Abstract
Transglutaminase 2 (TG2) is a distinctive member of the family of Ca2+-dependent enzymes recognized mostly by their abilities to catalyze the posttranslational crosslinking of proteins. TG2 uniquely binds and hydrolyzes GTP; binding GTP inhibits its crosslinking activity but allows it to function in signal transduction (hence the Gh designation). The core domain of TG2 (residues 139–471, rat) comprises the papain-like catalytic triad and the GTP-binding domain (residues 159–173) and contains almost all of the conserved tryptophans of the protein. Examining point mutations at Trp positions 180, 241, 278, 332, and 337 showed that, upon binding 2′-(or 3′)-O-(N-methylanthraniloyl)GTP (mantGTP), the Phe-332 mutant was the weakest (35% less than wild type) in resonance energy transfer from the protein (λexc, max = 290 nm) to the mant fluorophore (λem = 444 nm) and had a reduced affinity for mantGTP. Trp-332, situated near the catalytic center and the nucleotide-binding area of TG2, may be part of the allosteric relay machinery that transmits negative effector signals from nucleotide binding to the active center of TG2. A most important observation was that, whereas no enzyme activity could be detected when Trp-241 was replaced with Ala or Gln, partial preservation of catalytic activity was seen with substitutions by Tyr > Phe > His. The results indicate that Trp-241 is essential for catalysis, possibly by stabilizing the transition states by H-bonding, quadrupole–ion, or van der Waals interactions. This contrasts with the evolutionarily related papain family of cysteine proteases, which uses Gln-19 (papain) for stabilizing the transition state.
Transglutaminase 2 [here designated TG2 (EC2.3.2.13), but also identified as the GTP-binding protein, Gh] differs from its relatives in the family of Ca2+-dependent protein crosslinking enzymes mainly by its affinity for nucleotides (1–3) and for fibronectin (4–8). GTP-binding and hydrolysis enable TG2 to act as a G protein in signal transduction (Gh; ref. 9); GTP also serves as a potent allosteric inhibitor that suppresses the Ca2+-activated crosslinking activities of the enzyme. Independently of any of its catalytic functions and GTP-binding activity, TG2 binds fibronectin with very high affinity (7). Thus, when expressed on cell surfaces, TG2 plays an important role in organizing the extracellular matrix as an integrin-binding adhesion coreceptor (10, 11).
Crystal structures are available for two related proteins: the A subunit of the coagulation factor XIII zymogen (fXIIIA; ref. 12) and the sea bream liver TG (13). Although neither of these is regulated by GTP (personal information from Kohki Ishikawa regarding the latter enzyme), sequence homologies between TG2 (rat) and these proteins suggest that TG2/Gh also would be organized into four domains: a β sandwich (residues 1–138) followed by a large α/β catalytic core (139–471) and two β barrels [barrel 1 (472–584) and barrel 2 (585–686)]. The N-terminal (≈28 kDa) region of TG2 is responsible for binding fibronectin (8) whereas the C-terminal segment of barrel 2 could interact with phospholipase in signal transduction (14). The core domain comprises the papain-like catalytic center (15) and the nucleotide-binding residues (16). Like the monomeric but unlike the heterotrimeric G proteins, TG2 was shown to bind 2′-(or 3′)-O-(N-methylanthraniloyl)GTP (mantGTP) with high affinity (17). Binding was evidenced also by the appearance of an energy transfer band from one or more Trp residues of the protein to the mant fluorophore in this nucleotide. Because the core domain contains almost all of the tryptophans conserved throughout the TG family, we evaluated the nucleotide-binding properties and the transamidating activities of candidate tryptophan point mutants of TG2. Specifically, W180, 241, 278, 332, and 337 were targeted because they surround S171, a residue identified to be critical for GTP-binding. On a model of TG2 based on the fXIIIA zymogen crystal structure (16), all of the mutated residues are within 20Å (range 9.3–18.7Å) of both S171 and the active-site cysteine C277. We show that no single Trp residue alone accounts for the observed energy transfer from the protein to the nucleotide but some of the tryptophan mutants displayed greatly diminished mantGTP-binding affinities. Another important finding was that residue W241—that is highly conserved among the TGs but is lacking in the catalytically inactive family member of human red cell, band 4.2—is critical for transamidation by TG2.
Materials and Methods
Constructs.
Site-directed mutants of rat TG2/Gh cDNA were generated in a temperature-cycling reaction with Pfu DNA polymerase by using two complementary oligonucleotide primers containing the desired mutation and glutathione S-transferase (GST)-TG2/pGEX2T (18) as the template. The parental strand was removed by DpnI digestion before transformation into Escherichia coli strain DH5α. Mutations were verified by DNA sequence analysis.
Purification of Recombinant Proteins.
Wild-type (wt) TG2 and mutants were expressed as thrombin-cleavable glutathione S-transferase fusion proteins (18). Protein expression was induced overnight at 30°C with 100 μM isopropyl β-d-thiogalactoside, and the resulting TG2 proteins were purified from the soluble fraction after cell lysis by affinity chromatography by using a glutathione-Sepharose 4B resin. TG2 or mutants were liberated from the resin-bound glutathione S-transferase moiety by thrombin (Sigma) cleavage for 2 h at 25°C, as described (16). Eluted TG2 proteins were purified by using a MonoQ 5/5 anion exchange column at 25°C, with a 50–600 mM NaCl linear gradient over 30 min at 0.5 ml/min. The peak TG2 fractions were concentrated by using centrifugal concentrators (Millipore) to 1–2 mg/ml and after dialysis into imidazole buffer (20 mM imidazole-HCl/100 mM NaCl/1 mM EDTA/20% glycerol, pH 7.5) they were stored at 4°C.
The protein preparations were examined by SDS/PAGE (19), and by nondenaturing electrophoresis in agarose before and after the addition of GTPγS (4 mM) as described (20).
Nucleotide-binding constants were measured by titrating changes of fluorescence anisotropy with mantGTP (17).
TG activity was measured as described previously (16). In brief, a 50-μl reaction mixture of 5 μM 1,4-[3H]putrescine (15–50 Ci/mmol; NEN) and 2 mM putrescine, containing also 8 mg/ml N,N′-dimethylcasein, 20 mM DTT, 10 mM MgCl2, 0 (basal) or 2 mM (maximal) CaCl2, and the amounts of TG2 enzyme indicated in the figures, was incubated at 37°C for 10 min (wt TG2) or 40 min (TG2 mutants). Reactions were stopped by the addition of 400 μl of ice-cold 50% trichloroacetic acid. The resulting pellets were dissolved (100 μl of 0.1 N NaOH) and counted in 5 ml of ReadySafe (NEN).
Results and Discussion
All targeted Trp residues (Fig. 1) were replaced by Ala (that has only a minimal hydrophobic side chain) or Phe (that mimics Trp hydrophobicity and volume). Recoveries for the W180A, W332A, and W337A mutants were too low to allow functional analysis. Interestingly, the W241A and W241F mutants demonstrated marked loss of TG activity. These mutants and the additionally constructed Q and H 241 substitutions were thus subjected to detailed analysis. Except for the presence of occasional minor fragments, possibly because of proteolytic breakdown, all preparations showed good homogeneity by SDS/PAGE criteria (Fig. 2A) for a protein with an Mr value of ≈77 kDa (the calculated Mr for rat TG2 is 76,755). In preliminary experiments, no single Trp mutation abrogated energy transfer to mantGTP but, judged by the areas of the transfer bands (λexc, max = 290 nm, λem = 444 nm; ref. 17), W332F was most deficient (35% less than wt) in this regard (data not shown).
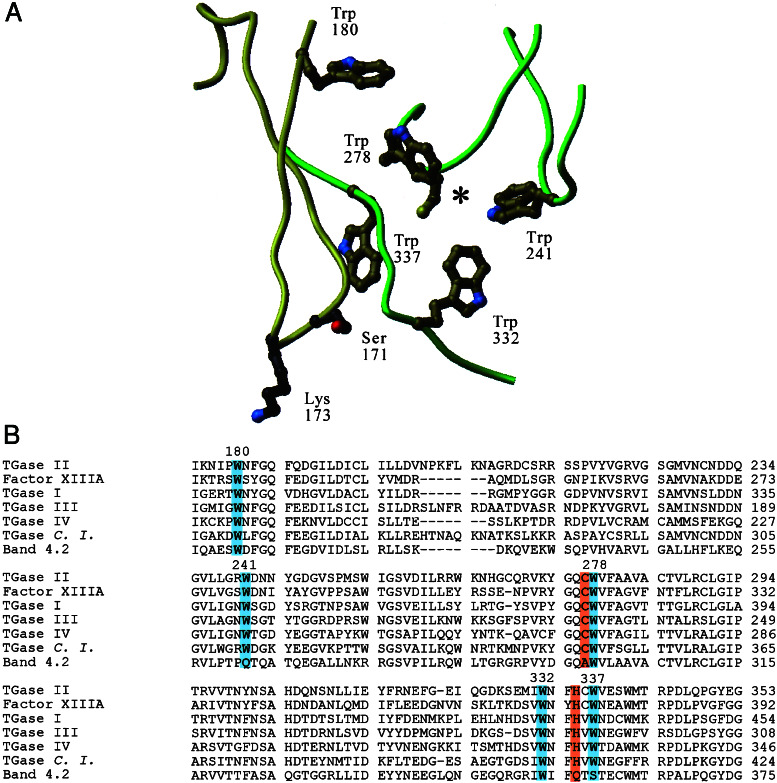
Figure 1.
Conserved tryptophan residues in a model of TG2 based on the fXIIIA crystal structure. (A) Location of conserved tryptophan residues situated within 20Å of Ser-171, a residue determined to be critical for GTP-binding, and within 20Å of the active-site cysteine, Cys-277 (*). Lys-173, another residue involved in GTP binding, is also shown. The GTP-binding loop (residues 159–173), previously identified (16), is shown in yellow. (B) Alignment of human TGases I–IV, fXIIIA, and band 4.2, and the TG-like protein from Ciona intestinalis (C.I.; ref. 26), showing some of the Trp residues conserved throughout evolution. The mutated tryptophans are highlighted in blue, and the active-site cysteine and histidine residues are shown in orange. The Asp of the catalytic triad is not shown. Note the catalytically inactive band 4.2 not only lacks the active-site cysteine and histidine (shown) and aspartic acid (not shown) but also the Trp at the 241-equivalent position.
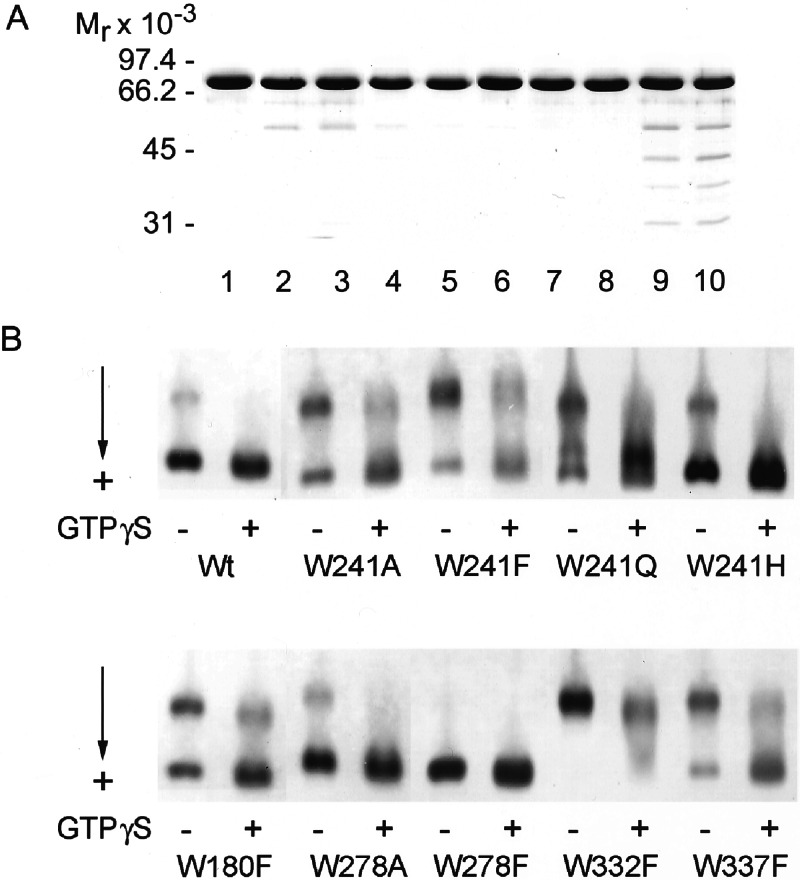
Figure 2.
Electrophoretic profiles of TG2 (rat, recombinant) and core domain tryptophan mutants. (A) SDS/PAGE, lanes 1–10: wt, W241A, W241F, W241Q, W241H, W180F, W278A, W278F, W332F, and W337F. Approximately 4 μg of proteins were applied to each lane. Mr markers are shown on the left. (B) Nondenaturing electrophoresis was carried out in 2% agarose without and with the addition of 4 mM GTPγS/Mg2+, as described (20). Approximately 8 μg of proteins were applied to each lane. The W241Y mutant gave the same response with GTPγS as wt.
As evidenced by electrophoretic mobilities under nondenaturing conditions, in terms of surface charge distribution and hydrodynamic properties, all mutant proteins seemed to have assumed one or both of the known native conformations (s = slow and f = fast) that characterize the human red cell and guinea pig liver TG2s (20). Although different proportions of the f and s forms were found in the various preparations (Fig. 2B), both of these were present in the wt recombinant TG2 and also in W241A, W241F, W241Q, W241H, W180F, W278A, and W337F. However, the W278F mutant was almost entirely in the f and W332F almost exclusively in the s form. As shown earlier, f represents a nucleotide-bound TG2 [with slight differences in the mobilities of the fast species (f1, f2, f3) depending on whether a nucleotide mono-, di-, or triphosphate is bound; 20]. By admixture of GTPγS, the s components in some of the recombinant proteins, i.e., wt and W278A, converted more fully to f than in others, whereas the mobility of s in W332F was barely affected by the addition of the nucleotide. The observation suggests that significant differences exist in the GTP affinities of the mutants.
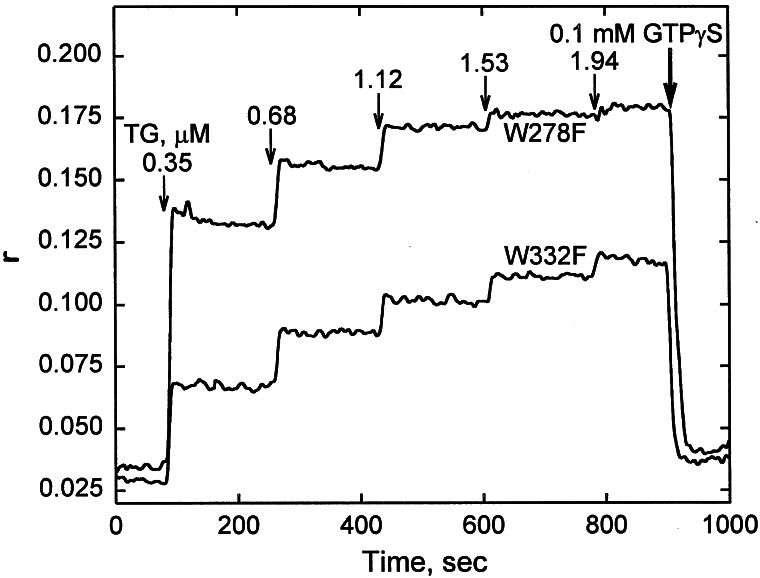
To strengthen this qualitative assessment, titration with mantGTP was used to evaluate binding constants for two mutant proteins that showed the greatest difference in electrophoretic mobilities: the W278F pure f form and W332F, the pure s form. As detected by fluorescence depolarization (anisotropy r values are plotted on the ordinate of Fig. 3), incremental additions of TG2s (to 0.35, 0.68, 1.12, 1.53, and 1.94 μM) to the mantGTP/Mg2+ (0.1 μM) originally present in the mixture, produced much larger changes in the titrations with W278F than with W332F. Binding constants were calculated as before (20): K ≈ 3 × 107 M−1 for W278F and K ≈ 4 × 106 M−1 for W332F.
Figure 3.
Comparison of mantGTP/Mg2+-binding between TG2 mutants W278F and W332F. The titration was carried out by changes in fluorescence anisotropy (ordinate) when aliquots of the proteins were injected stepwise (vertical arrows, to the concentrations marked) into 0.1 μM starting solution of the nucleotide. At the end of the experiment, 0.1 mM GTPγS/Mg2+ was injected (heavy vertical arrow). For details, see ref. 17.
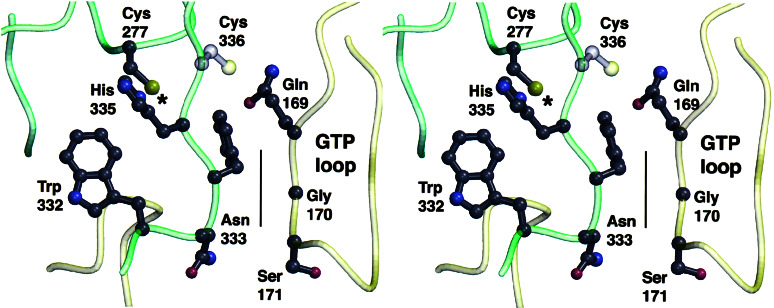
The structural relationship between W332 and the 15-aa loop comprising residues 159–173 that represents the nucleotide-binding domain of TG2/Gh (16) is demonstrated in Fig. 4. By being adjacent to the β-strand that abuts the active site of TG2, W332 is situated strategically just below the catalytic residues and is directed toward the active site. The residues of the interface between the β-strand and the nucleotide-binding loop (marked by the line in Fig. 4) are highly conserved, and this interface may be critical for the allosteric communication between the active center of TG2 and its GTP-binding domain. Because of its close association to the interface, the W332F mutation may cause a conformational change in the 333–336 strand that would disrupt this interface. It may be of interest that greatly reduced nucleotide-binding affinities similar to W332F were measured also for the active center mutants of TG2/Gh: C277A and C277S (data not shown).
Figure 4.
The structural relationship between Trp-332 and the putative GTP-binding loop (16) modeled on the coordinates of the sea bream transglutaminase (13). Gly-170 of the loop is highly conserved and located at the center of a packing interface indicated by the line that occurs between residues 169 and 171 of the loop and one strand of the β-sheet that abuts the active site. * marks the catalytic Cys-277.
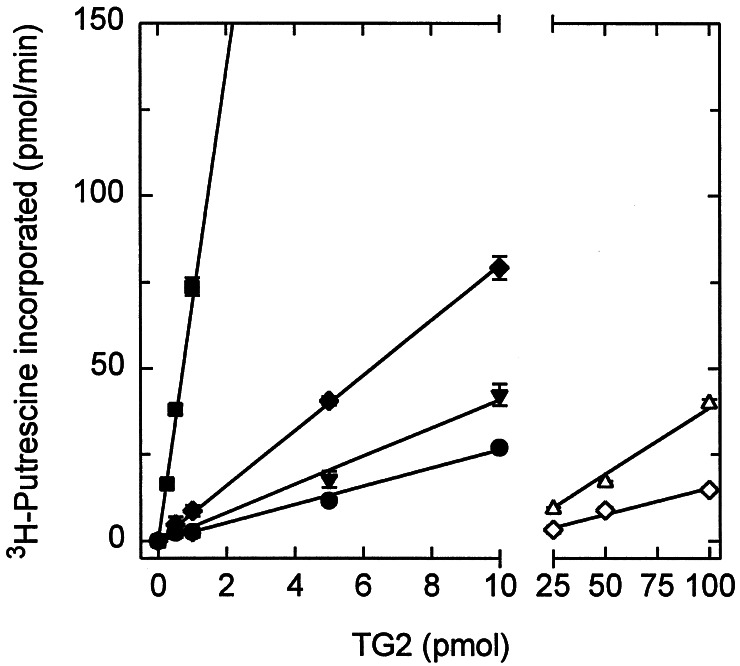
Transamidase activities of the TG2 mutants were compared to the wt enzyme by the rates of incorporation of [3H]putrescine into N,N-dimethylcasein (16). Although some of the mutant enzymes were near equal or even better (W278F) in activities than wt and others less so, our attention was drawn to four mutants of W241 because these showed the greatest loss of catalytic activity. In fact, as presented in Fig. 5, hardly any enzyme activity could be detected with W241Q and W241A. Most interestingly, however, low but nevertheless definite and readily measurable catalytic activities were seen with W241F and particularly with W241H. Based on these observations, we also constructed a W241Y mutant, whose Tyr side chain, like that of the native Trp, has aromatic and H-bonding character. Although the activity of this mutant also was quite impaired, it was at least as good or even a somewhat better enzyme than W241H and F. Specific activities were calculated approximately as 68 pmol putrescine/min/pmol of TG2 for the wt, 8 for the W241Y, 4 for the W241F, 3 for the W241H, 0.4 for the W241A, and 0.2 for the W241Q proteins.
Figure 5.
TG activities were measured for wt TG2 (■) and for the W241 mutants (Y, ♦; F, ▾; H, ●; A, ▵; Q, ⋄) by incorporation of [3H]putrescine into N,N′-dimethylcasein (16). Activity was determined by using 0.25, 0.5, 1, or 5 pmol for wt; 0.5, 1, 5, or 10 pmol for W241F, W241H, and W241Y; and 25, 50, or 100 pmol for W241A and W241Q. In this assay, activities for W241Q and A were detectable only at much higher concentrations of the TGs than for W241Y/F/H. Values shown are the means ±1 SE (error bars) of triplicate determinations (note error bars are not visible when smaller than the symbols).
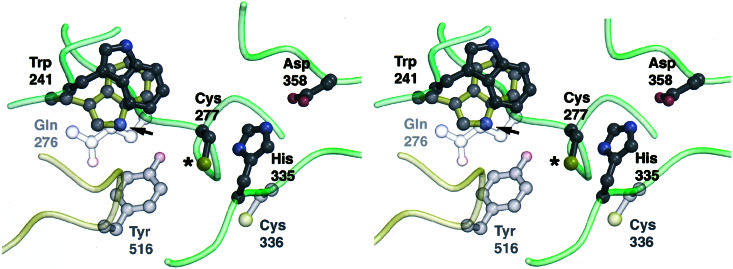
Trp-241 is a highly conserved residue in TGs, and W241 rat corresponds to W279 of human fXIIIA (12) and to W236 of the sea bream TG (13). In the crystal structures (1ggt.pdb and 1g0d.pdb), conformations for this tryptophan differ by a rotation of 180° of the side chain χ2 torsion angle without, however, any difficulty of being accommodated in the two structures (Fig. 6). If, as has been suggested for W279 in fXIIIA (12), the W241 rat TG2/Gh was to play an H-bonding role in stabilizing the transition state during catalysis, it would have to assume an orientation in which the indole nitrogen (marked by the arrow) is directed toward the putative oxyanion intermediate. In addition, the aromatic Trp side chain may also contribute to stabilizing the transition state by quadrupole–ion or van der Waals interaction. Preservation of appreciable TG activity of the W241Y/F/H mutants, relative to W241A/Q, suggests that the hydrogen bonding, electrostatic, or packing interactions of the side chains of the former can to some degree substitute for the properties of the Trp ring. The essentially total loss of activity of the W241A mutant indicates that the previously postulated interaction of the oxyanion intermediate with the backbone amide nitrogen of Cys-277 (15) would not be sufficient by itself to stabilize the transition state.
Figure 6.
Position of the highly conserved tryptophan 241 in the rat TG2 active site, modeled on the coordinates of sea bream TG (1g0d.pdb; ref. 13). Also shown is a superpositioning of the Trp-241 equivalent of the FXIIIA crystal structure (arrow). Note that the Trp side chains are rotated at 180° in the two structures resulting in the divergent positioning of the indole nitrogens (blue). The three residues Asp-358, His-335, and Cys-277 (*) comprise the papain-like catalytic triad (15). Small shifts of the tryptophan side chain may be accommodated by the surface provided by the side chain of the highly conserved Gln-276 underneath it, positioned by a number of conserved hydrogen bond interactions. The two residues Cys-336 and Tyr-516 may play a regulatory role (13), as Cys-336 has the potential to interact directly with the active-site cysteine, and in the inactive protein, Tyr-516 is in a position to hydrogen-bond to the catalytic cysteine side chain (15).
Although there are substantial similarities between the kinetic pathways of TGs and the evolutionarily related papain family of enzymes in that both proceed by consecutive steps of acylation and deacylation, a number of major differences are also evident (21–24). Just as papain, TGs can operate in a hydrolytic mode and, just as the TGs, the papain family of proteases can also function well in transamidating and transesterification reactions. The difference lies in the fact that, in sharp contrast to papain, TGs show great specificities in the transfer reactions for the second substrates of amines or alcohols. In the case of TG, the thiolester acylenzyme intermediate forms a saturable Michaelis complex with the amines and alcohols before deacylation, whereas deacylation in the case of papain is simply a bimolecular reaction between the acylenzyme and the nucleophiles without any preequilibration. Our finding that the conserved Trp-241 in TG is critical for catalysis seems to point to yet another important divergence of this family of enzymes from the papain model, in which stabilization of the oxyanion intermediate is mediated by a conserved glutamine side chain, i.e., Gln-19 (25).
Acknowledgments
We thank Ms. Pauline T. Velasco for help with calculations of nucleotide-binding constants, Ms. Sara Holman and Dr. Tadeusz Marciniec for expert technical support, and Dr. Jiri Novotny for assistance with TG2/Gh modeling. This work was supported by National Institutes of Health Grant HL-02212 and by Grant 14–2000 from the National Health and Medical Research Council, Australia.
Abbreviations
- fXIIIA
a subunit of coagulation factor XIII zymogen
- mantGTP
2′-(or 3′)-O-(N-methylanthraniloyl)GTP
- TG
transglutaminase
- wt
wild type
References
- 1.Achyuthan K E, Greenberg C S. J Biol Chem. 1987;262:1901–1906. [PubMed] [Google Scholar]
- 2.Bergamini C M, Signorini M, Poltronieri L. Biochim Biophys Acta. 1987;916:149–151. doi: 10.1016/0167-4838(87)90222-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee K N, Birckbichler P J, Patterson M K., Jr Biochem Biophys Res Commun. 1989;162:1370–1375. doi: 10.1016/0006-291x(89)90825-5. [DOI] [PubMed] [Google Scholar]
- 4.Lorand L, Dailey J E, Turner P M. Proc Natl Acad Sci USA. 1988;85:1057–1059. doi: 10.1073/pnas.85.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner P M, Lorand L. Biochemistry. 1989;28:628–635. doi: 10.1021/bi00428a032. [DOI] [PubMed] [Google Scholar]
- 6.LeMosy E K, Erickson H P, Beyer W F, Jr, Radek J T, Jeong J-M, Murthy S N P, Lorand L. J Biol Chem. 1992;267:7880–7885. [PubMed] [Google Scholar]
- 7.Radek J T, Jeong J-M, Murthy S N P, Ingham K C, Lorand L. Proc Natl Acad Sci USA. 1993;90:3152–3156. doi: 10.1073/pnas.90.8.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong J-M, Murthy S N P, Radek J T, Lorand L. J Biol Chem. 1995;270:5654–5658. doi: 10.1074/jbc.270.10.5654. [DOI] [PubMed] [Google Scholar]
- 9.Nakaoka H, Perez D M, Baek K J, Das T, Husain A, Misono K, Im M-J, Graham R M. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 10.Akimov S S, Belkin A M. J Cell Sci. 2001;114:2989–3000. doi: 10.1242/jcs.114.16.2989. [DOI] [PubMed] [Google Scholar]
- 11.Akimov S S, Belkin A M. Blood. 2001;98:1567–1576. doi: 10.1182/blood.v98.5.1567. [DOI] [PubMed] [Google Scholar]
- 12.Yee V C, Pedersen L C, Le Trong I, Bishop P D, Stenkamp R E, Teller D C. Proc Natl Acad Sci USA. 1994;91:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi K, Ishikawa K, Yokoyama K, Ohtsuka T, Nio N, Suzuki E. J Biol Chem. 2001;276:12055–12059. doi: 10.1074/jbc.M009862200. [DOI] [PubMed] [Google Scholar]
- 14.Monsonego A, Friedmann I, Shani Y, Eisenstein M, Schwartz M. J Mol Biol. 1998;282:713–720. doi: 10.1006/jmbi.1998.2052. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen L C, Yee V C, Bishop P D, Le Trong I, Teller D C, Stenkamp R E. Protein Sci. 1994;3:1131–1135. doi: 10.1002/pro.5560030720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iismaa S E, Wu M-J, Nanda N, Church W B, Graham R M. J Biol Chem. 2000;275:18259–18265. doi: 10.1074/jbc.M000583200. [DOI] [PubMed] [Google Scholar]
- 17.Murthy S N P, Lorand L. Proc Natl Acad Sci USA. 2000;97:7744–7747. doi: 10.1073/pnas.140210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iismaa S E, Chung L, Wu M-J, Teller D C, Yee V C, Graham R M. Biochemistry. 1997;36:11655–11664. doi: 10.1021/bi970545e. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Murthy S N P, Lomasney J W, Mak E C, Lorand L. Proc Natl Acad Sci USA. 1999;96:11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorand L, Chou C-H J, Simpson I. Proc Natl Acad Sci USA. 1972;69:2645–2648. doi: 10.1073/pnas.69.9.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis C G, Stenberg P, Brown K L, Baron A, Chen K, Gray A, Simpson I, Lorand L. Biochemistry. 1974;13:3257–3262. doi: 10.1021/bi00713a012. [DOI] [PubMed] [Google Scholar]
- 23.Stenberg P, Curtis C G, Wing D, Tong Y S, Credo R B, Gray A, Lorand L. Biochem J. 1975;147:155–163. [PMC free article] [PubMed] [Google Scholar]
- 24.Parameswaran K N, Lorand L. Biochemistry. 1981;20:3703–3711. doi: 10.1021/bi00516a006. [DOI] [PubMed] [Google Scholar]
- 25.Menard R, Carriere J, Laflamme P, Plouffe C, Khouri H E, Vernet T, Tessier D C, Thomas D Y, Storer A C. Biochemistry. 1991;30:8924–8928. doi: 10.1021/bi00101a002. [DOI] [PubMed] [Google Scholar]
- 26.Cariello L, Ristoratore F, Zanetti L. FEBS Lett. 1997;408:171–176. doi: 10.1016/s0014-5793(97)00342-6. [DOI] [PubMed] [Google Scholar]