | Celera | LDU | Flanking | 210* | 189 | 0.9952 | 0.0923 | (0.0505) | 0.9783 | 4.69 |
| 4 | Celera | LDU | Interval | 210* | 189 | 0.8724 | 0.0986 | (0.0505) | 0.8693 | 3.52 |
| 5 | Local | kb | — | 231 | 229 | 0.0689 | 0.0103 | (0.0501) | 0.9293 | 8.13 |
| 6 | Local | LDU | Flanking | 231 | 209 | 0.6836 | 0.0499 | (0.0501) | 1.0000 | 5.62 |
| 7 | Local | LDU | Interval | 231 | 209 | 1.0654 | 0.1089 | (0.0501) | 0.8721 | 3.41 |
Omitting B41.
df = no. of pairs − no. of estimated parameters.
Table 4.
LD maps of SNPs in 19p13.3 for Celera sequence
| SNP | Local kb | Celera kb* | Flanking LD
|
Interval LD
|
||
|---|---|---|---|---|---|---|
| ɛ | LDU | ɛ | LDU | |||
| A70 | 365 | 0 | — | 0.00 | — | 0.00 |
| A68 | 200 | 284 | 0.0079 | 2.24 | 0.0076 | 2.17 |
| A69 | 210 | 310 | 0.0147 | 2.62 | 0.0076 | 2.36 |
| A61 | 0 | 476 | 0.0146 | 5.05 | 0.0294 | 7.26 |
| A72 | 560 | 819 | 0.0327 | 16.28 | 0.0356 | 19.45 |
| A74 | 580 | 848 | 0.0422 | 17.49 | 0.0417 | 20.65 |
| A75 | 583 | 856 | 0.0645 | 18.03 | 0.0000 | 20.65 |
| A76 | 583 | 856 | 0.0696 | 18.04 | 0.0000 | 20.65 |
| A77 | 585 | 858 | 0.0823 | 18.20 | 0.0000 | 20.65 |
| A78 | 587 | 860 | 0.0887 | 18.35 | 0.1117 | 20.85 |
| A79 | 589 | 862 | 0.0741 | 18.47 | 0.0000 | 20.85 |
| A81 | 595 | 873 | 0.0891 | 19.45 | 0.3163 | 24.34 |
| A82 | 605 | 882 | 0.1699 | 20.92 | 0.0356 | 24.65 |
| A83 | 617 | 896 | 0.0854 | 22.11 | 0.1820 | 27.18 |
| A85 | 617 | 896 | 0.0422 | 22.11 | 0.0000 | 27.18 |
| A86 | 617 | 896 | 0.0414 | 22.11 | 0.0000 | 27.18 |
| A87 | 620 | 899 | 0.0458 | 22.26 | 0.0000 | 27.18 |
| A88 | 620 | 899 | 0.0610 | 22.26 | 0.0000 | 27.18 |
| A93 | 650 | 917 | 0.0219 | 22.65 | 0.0181 | 27.51 |
| A98 | 660 | 921 | 0.0304 | 22.79 | 0.0574 | 27.77 |
| A99 | 695 | 1019 | 0.0360 | 26.32 | 0.0574 | 33.39 |
| B41 | 300 | 1928 | — | — | — | — |
Rounded.
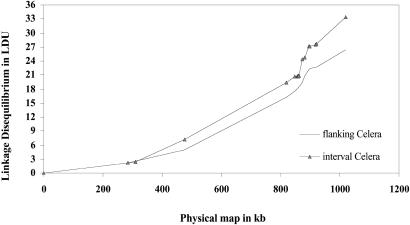
To analyze alternative estimates of ɛi we took the kb-based estimates of M with the predicted values of Ls under H0. The interval between A99 and B41 in the Celera map is so large that ɛi was indeterminate. Omitting B41, the fit measured by V is much better to the LD map (models 3 and 4) than to the physical map (model 2). The interval estimate gives a better fit than the flanking estimate. By using σɛ to measure goodness of fit, the Celera map is superior for the interval estimate but not for the flanking estimate.
Adopting the Celera map, with a swept radius of 1/ɛ = 17 kb, the value of ɛ peaks around A82 (Table 4, Fig 1). This peak is more clearly delineated by the interval LD, although the flanking estimate is similar. The map length is 33.39 LDU and 1,019 kb, their ratio corresponding to ɛ = 0.0328. This is substantially less than the kb-derived values in Table 3, which give ɛ = 0.0583, but is still much greater than estimates for other regions. The kb/cM ratio is 573 for chromosome 19 (21) and 441 for the 19p13.3 region. Taking the lesser estimates for ɛ and z, the corresponding estimate of time since the last bottleneck is t = 100zɛ, or at least 1,446 generations, which is larger than other regions have given (3). To reduce t to a typical value of 300 would require z no greater than 100, implying 10 cM/Mb. Such a high recombination rate has not been observed over distances as great as 1 Mb. Therefore, the elevated value of ɛ suggests an unusually long time as well as a high recombination rate.
Figure 1.
LD maps for 19p13.3.
A striking feature of the data is that the estimate of ɛ from the LD map does not equal unity, as we verified that it does if the estimate of ɛi is constant over all intervals or if ɛi and di vary independently in the sample, as they presumably would if the number of SNPs were very large. However, when the number of SNPs is small, the correlation between ɛι and di is expected to vary symmetrically around 0, and therefore interpolation of small LD maps into a standard map should scale LDU to restore the relation Σɛidi/Σdi = ɛ. For example, the LD map in Table 4 should be multiplied by ɛd/Σɛidi = (0.0583) (1019)/33.39, which changes the scale but not the shape of Fig. 1. We omit this refinement pending a trustworthy sequence and a dense marker map.
Despite the complexity introduced by uncertain sequence, there is good agreement for ɛ between the Celera and local maps (models 1 and 5), which may properly be compared because they have the same distribution of Kρ, reflected by the same predicted value Ls. Such comparisons for H1 and exclusion of the same SNPs also agree, although ɛ is reduced to 0.029 for Celera and 0.031 for Golden Path in maps of the 8 SNPs located in all draft sequences (data not shown). These SNPs are proximal in Fig. 1, where ɛ is minimal.
Chromosome 3
The 400 individuals are unaffected parents for a psoriasis study in southwest Sweden (11). There are four higher-order symbols in dbSNP: A = ss3173, B = ss2992, C = ss4250, and D = ss2, generating symbols like A382, B188, D665, etc. Primers for 6 SNPs could not be located in the Celera sequence, and 3 primers were not found in Golden Path. The estimate of M in Table 5 is consistently small, suggesting polyphyletic origin, perhaps caused by gene conversion. If so, the conversion probability is regionally specific, a phenomenon not previously encountered or easily explained. The Celera map (model 1) has the smallest value of V, whereas Golden Path (model 2) has the smallest σɛ and a larger number of SNPs, making comparison of V invalid. Estimates of ɛ exceed most other regions (1–3), but are much less than for 19p13.3. The swept radius is 205 kb.
Table 5.
Application of the Malecot model to 3q21 under H0 (parameters specified by hypothesis in parentheses)
| Model | Sequence | Unit | Estimate of ɛi | No. of pairs | df* | ɛ | σɛ | L | M | V |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Celera | kb | — | 231 | 229 | 0.0092 | 0.0041 | (0.0705) | 0.2914 | 1.86 |
| 2 | Golden Path | kb | — | 300 | 298 | 0.0049 | 0.0010 | (0.0706) | 0.2672 | 2.85 |
| 3 | Golden Path | LDU | Flanking | 300 | 275 | 0.6854 | 0.1037 | (0.0706) | 0.3215 | 2.90 |
| 4 | Golden Path | LDU | Interval | 300 | 275 | 1.0262 | 0.1898 | (0.0706) | 0.3393 | 2.37 |
| 5 | Local | kb | — | 378 | 376 | 0.0063 | 0.0011 | (0.0695) | 0.3343 | 3.78 |
| 6 | Local | LDU | Flanking | 378 | 350 | 0.5603 | 0.0769 | (0.0695) | 0.3704 | 3.73 |
| 7 | Local | LDU | Interval | 378 | 350 | 1.4383 | 0.2771 | (0.0695) | 0.4331 | 3.01 |
df = no. of pairs − no. of estimated parameters.
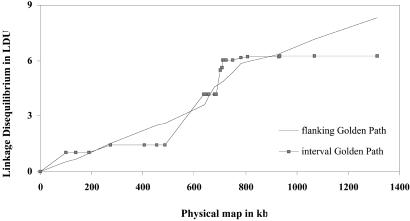
Among LD maps for Golden Path the smallest V is given by the interval estimate. For the Golden Path and local sequences the LD map has a much smaller value of V than the physical map. Golden Path was chosen as a compromise between number of markers and reliability for Table 6. The map in LDU has distortion for the flanking estimate as discussed above for chromosome 19. However, the interval estimate is not distorted and shows blocks of conserved LD more clearly (Fig. 2). The transition between blocks extends over many kb in contrast with tight clustering in recombination hot spots (15). Three possible explanations are discussed below.
Table 6.
LD maps of SNPs in 3q21 for Golden Path sequence
| SNP | Local kb | Celera kb | Golden Path kb* | Flanking LD
|
Interval LD
|
||
|---|---|---|---|---|---|---|---|
| ɛ | LDU | ɛ | LDU | ||||
| B188 | 1,000 | 1,423 | 0 | — | 0.00 | — | 0.00 |
| B193 | 1,050 | 1,520 | 101 | 0.0053 | 0.54 | 0.0104 | 1.05 |
| B195 | 920 | 1,557 | 137 | 0.0036 | 0.67 | 0.0000 | 1.05 |
| B194 | 860 | 1,610 | 189 | 0.0062 | 1.00 | 0.0000 | 1.05 |
| A382 | 830 | 1,696 | 275 | 0.0063 | 1.54 | 0.0046 | 1.44 |
| B197 | 720 | 1,767 | 405 | 0.0055 | 2.26 | 0.0000 | 1.44 |
| D665 | 650 | 1,920 | 455 | 0.0054 | 2.53 | 0.0000 | 1.44 |
| B196 | 790 | 1,853 | 486 | 0.0029 | 2.62 | 0.0000 | 1.44 |
| B207 | 495 | 157 | 637 | 0.0066 | 3.61 | 0.0181 | 4.18 |
| B206 | 490 | 151 | 643 | 0.0179 | 3.71 | 0.0000 | 4.18 |
| A380 | 520 | 183 | 655 | 0.0303 | 4.08 | 0.0000 | 4.18 |
| C116 | 470 | 139 | 678 | 0.0216 | 4.58 | 0.0000 | 4.18 |
| B205 | 465 | 133 | 684 | 0.0069 | 4.62 | 0.0000 | 4.18 |
| A379 | 450 | 116 | 701 | 0.0089 | 4.77 | 0.0780 | 5.49 |
| B202 | 445 | 110 | 707 | 0.0143 | 4.86 | 0.0234 | 5.64 |
| B204 | 440 | 105 | 712 | 0.0028 | 4.87 | 0.0803 | 6.05 |
| B203 | 435 | 101 | 716 | 0.0035 | 4.89 | 0.0000 | 6.05 |
| B191 | 430 | 94 | 723 | 0.0208 | 5.03 | 0.0000 | 6.05 |
| B201 | 400 | 66 | 751 | 0.0120 | 5.37 | 0.0000 | 6.05 |
| B189 | 300 | 35 | 782 | 0.0161 | 5.86 | 0.0038 | 6.17 |
| B200 | 250 | — | 808 | 0.0035 | 5.95 | 0.0019 | 6.21 |
| B187 | 200 | — | 929 | 0.0033 | 6.35 | 0.0000 | 6.21 |
| B199 | 100 | — | 932 | 0.0068 | 6.37 | 0.0082 | 6.24 |
| B190 | 360 | 0 | 1067 | 0.0059 | 7.16 | 0.0000 | 6.24 |
| B198 | 0 | 3,338 | 1,313 | 0.0047 | 8.31 | 0.0000 | 6.24 |
| A383 | 390 | — | — | — | — | — | — |
| A381 | 600 | — | — | — | — | — | — |
| B192 | 690 | — | — | — | — | — | — |
Rounded.
Figure 2.
LD maps for 3q21.
Discussion
Errors in distance and order are present in genetic maps constructed before a draft of the genome sequence was available, but their effect has been blunted by the low density of microsatellites used for these scans. On the contrary, errors in the draft sequences are frequent (20) and consequential for LD mapping. We have examined alternative maps, but all LD mapping must be taken cautiously until sequences are verified. Recently, two releases of the Golden Path sequence moved the FRAXE region 75 Mb from its location in Xq28. That error has been corrected, but draft sequences continue to have many gaps and errors, and there is no international effort to integrate the sequence with an accurate LD map. Both a trustworthy sequence and an accurate LD map are indispensable for efficient positional cloning.
Controversy about LD mapping and its alternatives extends to differences among populations, reflected by the parameters ɛ and t, which are specific to the metric fitted by the Malecot model. We have shown that ρ is more efficient than alternative metrics that continue to be used (3). Estimates of ɛ derived from an evolutionary model for ρ have the least confounding with sample size and are most robust to allele frequencies. One alternative is kinship ϕ, a prediction of the squared correlation coefficient r2 with an unbiased estimate of (χ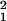 − 1)/(N − 1) for a sample of N haplotypes (22). It is usually justified by an equilibrium between drift (measured by effective size Ne, assumed constant) and recombination θ as duration t approaches infinity. Under these strong assumptions, the expected value of ϕ is 1/(1 + 4Neθ), with information estimated for a noncentral χ2 distribution. However, effective population size is not constant and duration is not infinite. A close approach to the general theory requires distance greater than the swept radius 1/ɛ, where all metrics are indistinguishable from their asymptote L (23, 24). Positional cloning by allelic association, especially for major genes with a short history (1, 25), depends on the relation of LD to time within distances less than the swept radius (3). The relation with time is supported by archaeology and history and captured by the Malecot model, but lost when duration approaches infinity. Constancy of effective population size is not assumed by the Malecot model, but is required by asymptotic theory. There is an intermediate range of θ, perhaps between 0.02 and 0.10, where equilibrium is approached in a few centuries and therefore ρ may be more closely related to Ne than to t as ρ ∼ (1 − L)/(1 + 2Neθ) + L, but the excess over L in this range is small. On present evidence it does not seem useful to pursue alternatives to the Malecot model for ρ.
− 1)/(N − 1) for a sample of N haplotypes (22). It is usually justified by an equilibrium between drift (measured by effective size Ne, assumed constant) and recombination θ as duration t approaches infinity. Under these strong assumptions, the expected value of ϕ is 1/(1 + 4Neθ), with information estimated for a noncentral χ2 distribution. However, effective population size is not constant and duration is not infinite. A close approach to the general theory requires distance greater than the swept radius 1/ɛ, where all metrics are indistinguishable from their asymptote L (23, 24). Positional cloning by allelic association, especially for major genes with a short history (1, 25), depends on the relation of LD to time within distances less than the swept radius (3). The relation with time is supported by archaeology and history and captured by the Malecot model, but lost when duration approaches infinity. Constancy of effective population size is not assumed by the Malecot model, but is required by asymptotic theory. There is an intermediate range of θ, perhaps between 0.02 and 0.10, where equilibrium is approached in a few centuries and therefore ρ may be more closely related to Ne than to t as ρ ∼ (1 − L)/(1 + 2Neθ) + L, but the excess over L in this range is small. On present evidence it does not seem useful to pursue alternatives to the Malecot model for ρ.
Choice of population influences ɛ and t: pedigrees, villages, provinces, and countries have different values, but the origins of samples used for studies of human diversity are rarely specified with precision. Populations with few founders and small values of t are expected and observed to have small values of ɛ (26, 27), but the magnitude of this effect is uncertain. Slatkin (28) argued that a stable population should have smaller values of ɛ (i.e., more LD) than an expanding population, but his argument was based on simulations with different effective sizes. Population genetic theory shows that the effective size is the harmonic mean of values over t generations, which is unaffected by the order of those values and is therefore not systematically different for stable and expanding populations (3).
The data in Table 2 are ambiguous in the absence of sample definition and they do not justify enthusiasm about high LD in isolates. However, part of this material has been used to support the opposite conclusion (23). These authors reexamined a small subset of the data (9), selected by the largest values of r2 = χ2/N in a Finnish sample (regardless of distance), among SNP pairs informative in the Sardinian sample, where many markers were not typed. Even in these unrepresentative data we were unable to confirm high values of LD at large distances in isolates relative to the Centre d'Etude du Polymorphisme Humain (CEPH) sample, which is a mixture from four populations (French, Utahan, Venezuelan, and Amish). Data on the Ashkenazi sample from which 17 pairs were chosen (23) will be awaited with interest. Their metric was the ratio of r2 in two samples, which is far too skewed for a parametric test. On the evidence, a conspicuously high value of LD in isolates has not been demonstrated.
Recent months have seen advocacy of several approaches to allelic association of markers. One strategy is to pool DNA within normal and affected groups; this sacrifices Hardy–Weinberg quality control, allowance for population substructure, and information about LD in the candidate region. At the opposite extreme, pairwise LD is ignored and all emphasis is placed on haplotype frequencies, perhaps determined in somatic cell hybrids that are monosomic for the chromosome of interest (5). Primers must be long enough to be specific for human markers, and it is unjustifiably assumed that information about haplotype frequencies is equal to information about positional cloning. This faith has given rise to the concept of “haplotype mapping” (29, 30). Among sequences of the same length, the number of common haplotypes is minimal under low recombination and therefore high LD. A selective sweep has the same effect but is presumably infrequent. An LD map allows selection of markers at distances approaching their swept radius 1/ɛ, and therefore at low density in regions of high LD. Although haplotypes are useful in the positional cloning endgame to identify causal SNPs within a significant candidate region, the proposal that researchers can restrict their studies to SNPs that differentiate the few common haplotypes is misguided for causal SNPs in rare haplotypes (24) and impractical in regions of low LD. For example, positional cloning in the 19q13.3 cold spot examined here requires SNPs at high density regardless of the small haplotypes with which they are associated. To distinguish causal from associated SNPs an even greater density is required. Fortunately, LD declines with distance even within a tagged haplotype (15). Haplotypes are population-specific, and therefore a “haplotype map” must be racially biased (29). On the contrary, a standard LD map contains no information that could conceivably stigmatize any population and thus is ethnically as blind as the linkage map, although effective use requires estimates of population-specific duration.
Pairwise LD can be represented in a triangular matrix (trimat) of ordered SNPs that confounds intensity of LD with distance (and with sample size and allele frequencies if intensity is measured by statistical significance). The suggestion that correlated SNPs “probably constitute a haplotype” (29) is a misleading way of saying that the set of haplotypes defined by those SNPs might be interesting. An LD map conveys the same information without confounding. Haplotype annotation contributes to evolutionary studies and in ways yet to be developed may increase resolution of LD mapping, but we should not on that account fail to recognize that an LD map is not a haplotype map, and a haplotype map is an oxymoron unless defined by changes of slope in an LD map. Such changes tend to transform the logarithmic likelihood from its predicted parabola under the Malecot model to a superposed curve with inflexions more like a step pyramid, the steps corresponding to recombination events and perhaps to hot spots of recombination. A causal SNP in a relatively flat terrace is distinguishable from neighboring predictive SNPs by mutation, gene conversion, and recombination that subdivide the deeper clades (15), if not in an isolate then in an older population with other recombination events and different haplotypes. Major alleles with short duration are most associated with particular haplotypes and therefore show the greatest deviation from the Malecot model, which nevertheless determines their location well (1, 6). Older oligogenes must be less sensitive to inflexions in the LD map, which may mirror recombination more accurately than the few families on which the linkage map is currently based. LD maps will certainly increase the efficiency of positional cloning, for which the physical map is an unreliable measure of distance (31).
For the HLA region male meiosis shows tight clustering of recombinants in hot spots with an SD of 300 bp (15). In contrast, LD maps show larger intervals between blocks of conserved LD (9, 15, 32). There are at least three possible explanations. First, current LD maps are not at high resolution and therefore confound tight clustering with flanking sequences that have less recombination. Second, the recombinational hot spots are clustered and therefore pooled in low-resolution LD maps. Third, sequences predisposed to high recombination have not been identified in humans and may depend on position in an isochore or other chromosome structure that can be altered by insertions or deletions in flanking sequences; in that event, the location of a hot spot can vary during hominid evolution. These uncertainties will remain until there is a high-resolution LD map of the genome with haplotype annotation.
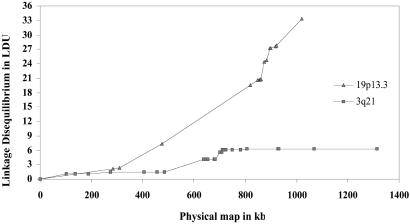
Because we cannot foresee the impact of these developments, this article is limited to elaboration of methods and their application to two datasets. Interval estimates of ɛi give the best fit. It seems that the 19p13.3 region is a cold spot for LD with a swept radius of 17 kb, whereas the 205-kb swept radius of the 3q21 region is more typical and demonstrates the alternation of hot and cold spots that is expected when markers are closer than the swept radius (Fig. 3). Although an artifact of errors in the draft sequences cannot be excluded, all analyses of alternative sequences are consistent. The apparent association between high recombination and long duration remains to be explained. It could be spurious, because the distribution of these estimates along the map is as yet unknown. However, there is a connection between the two phenomena, because high recombination reduces ρ and therefore makes loss of the rarest haplotype less likely.
Figure 3.
Interval LD maps for 19p13.3 and 3q21.
Although the magnitude of any association between θ and t and consistency among ethnic groups are uncertain, a plausible but untested hypothesis is that different populations have LD maps in substantial agreement except for a scalar representing duration. Such uncertainties raise obvious problems for construction of a standard LD map that cannot be resolved without critical data. Ten years ago these problems could not have been imagined. Ten years from now, they will have been solved if they are appropriately pursued in reliable sequences. The intimate relation between an LD map and haplotyping annotation not only makes them complementary, but assures that they are provided by the same dataset.
Acknowledgments
This analysis of data generated by GlaxoSmithKline was supported by grants from the Medical Research Council.
Abbreviations
- SNP
single-nucleotide polymorphism
- cM
centimorgan
- LD
linkage disequilibrium
- LDU
LD unit
References
- 1.Collins A, Morton N E. Proc Natl Acad Sci USA. 1998;95:1741–1745. doi: 10.1073/pnas.95.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins A, Lonjou C, Morton N E. Proc Natl Acad Sci USA. 1999;96:15173–15177. doi: 10.1073/pnas.96.26.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morton N E, Zhang W, Taillon-Miller P, Ennis S, Kwok P-Y, Collins A. Proc Natl Acad Sci USA. 2001;98:5217–5221. doi: 10.1073/pnas.091062198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill W G. Heredity. 1974;33:229–239. doi: 10.1038/hdy.1974.89. [DOI] [PubMed] [Google Scholar]
- 5.Douglas J A, Boehnke M, Gillanders E, Trent J M, Gruber S B. Nat Genet. 2001;28:361–364. doi: 10.1038/ng582. [DOI] [PubMed] [Google Scholar]
- 6.Thompson E A, Deeb S, Walker D, Motulsky A G. Am J Hum Genet. 1988;42:113–124. [PMC free article] [PubMed] [Google Scholar]
- 7.Ennis S, Collins A, Tapper W J, Murray A, Macpherson J N, Morton N E. Ann Hum Genet. 2001;65:503–504. doi: 10.1017/s000348000100879x. [DOI] [PubMed] [Google Scholar]
- 8.Eaves I A, Merriman T R, Barber R A, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd J A. Nat Genet. 2000;25:320–323. doi: 10.1038/77091. [DOI] [PubMed] [Google Scholar]
- 9.Taillon-Miller P, Bauer-Sardina I, Saccone N L, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice J P, Kwok P-Y. Nat Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy L C, Hosford D A, Riley J H, Bird M I, White N J, Hewett D R, Peroutka S J, Griffiths L R, Boyd P R, Lea R A, et al. Genomics. 2001;78:135–149. doi: 10.1006/geno.2001.6647. [DOI] [PubMed] [Google Scholar]
- 11.Hewett, D., Samuelsson, L., Polding, J., Enlund, F., Cantone, K., See, C. G., Smart, D., Chadha, S., Inerot, A., Enerback, C., et al. (2002) Genomics, in press. [DOI] [PubMed]
- 12.Reich D E, Cargill M, Bolk S, Ireland J, Sabeti P C, Richter D J, Lavery T, Kouyoumjian R, Farhadian S F, Ward R, Lander E S. Nature (London) 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 13.Gomes I, Collins A, Lonjou C, Thomas N S, Wilkinson J, Watson M, Morton N. Ann Hum Genet. 1999;63:535–538. doi: 10.1017/S0003480099007824. [DOI] [PubMed] [Google Scholar]
- 14.Collins A, Teague J, Keats B J, Morton N E. Genomics. 1996;36:157–162. doi: 10.1006/geno.1996.0436. [DOI] [PubMed] [Google Scholar]
- 15.Jeffreys A J, Kauppi L, Neumann R. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 16.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 17.Teague J W, Collins A, Morton N E. Proc Natl Acad Sci USA. 1996;93:11814–11818. doi: 10.1073/pnas.93.21.11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 19.Olivier M, Aggarwal A, Allen J, Ahmendras A A, Bajorek E S, Beasley E M, Brady S D, Bushard J M, Bustos V I, Chu A, et al. Science. 2001;291:1298–1302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- 20.Semple C A. Genome Biol. 2001;2:2001.1–2001.6. doi: 10.1186/gb-2001-2-6-reviews2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins A, Frezal J, Teague J, Morton N E. Proc Natl Acad Sci USA. 1996;93:14771–14775. doi: 10.1073/pnas.93.25.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton N E, Wu S S. Am J Hum Genet. 1988;42:173–177. [PMC free article] [PubMed] [Google Scholar]
- 23.Shifman S, Darvasi A. Nat Genet. 2001;28:309–310. doi: 10.1038/91060. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard J K. Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Nat Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 26.Gordon D, Simonic I, Ott J. Genomics. 2000;66:87–92. doi: 10.1006/geno.2000.6190. [DOI] [PubMed] [Google Scholar]
- 27.Kruglyak L. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 28.Slatkin M. Genetics. 1994;137:331–336. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helmuth L. Science. 2001;293:583–585. doi: 10.1126/science.293.5530.583b. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein D B. Nat Genet. 2001;29:109–111. doi: 10.1038/ng1001-109. [DOI] [PubMed] [Google Scholar]
- 31.Lonjou C, Collins A, Ajioka R S, Jorde L B, Kushner J P, Morton N E. Proc Natl Acad Sci USA. 1998;95:11366–11370. doi: 10.1073/pnas.95.19.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly M J, Rioux J D, Schaffner S F, Hudson T J, Lander E S. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]