Abstract
Evidence of the existence of major prostate cancer (PC)–susceptibility genes has been provided by multiple segregation analyses. Although genomewide screens have been performed in over a dozen independent studies, few chromosomal regions have been consistently identified as regions of interest. One of the major difficulties is genetic heterogeneity, possibly due to multiple, incompletely penetrant PC-susceptibility genes. In this study, we explored two approaches to overcome this difficulty, in an analysis of a large number of families with PC in the International Consortium for Prostate Cancer Genetics (ICPCG). One approach was to combine linkage data from a total of 1,233 families to increase the statistical power for detecting linkage. Using parametric (dominant and recessive) and nonparametric analyses, we identified five regions with “suggestive” linkage (LOD score >1.86): 5q12, 8p21, 15q11, 17q21, and 22q12. The second approach was to focus on subsets of families that are more likely to segregate highly penetrant mutations, including families with large numbers of affected individuals or early age at diagnosis. Stronger evidence of linkage in several regions was identified, including a “significant” linkage at 22q12, with a LOD score of 3.57, and five suggestive linkages (1q25, 8q13, 13q14, 16p13, and 17q21) in 269 families with at least five affected members. In addition, four additional suggestive linkages (3p24, 5q35, 11q22, and Xq12) were found in 606 families with mean age at diagnosis of ⩽65 years. Although it is difficult to determine the true statistical significance of these findings, a conservative interpretation of these results would be that if major PC-susceptibility genes do exist, they are most likely located in the regions generating suggestive or significant linkage signals in this large study.
Introduction
Familial clustering of prostate cancer (PC [MIM 176807]) has been consistently recognized for many years (reviewed by Isaacs and Xu [2002]). Segregation analyses and twin studies strongly suggest that genetic factors explain at least some of the familial aggregation of PC (reviewed by Schaid [2004]). Research groups worldwide have recruited families with multiple members with PC and have performed linkage analyses to search for PC-susceptibility genes. More than a dozen genomewide screens have been performed (Easton et al. 2003), and numerous regions have been suggested as harboring hereditary PC (HPC) genes. Furthermore, several genes in regions linked to PC have been proposed as candidate HPC genes, notably ELAC2 (MIM 605367), RNASEL (MIM 180435), and MSR1 (MIM 153622) (Tavtigian et al. 2001; Carpten et al. 2002; Xu et al. 2002).
Despite these extensive efforts, linkage findings suggested by individual groups and proposed associations with variants in candidate genes have not been reproducibly replicated by other groups. The difficulties in mapping PC genes have been widely discussed (Isaacs and Xu 2002; Edwards and Eeles 2004; Ostrander et al. 2004; Schaid 2004). Briefly, it is likely that multiple genes predispose to PC and that no single gene is sufficiently important to provide a reliable linkage signal when a small number of families are analyzed. PC linkage may be further complicated by phenocopies, particularly given the high prevalence of the disease and widespread use of prostate-specific antigen screening. These difficulties are inherent to PC-linkage studies, and, although they cannot be completely overcome, several approaches can be used to reduce their impact. One approach is to study a much larger number of families, which should improve the statistical power to detect regions containing genes that are mutated in a small proportion of families. Another approach is to study subsets of families with PC that are more likely both to segregate mutations in genes conferring a strong PC risk and to have a reduced number of phenocopies, such as those with a large number of affected members and/or affected members with early ages at diagnosis.
The International Consortium for Prostate Cancer Genetics (ICPCG) was formed to facilitate the task of PC–susceptibility gene identification through the combined analyses of linkage data from families with PC. In the present study, we describe the results from a combined genomewide screen for PC-susceptibility genes among 1,233 PC-affected families within the ICPCG, the largest study of its kind to date.
Methods
Ascertainment of Families
The overall ICPCG study population was described in detail elsewhere (Schaid et al. 2005). All members of the ICPCG recruited their study population, supported through their own research funding. Ten ICPCG groups participated in this combined genomewide screen, ACTANE (Anglo/Canadian/Texan/Australian/Norwegian/European Union Biomed), BC/CA/HI (British Columbia, California, and Hawaii), Johns Hopkins University (JHU), Mayo Clinic, University of Michigan, PROGRESS (Prostate Cancer Genetic Research Study, Fred Hutchinson Cancer Research Center), University of Tampere in Finland, University of Ulm in Germany, University of Umeå in Sweden, and University of Utah. There were 1,233 PC pedigrees in this combined analysis. The research protocols and informed consent procedures were approved by each group’s institutional review board.
Definition of Affection Status and Classification of Pedigrees
Affected individuals were defined as “those men affected with PC who had been confirmed by either medical records or death certificates.” Affected individuals without either medical records or death-certificate confirmation were considered as having unknown affection status (hence, instances of self-reported PC and of PC status that was based solely on family-history interviews were considered of unknown status). Because of this restricted definition, some pedigrees had fewer affected men than were previously reported in publications by the respective groups. All men without a diagnosis of PC were coded as having unknown affection status, regardless of whether they had undergone screening for PC. Hence, all analyses were based on the sharing of marker genotypes among affected individuals, with no consideration of the phenotype for the remaining subjects. Although such an approach may result in some loss of power, it provided a uniform approach across all participating groups.
Genotyping and Consensus Genetic Map
Various methods were used by different groups to genotype microsatellite markers in their respective genomewide screen, as described in detail elsewhere (Hsieh et al. 2001; Cunningham et al. 2003; International ACTANE Consortium 2003; Janer et al. 2003; Lange et al. 2003; Schleutker et al. 2003; Wiklund et al. 2003, Xu et al. 2003; Maier et al. 2005; Camp et al., in press). Different sets of genomewide-screen markers were used by these 10 groups (see individual references for complete description of markers), with a range of information contents of 0.38–0.57 across the various groups and a total of 1,322 markers. To facilitate a combined linkage analysis, we generated a consensus map by aligning all these markers to the draft human reference sequence (physical position) on the basis of the Human hg13 assembly (released November 14, 2002). Ten of these markers could not be uniquely located in the human reference sequence and were dropped from the combined analysis. The genetic position of the aligned markers was primarily determined on the basis of the deCode map (Kong et al. 2002). Among the 1,312 mapped markers, we were able to find the deCode genetic position for 964 markers. For the remaining 348 markers, for which only physical position was available, we estimated their genetic positions by interpolation based on the flanking markers for which both physical positions and deCode positions are available.
Linkage-Analysis Methods
The combined linkage analysis was performed in two stages. In the first stage, the linkage analyses were performed by each of the 10 groups, by use of the same definition of affection status and parametric model and the same linkage programs and options. All linkage results were based on multipoint calculations implemented in the Genehunter-Plus software (Kruglyak et al. 1996; Kong and Cox 1997). Because of their large size, the Utah pedigrees were selected from the larger set of all Utah pedigrees with at least four subjects with PC with no more than two meioses separating them (Camp et al. 2005); these pedigrees were then further trimmed to allow analysis by Genehunter-Plus. The analysis performed by each group was implemented by scripts provided by the ICPCG Data Coordinating Center (DCC), to facilitate consistency and automation. Although different genomewide screen markers were used among groups, the marker position was determined on the basis of the consensus map described above; therefore, the length of each chromosome was the same across all the groups. The linkage was evaluated at a resolution of 1 cM for each chromosome. The output files containing pedigree-specific linkage information at every cM across the genome were sent to the DCC. In the second stage, a combined analysis was performed at the DCC. For nonparametric linkage analysis, the combined allele-sharing LOD score was evaluated for each chromosome on the basis of the family-specific allele sharing at each centimorgan by use of the computer program ASM (Kong and Cox 1997). For the parametric linkage analysis, the combined LOD score with the assumption of heterogeneity (HLOD) was evaluated for each chromosome, on the basis of the family-specific LOD score at each centimorgan, by use of the computer program HOMOG (Ott 1999). Throughout the present study, we used LOD scores to describe HLODs for the results of parametric analyses and allele sharing LODs for nonparametric analyses.
The allele frequencies used were population specific; that is, for each marker, allele frequencies were estimated by counting alleles across all families within each individual group, without consideration of genetic relationships. Although not fully efficient, this provides straightforward, unbiased allele-frequency estimates. Because few families within any participating group had a known nonwhite racial background, allele frequencies were estimated from the pool of all data within a group, without consideration of race. Both nonparametric and parametric linkage analyses were performed. Allele-sharing nonparametric linkage analysis was performed, because that did not require specification of a model and would be expected to have good power against a wide range of alternative models. The linear allele-sharing model was implemented using ASM (Kong and Cox 1997). Families were weighted equally, and the score function “all” was used, which provides more evidence of linkage than does the “pairs” option whenever most affected individuals in a pedigree share the same allele that is identical by descent. For the parametric linkage analyses, a dominant model and a recessive model were used. The dominant model was similar to the one used to map HPC1 (MIM 601518) (Smith et al. 1996). The frequency of the susceptibility allele was assumed to be 0.003, with a penetrance of 0.001 for noncarriers and 1.0 for carriers. Unaffected subjects were coded as having noninformative phenotypes. The recessive model was similar to the dominant model, except that the susceptibility-allele frequency was set to 0.15 and the penetrance for heterozygous carriers was set equal to the penetrance for homozygous noncarriers. Stratified linkage analyses were also performed in two predetermined subsets of families: 269 families with at least five affected members, and 606 families with family mean age at diagnosis of ⩽65 years. The planned analyses were developed and approved by members of the ICPCG.
We summarized our linkage results on the basis of the proposed guidelines for reporting linkage results of a genomewide screen: a cutoff LOD score of 3.30 as “significant” evidence of linkage and a cutoff LOD score of 1.86 as “suggestive” evidence of linkage (Lander and Kruglyak 1995). On the basis of asymptotic arguments, a LOD score of 3.30 is expected to occur 0.05 times in a genome screen that makes use of a fully informative marker set, and a LOD score of 1.86 is expected to occur once by chance.
Results
Analyses of All Families
Table 1 summarizes the characteristics of the 1,233 PC-affected families from 10 different ICPCG groups that were included in the analysis. Fifty-one percent of families had a mean age at onset of <65 years; 22% had five or more affected family members.
Table 1.
Characteristics of Families
|
Mean Ageat Diagnosisa(years) |
No. of Affected Members |
Raceb |
|||||||
| ICPCG Member | ⩽65 | >65 | 2 | 3 | 4 | ⩾5 | White | Black | Total No. of Families |
| ACTANE | 41 | 21 | 18 | 32 | 11 | 3 | 64 | 0 | 64 |
| BC/CA/HI | 41 | 57 | 24 | 54 | 16 | 4 | 83 | 7 | 98 |
| JHU | 95 | 93 | 2 | 26 | 47 | 113 | 169 | 17 | 188 |
| Mayo Clinic | 72 | 87 | 70 | 58 | 21 | 10 | 158 | 0 | 159 |
| PROGRESS | 141 | 113 | 38 | 107 | 66 | 43 | 240 | 8 | 254 |
| University of Michigan | 103 | 73 | 55 | 76 | 29 | 16 | 158 | 16 | 176 |
| University of Tampere | 3 | 7 | 0 | 2 | 5 | 3 | 10 | 0 | 10 |
| University of Ulm | 84 | 55 | 60 | 42 | 29 | 8 | 139 | 0 | 139 |
| University of Umeå | 10 | 40 | 0 | 13 | 17 | 20 | 50 | 0 | 50 |
| University of Utah | 16 |
79 |
18 |
14 |
14 |
49 |
95 |
0 |
95 |
| Total | 606 | 625 | 285 | 424 | 255 | 269 | 1,166 | 48 | 1,233 |
Information about family mean age at diagnosis was not available for two families.
Nineteen families are from other ethnic groups, such as Asian, Hispanic, or Native American.
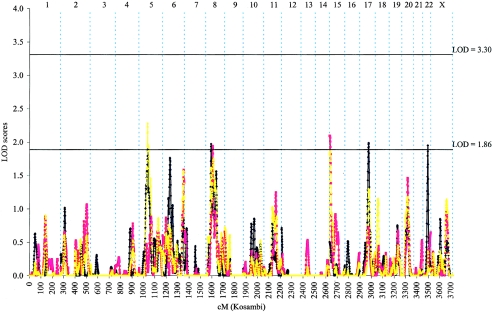
We first performed a combined genomewide linkage analysis of the complete set of 1,233 PC-affected families, using parametric and nonparametric approaches. Although no significant evidence of linkage was observed in the genome, evidence of suggestive PC linkage wasobserved at five chromosomal regions, 5q12, 8p21, 15q11, 17q21, and 22q12 (fig. 1 and table 2). The highest overall LOD score in the genome was 2.28 from the nonparametric analysis, found near marker D5S2858 on 5q12 (77 cM from pter). The linkage results for each individual family collection for each of these five chromosomal regions are shown in table 3. As seen in table 3, with the exception of 17q21, the LOD scores for each of the highlighted regions are higher in the combined analysis than those observed in any individual group, reaching a suggestive level of evidence only in the combined family data.
Figure 1.
Combined genomewide screen for PC-susceptibility genes with use of nonparametric and parametric multipoint linkage analyses among the entire set of 1,233 PC-affected families recruited from 10 ICPCG members. LOD scores obtained from parametric analysis with use of a dominant model (blue line), a recessive model (red line), and nonparametric analysis (yellow line) are plotted by individual chromosome for the whole genome.
Table 2.
Chromosomal Regions with Suggestive Evidence of Linkage
|
1-LOD Drop Interval |
||||||
| Population and Region | Distance from ptercM | Nearest Marker | Analysis Type | LOD | Genetic(cM) | Physical(Mb) |
| Primary analysis: entire set of families (N=1,233): | ||||||
| 5q12 | 77 | D5S2858 | Nonparametric | 2.28 | 66–96 | 43–78 |
| 8p21 | 46 | D8S1048 | Dominant | 1.97 | 39–52 | 22–32 |
| 15q11 | 1 | D15S817 | Recessive | 2.10 | 0–14 | 0–25 |
| 17q21 | 77 | D17S1820 | Dominant | 1.99 | 66–85 | 35–54 |
| 22q12 | 42 | D22S283 | Dominant | 1.95 | 35–47 | 29–37 |
| Secondary analysis: subset of families with at least five affected family members (n=269): | ||||||
| 1q25 | 184 | D1S2818 | Nonparametric | 2.62 | 170–198 | 165–196 |
| 8q13 | 81 | D8S543 | Recessive | 2.41 | 75–90 | 66–75 |
| 13q14 | 56 | D13S1807 | Recessive | 2.27 | 42–67 | 39–71 |
| 16p13 | 34 | D16S764 | Nonparametric | 1.88 | 19–46 | 9–23 |
| 17q21 | 77 | D17S1820 | Dominant | 2.04 | 66–83 | 39–53 |
| 22q12 | 42 | D22S283 | Dominant | 3.57 | 32–50 | 27–42 |
| Secondary analysis: subset of families with mean age at diagnosis of ⩽65 years (n=606): | ||||||
| 3p24 | 57 | D3S2432 | Dominant | 2.37 | 47–69 | 28–49 |
| 5q35 | 179 | D5S1456 | Dominant | 2.05 | 166–193 | 162–174 |
| 11q22 | 102 | D11S898 | Recessive | 2.20 | 89–112 | 81–111 |
| Xq12 | 80 | DXS7132 | Dominant | 2.30 | 62–90 | 40–85 |
Table 3.
Support for Linkage from Each Group at Chromosomal Regions with Suggestive Linkage
|
LOD Score by Chromosomal Region and Model |
|||||
| Population | 5q12 (77 cM)Nonparametric | 8p21 (46 cM)Dominant | 15q11 (1 cM)Recessive | 17q21 (77 cM)Dominant | 22q12 (42 cM)Dominant |
| All groups (N=1,233) | 2.28 | 1.97 | 2.10 | 1.99 | 1.95 |
| ACTANE (n=64) | .00 | .00 | .00 | .00 | .00 |
| BC/CA/HI (n=98) | 1.17 | .00 | .26 | .76 | .00 |
| JHU (n=188) | .29 | .16 | .95 | .72 | 1.28 |
| Mayo Clinic (n=159) | .32 | .09 | .00 | .00 | 1.10 |
| PROGRESS (n=254) | .25 | 1.64 | .64 | .01 | .00 |
| University of Michigan (n=176) | .01 | .28 | 1.06 | 3.07 | .13 |
| University of Tampere (n=10) | .00 | .19 | .00 | .42 | .02 |
| University of Ulm (n=139) | .38 | .77 | .04 | .00 | .00 |
| University of Umeå (n=50) | 1.62 | .00 | .87 | .00 | .00 |
| University of Utah (n=95) | .27 | .17 | .00 | .03 | 1.47 |
Analyses of Subsets of Families
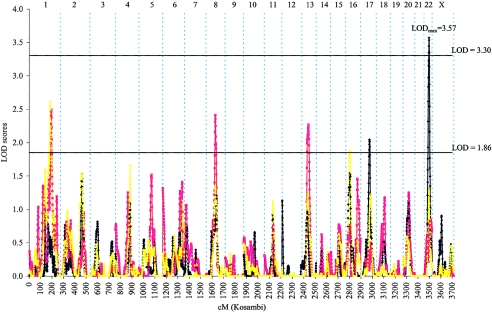
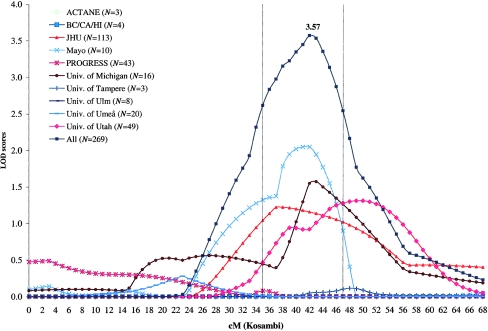
We also performed linkage analyses in subsets of families that might be more likely to segregate genes conferring strong PC risk: families with at least five affected members or with family mean age at diagnosis of ⩽65 years. As hypothesized, we found stronger evidence of linkage among 269 families with at least five affected members—one region with significant evidence of linkage and four additional regions with suggestive evidence of linkage (fig. 2 and table 2). The strongest evidence of linkage in the genome was found at 22q12 with use of the dominant model, with LOD score of 3.57 at 42 cM (near marker D22S283). This LOD score exceeded the criterion of significant evidence of linkage in the genomewide screen. Evidence of linkage at this region was provided by multiple ICPCG groups (fig. 3). Of the 10 groups, 4 had a LOD score >1.0 at this region, including a LOD score of 2.05 from the Mayo group, a LOD score of 1.57 from the Michigan group, a LOD score of 1.31 from the Utah group, and a LOD score of 1.22 from the JHU group. It is noted that linkage evidence at this region was observed in the complete set of 1,233 families (LOD score of 1.95 at 42 cM) and was strengthened in this subset. When families with at least five affected family members were removed from the analysis, no evidence of linkage at this region was found in the remaining 964 families. In addition to the 22q12 region, five additional regions reached suggestive evidence of PC linkage in this subset of families with at least five affected family members (fig. 2 and table 2).
Figure 2.
Combined genomewide screen for PC-susceptibility genes with use of nonparametric and parametric multipoint linkage analyses among 269 families with at least five affected members recruited from 10 ICPCG members. LOD scores obtained from parametric analysis with use of a dominant model (blue line), a recessive model (red line), and nonparametric analysis (yellow line) are plotted by individual chromosome for the whole genome.
Figure 3.
Parametric linkage analysis of chromosome 22 among families with at least five affected members with use of the dominant model. LOD scores are plotted for each of the 10 ICPCG groups.
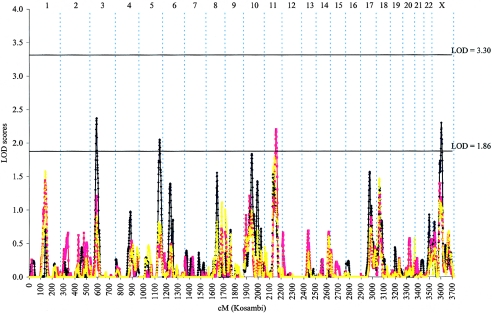
For 606 families with family mean age at diagnosis of ⩽65 years, suggestive evidence of PC linkage was found at four chromosomal regions (fig. 4 and table 2), with the highest LOD score of 2.37 near marker D3S2432 at 3p24 (57 cM). The four PC linkages identified in this subset of families were unique to the early-age-at-diagnosis subset. No evidence of linkage at these four regions was observed in the complete set of 1,233 families.
Figure 4.
Combined genomewide screen for PC-susceptibility genes with use of nonparametric and parametric multipoint linkage analyses among 606 families with family mean age at diagnosis ⩽65 years recruited from 10 ICPCG members. LOD scores obtained from parametric analysis with use of a dominant model (blue line), a recessive model (red line), and nonparametric analysis (yellow line) are plotted by individual chromosome for the whole genome.
Discussion
We have described results from the largest PC genomewide screen reported to date, with combined linkage data from 1,233 PC-affected families collected by 10 different groups in the ICPCG. From the primary analysis of the entire set of the families, we identified five chromosomal regions (5q12, 8p21, 15q11, 17q21, and 22q12) with suggestive evidence of linkage. With one exception (i.e., 17q21), the threshold for suggestive evidence of linkage was reached only in the combined analysis, which emphasizes the advantage of this combined approach.
Importantly, we found significant evidence of linkage at the 22q12 region in 269 families with at least five affected members, a subset of PC-affected families that is more likely to segregate mutations in genes conferring a strong PC risk. Suggestive evidence of linkage at five other regions (1q25, 8q13, 13q14, 16p13, and 17q21) was also observed in this subset of families. In addition, four additional regions (3p24, 5q35, 11q22, and Xq12) were found to have suggestive evidence of PC linkage in 606 families with family mean age at diagnosis of ⩽65 years.
We recognize that many of the regions identified in this study may represent false positive findings due to multiple tests in a genomewide screen and that it is difficult to dissect true linkages from false signals. On the basis of the assumption of a fully informative marker map, LOD scores >3.30 or 1.86 would have been expected to occur 0.05 times and 1 time, respectively, in a single genomewide screen. Here, we performed nine genomewide screens (three in the primary analyses and six in the subgroup analyses); this needs to be considered when interpreting the results. However, these nine analyses are not independent. Using the method of Camp and Farnham (2001), we determined that the nine nonindependent analyses performed were equivalent to ∼5.2 independent genomewide screens. We therefore estimate that, after correcting for multiple testing, regions with a LOD score of >3.30 (significant evidence) in at least one analysis would be expected 0.25 times in 5.2 independent screens and regions with a LOD score of >1.86 (suggestive evidence), 5.2 times. Our empirical results (one observed LOD score of 3.57 and 13 regions with LOD scores ⩾1.86) therefore exceeded the expectation under the null hypothesis of no linkage. Furthermore, these thresholds may be unduly conservative for the less-than-informative real data used in these genomewide screens. Although it is difficult to determine the true statistical significance of these findings, a conservative interpretation of these results would be that if major PC-susceptibility genes do exist, they are most likely to be located in the regions generating suggestive or significant linkage signals. Therefore, results from this analysis are likely to be helpful in prioritizing any efforts to identify PC-susceptibility genes.
Lack of reproducibility among PC-linkage studies in recent years demonstrates the difficulties faced in the effort to identify PC-susceptibility genes with the linkage approach (Isaacs and Xu 2002; Edwards and Eeles 2004; Ostrander et al. 2004; Schaid 2004). One of the major difficulties is genetic heterogeneity due to multiple but incompletely penetrant PC-susceptibility genes. Each of these genes may be responsible for a small fraction of PC-affected families. In this study, we planned two approaches to address the impact that these difficulties have on identification of PC linkage. One approach was to perform linkage analysis in a large number of PC-affected families, to increase the statistical power to detect linkage. This approach led to the identification of five regions with evidence suggestive of linkage in the complete set of 1,233 families. However, the failure to identify significant linkage in the genome, even with this large number of families, indicates a substantial degree of genetic heterogeneity and suggests that this approach alone is insufficient to uncover a significant signal if it is present. Our second approach was to focus on subsets of families that are more likely to segregate highly penetrant mutations, including families with large numbers of affected individuals and/or early age at diagnosis. This latter approach appeared to be more effective; stronger evidence of linkage was found in several regions among these subsets of families than from the complete set of families. The most noteworthy finding was the considerable increase in evidence of linkage at 22q12—from a LOD score of 1.95 (suggestive linkage) in the complete set of families to a LOD score of 3.57 (significant linkage) in 269 families with at least five affected members. It is important to note that the large number of families in our combined study makes it possible to analyze sufficient numbers in each subset of families. Linkage studies in families with large numbers of affected individuals and/or early ages at diagnosis have proved to be effective in identifying breast cancer–susceptibility genes (Hall et al. 1990; Easton et al. 1993).
Three pieces of evidence from our study increase our confidence that the linkage at 22q12 is due to PC-susceptibility gene(s) at this region. First, the LOD score at this region reached the criterion of significant linkage. The chance of observing this magnitude of LOD score in the genome under a null hypothesis of no linkage is <0.25 times in our study. Second, this linkage was identified in the families with at least five affected members, a subset of families that is more likely to segregate mutations in genes conferring strong PC risk. Third, the evidence of linkage at this region was supported by multiple individual groups; of six groups with ⩾10 such large families, four had LOD scores >1 in this interval. The relatively good reproducibility of this linkage finding is an unusual observation in PC-linkage studies (Easton et al. 2003). More than 129 known genes are in the 1-LOD drop interval (29–37 Mb). An important candidate gene, CHEK2 (MIM 604373), is outside the interval, at ∼27 Mb.
Because the mode of inheritance for PC is uncertain, we performed linkage analysis using both parametric (dominant or recessive) and nonparametric methods. In general, evidence of PC linkage was consistently provided by both parametric and nonparametric methods, although with different strengths at different regions. Parametric analysis will have better power to detect linkage when an assumed genetic model approximates the underlying mode of inheritance of a disease susceptibility gene (Clerget-Darpoux et al. 1986a, 1986b; Lio and Morton 1997). Nonparametric analysis, by assessment of allele sharing among affected individuals within a pedigree, may have better power when the underlying genetic model cannot be specified with any confidence (Whittemore and Halpern 1994).
Most of the linkage regions identified in the present study are broad. The information content of the marker sets used in these analyses is generally low, particularly since most of our families are small and often do not include genotypes of all parents. Further genotyping at a higher density, with use of either microsatellite markers or SNPs, should improve informativeness. An additional approach currently under way by the ICPCG incorporates clinical and pathological tumor variables in the assignment of affected status, to emphasize clinically aggressive disease in this large data set. Hopefully, these approaches should help to confirm or refute the evidence of linkage and narrow the regions of interest.
During the last decade, tremendous effort has been put forth to identify major susceptibility genes for PC. Linkage studies with smaller numbers of PC-affected families have identified and implicated many chromosomal regions that might harbor PC-susceptibility genes. The large number of different regions that have been implicated—and the general lack of reproducibility among these studies—has provided a tenuous foundation for subsequent PC-gene identification. In this context, results from the current study, with a very large number of families in the overall analysis, provides a strong basis for prioritizing regions for PC-gene identification.
Acknowledgments
We express our gratitude to the many families who participated in this study and to the many urologists who kindly assisted us by providing information and access to their patients. The ICPCG is supported by U.S. Public Health Service (USPHS) National Institutes of Health (NIH) grant CA89600. Additional support to participating groups or members within groups is as follows. ACTANE Group: Genotyping and statistical analysis for this study and recruitment of U.K. families was supported by Cancer Research U.K. Additional support was provided by the Prostate Cancer Charitable Trust (now Prostate Cancer Research Foundation), The Times Christmas Appeal, and the Institute of Cancer Research. Genotyping was conducted in the Jean Rook Gene Cloning Laboratory, which is supported by BREAKTHROUGH Breast Cancer–Charity 328323. The funds for the ABI 377 used in this study were generously provided by the legacy of the late Marion Silcock. We thank Mrs. Sheila Seal and Mrs. Anita Hall for kindly storing and logging the samples that were provided. D.F.E. is a principal research fellow of Cancer Research U.K. Recruitment of Australian PC-affected families was funded by National Health and Medical Research Council grant 940934 and was further supported by Tattersall’s and the Whitten Foundation; infrastructure was provided by the Cancer Council Victoria. We acknowledge the work of study coordinator Margaret Staples; the research team of Bernadette McCudden, John Connal, Richard Thorowgood, Chris Costa, Melodie Kevan, and Sue Palmer; and Jolanta Karpowicz, for DNA extractions. The Texas study of familial PC was initiated by the Department of Epidemiology, M. D. Anderson Cancer Center. M.B. was supported by NCI post-doctoral fellowship in Cancer Prevention R25. BC/CA/HI Group: USPHS grant CA67044. JHU Group: USPHS grants CA58236 (to W.B.I.), CA95052-01 (to J.X.), and CA106523-01A1 (to J.X.). Mayo Clinic Group: USPHS grant CA72818. Michigan Group: USPHS grant CA079596. PROGRESS Group: USPHS grants CA78835 (to E.A.O.) and CA080122 (to J.L.S.) and support from the Prostate Cancer Foundation and the Fred Hutchinson Cancer Research Center (University of Washington Markey Center). Tampere Group: Medical Research Fund of Tampere University Hospital, Reino Lahtikari Foundation, Finnish Cancer Organizations, Sigrid Juselius Foundation, and Academy of Finland grant 201480. Ulm Group: Deutsche Krebshilfe grant 70–3111-V03. Umeå Group: Grants from the Swedish Cancer Society (Cancerfonden) and Stiftelsen för Strategisk Forskning. Utah Group: NIH NCI grant R01 CA90752 (to L.C.A.), a subcontract from JHU with funds provided by NIH NCI grant R01 CA89600 (to L.C.A.), and NIH grant K07 CA98364 (to N.C.). Data collection for this publication was assisted by the Utah Cancer Registry, supported by NIH Contract NO1-PC-35141 and Surveillance, Epidemiology and End Results Program, with additional support from the Utah Department of Health, the University of Utah, and Public Health Services research grant M01-RR00064 from the National Center for Research Resources. Partial support for all data sets within the Utah Population Database was provided by the University of Utah Huntsman Cancer Institute. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through federal NIH contract N01-HG-65403 (to J.H.U.). Genotyping for the JHU, Michigan, Tampere, and Umeå groups was performed by Elizabeth Gillanders, MaryPat Jones, Derk Gildea, Erica Riedesel, Julie Albertus, Diana Freas-Lutz, Carol Markey, John Carpten, and Jeff Trent at the National Human Genome Research Institute, NIH. Other investigators who contributed to this work: ACTANE: United Kingdom (Sutton): Rifat Hamoudi, Audrey Ardern-Jones, Christine Southgate, Anna Dowe, Kim Coleman, David Dearnaley, The Cancer Research U.K./British Prostate Group U.K. Familial Prostate Cancer Study Collaborators, British Association of Urological Surgeons’ Section of Oncology, Translational Cancer Genetics Team, Molecular Genetics Team, Section of Cancer Genetics; Institute of Cancer Research, Royal Marsden NHS Trust Foundation Hospital. United Kingdom (Cambridge): M. Dawn Teare, Cancer Research U.K. Genetic Epidemiology Unit, Strangeways Research Labs. Australia: Dallas English, Gianluca Severi, Melissa Southey, Cancer Epidemiology Centre, The Cancer Council Victoria; University of Melbourne, Centre for Genetic Epidemiology. Canada: Nancy Hamel, Division of Medical Genetics, Research Institute of the McGill University Health Centre, Montreal; Steven Narod, Centre for Research in Women’s Health, University of Toronto, Toronto. Texas: Chris Amos, M. D. Anderson Cancer Centre, Houston. Norway (Oslo): Ketil Heimdal Unit of Medical Genetics, Norwegian Radium Hospital, Oslo. Norway (Ullevaal): Nicolai Wessel, Tone Andersen, Department of Oncology, Ullevaal University Hospital, Oslo. EU Biomed: The EU Biomed Prostate Cancer Linkage Consortium, Cancer Research U.K. Genetic Epidemiology Laboratory, St. James’ University Hospital, Leeds. JHU: Piroska Bujnovszky, Tanya Ray, Vivian Bailey, Mary Buedel, and Dawn Steinberg. Utah: Alan Thomas, Lewis Ershler, and Kim Nguyen.
Author affiliations.—Center for Human Genomics, Wake Forest University School of Medicine, Winston-Salem, NC (J.X., L.D., B.-L.C., T.S.A., A.R.T., and D.A.M.); Institute of Cancer Research and Royal Marsden National Health Service Trust Foundation Hospital, Sutton, United Kingdom (R.A.E., S.E., J.M., S.B., and Q.H.); Cancer Research U.K. Genetic Epidemiology Unit, Strangeways Research Labs, Cambridge, United Kingdom (D.F.E. and C.E.); Program in Cancer Genetics, Departments of Oncology and Human Genetics, McGill University, Montreal (W.D.F.); Cancer Genomics Laboratory, Centre hospitalier de l'Universite Laval Research Centre, Sainte-Foy, Quebec (J. Simard); University of Washington Medical Center (M.B. and G.P.J.) and Department of Medical Genetics, School of Public Health and Community Medicine, University of Washington (M.B. and G.P.J.), Divisions of Human Biology (D.M.F. and E.A.O.) and Public Health Sciences, Fred Hutchinson Cancer Center (S.K. and J.L.S.), and Institute for System Biology (K.D., M.J., and L.H.), Seattle; Cancer Epidemiology Centre, Cancer Council Victoria (G.G.G.), and Centre for Genetic Epidemiology, University of Melbourne (J.L.H.), Carlton, Australia; Unit of Medical Genetics, Norwegian Radium Hospital, Oslo (L.M. and P.M.); Cancer Research U.K. Genetic Epidemiology Laboratory, St. James’ University Hospital, Leeds (T.B.); University of Southern California, Los Angeles (C.-l.H.); Stanford University School of Medicine, Stanford (J.H., R.N.B., and A.S.W.); Northern California Cancer Center, Union City and Stanford (I.O.-G.); Department of Urology, Johns Hopkins Medical Institutions (C.M.E., M.G., S.D.I. P.C.W., K.E.W., and W.B.I), and Inherited Disease Research Branch, National Human Genome Research Institute, NIH (J.B.-W.), Baltimore; Mayo Clinic, Rochester, MN (S.N.T., S.K.M., J.M.C., K.E.Z., S.H., and D.J.S.); Cancer Genetics Branch, National Human Genome Research Institute, (E.A.O.), and National Cancer Institute (NCI) (D.S.), NIH, Bethesda; Department of Genetics, University of North Carolina, Chapel Hill (E.M.L.); University of Michigan, Ann Arbor (J.L.B.-D., C.E.M., and K.A.C.); University of Tampere and Tampere University Hospital, Tampere, Finland (T.I., H.F., M.P.M. T.L.T., and J. Schleutker); Fox Chase Cancer Center, Division of Population Science, Philadelphia (A.B.-B.); Abteilung Humangenetik, Universität Ulm (C.M., J.J.H., and W.V.), and Urologische Universitätsklinik und Poliklinik, Abteilung für Urologie und Kinderurologie (K.H. and T.P.), Ulm, Germany; Department of Radiation Sciences, Oncology, University of Umeå, Umeå, Sweden (F.W., M.E., E.S., B.-A.J., and H.G.); Division of Genetic Epidemiology, University of Utah, Salt Lake City (N.J.C., J.F., and L.C.A).
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PC, ELAC2, RNASEL, MSR1, HPC1, and CHEK2)
References
- Camp NJ, Farnham JM (2001) Correcting for multiple analyses in genomewide linkage studies. Ann Hum Genet 65:577–582 10.1046/j.1469-1809.2001.6560577.x [DOI] [PubMed] [Google Scholar]
- Camp NJ, Farnham JM, Cannon-Albright LA. Genome search for prostate cancer predisposition loci in Utah pedigrees. Prostate (in press) [DOI] [PubMed] [Google Scholar]
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, et al (2002) Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet 30:181–184 10.1038/ng823 [DOI] [PubMed] [Google Scholar]
- Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J (1986a) Effects of misspecifying genetic parameters in lod score analysis. Biometrics 42:393–399 [PubMed] [Google Scholar]
- Clerget-Darpoux F, Dizier MH, Bonaiti-Pellie C, Babron MC, Hochez J, Martinez M (1986b) Discrimination between genetic models for insulin dependent diabetes mellitus. Genet Epidemiol Suppl 1:313–318 10.1002/gepi.1370030747 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, McDonnell SK, Marks A, Hebbring S, Anderson SA, Peterson BJ, Slager S, French A, Blute ML, Schaid DJ, Thibodeau SN (2003) Genome linkage screen for prostate cancer susceptibility loci: results from the Mayo Clinic Familial Prostate Cancer Study. Prostate 57:335–346 10.1002/pros.10308 [DOI] [PubMed] [Google Scholar]
- Easton DF, Bishop DT, Ford D, Crockford GP (1993) Breast Cancer Linkage Consortium: genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am J Hum Genet 52:678–701 [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Schaid DJ, Whittemore AS, Isaacs WJ (2003) Where are the prostate cancer genes? A summary of eight genome wide searches. Prostate 57:261–269 10.1002/pros.10300 [DOI] [PubMed] [Google Scholar]
- Edwards SM, Eeles RA (2004) Unravelling the genetics of prostate cancer. Am J Med Genet C Semin Med Genet 129:65–73 10.1002/ajmg.c.30027 [DOI] [PubMed] [Google Scholar]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684–1689 [DOI] [PubMed] [Google Scholar]
- Hsieh C-l, Oakley-Girvan I, Balise RR, Halpern J, Gallagher RP, Wu AH, Kolonel LN, O’Brien LE, Lin IG, Van Den Berg DJ, Teh C-Z, West DW, Whittemore AS (2001) A genome screen of families with multiple cases of prostate cancer: evidence of genetic heterogeneity. Am J Hum Genet 69:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs WB, Xu J (2002) Prostate cancer. In: King RA, Totter JI, Motulsky AG (eds) The genetic basis of common diseases. Oxford University Press, New York [Google Scholar]
- International ACTANE Consortium (2003) Results of a genome-wide linkage analysis in prostate cancer families ascertained through the ACTANE consortium. Prostate 57:270–279 10.1002/pros.10301 [DOI] [PubMed] [Google Scholar]
- Janer M, Friedrichsen DM, Stanford JL, Badzioch MD, Kolb S, Deutsch K, Peters MA, Goode EL, Welti R, DeFrance HB, Iwasaki L, Li S, Hood L, Ostrander EA, Jarvik GP (2003) Genomic scan of 254 hereditary prostate cancer families. Prostate 57:309–319 10.1002/pros.10305 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson D, Sainz J, Jonsdottir G, Gudjonsson S, Richardsson B, Sigurdardottir S, Barnard B, Hallbeck B, Masson M, Shlien A, Palsson S, Frigge M, Thorgeirsson T, Gulcher J, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Lange EM, Gillanders EM, Davis CC, Brown WM, Campbell JK, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Giri V, Dimmer JB, Montie JE, Trent JM, Cooney KA (2003) Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan prostate cancer genetics project finds evidence for linkage on chromosome 17 near BRCA1. Prostate 57:326–334 10.1002/pros.10307 [DOI] [PubMed] [Google Scholar]
- Lio P, Morton NE (1997) Comparison of parametric and nonparametric methods to map oligogenes by linkage. Proc Natl Acad Sci USA 94:5344–5348 10.1073/pnas.94.10.5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier C, Herkommer K, Hoegel J, Vogel W, Paiss T (2005) A genomewide linkage analysis for prostate cancer susceptibility genes in families from Germany. Eur J Hum Genet 13:352–360 10.1038/sj.ejhg.5201333 [DOI] [PubMed] [Google Scholar]
- Ostrander EA, Markianos K, Stanford JL (2004) Finding prostate cancer susceptibility genes. Annu Rev Genomics Hum Genet 5:151–175 10.1146/annurev.genom.5.061903.180044 [DOI] [PubMed] [Google Scholar]
- Ott J (1999) Analysis of human genetic linkage. The Johns Hopkins University Press, Baltimore [Google Scholar]
- Schaid D (2004) The complex genetic epidemiology of prostate cancer. Hum Mol Genet 13:R103–R121 10.1093/hmg/ddh072 [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Chang BL, International Consortium for Prostate Cancer Genetics (2005) Description of the international consortium for prostate cancer genetics, and failure to replicate linkage of hereditary prostate cancer to 20q13. Prostate 63:276–290 10.1002/pros.20198 [DOI] [PubMed] [Google Scholar]
- Schleutker J, Baffoe-Bonnie AB, Gillanders E, Kainu T, Jones MP, Freas-Lutz D, Markey C, Gildea D, Riedesel E, Albertus J, Gibbs KD Jr, Matikainen M, Koivisto PA, Tammela T, Bailey-Wilson JE, Trent JM, Kallioniemi OP (2003) Genome-wide scan for linkage in Finnish hereditary prostate cancer (HPC) families identifies novel susceptibility loci at 11q14 and 3p25-26. Prostate 57:280–289 10.1002/pros.10302 [DOI] [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujinovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 10.1126/science.274.5291.1371 [DOI] [PubMed] [Google Scholar]
- Tavtigian S, Simard J, Teng D, Abtin V, Baumgard M, Beck A, Camp N, et al (2001) A strong candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27:172–180 10.1038/84808 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Wiklund F, Gillanders EM, Albertus JA, Bergh A, Damber JE, Emanuelsson M, Freas-Lutz DL, Gildea DE, Goransson I, Jones MS, Jonsson BA, Lindmark F, Markey CJ, Riedesel EL, Stenman E, Trent JM, Grönberg H (2003) Genome-wide scan of Swedish families with hereditary prostate cancer: suggestive evidence of linkage at 5q11.2 and 19p13.3. Prostate 57:290–297 10.1002/pros.10303 [DOI] [PubMed] [Google Scholar]
- Xu J, Gillanders EM, Isaacs SD, Chang BL, Wiley KE, Zheng SL, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Meyers DA, Walsh PC, Trent JM, Isaacs WB (2003) Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate 57:320–325 10.1002/pros.10306 [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Komiya A, Mychaleckyj JC, Isaacs SD, Hu JJ, Sterling D, et al (2002) Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet 32:321–325 10.1038/ng994 [DOI] [PubMed] [Google Scholar]