Abstract
Genetic recombination pathways and genes are well studied, but relatively little is known in plants, especially in lower plants. To study the recombination apparatus of a lower land plant, a recombination gene well characterized particularly in yeast, mouse, and man, the RAD51 gene, was isolated from the moss Physcomitrella patens and characterized. Two highly homologous RAD51 genes were found to be present. Duplicated RAD51 genes have been found thus far exclusively in eukaryotes with duplicated genomes. Therefore the presence of two highly homologous genes suggests a recent genome duplication event in the ancestry of Physcomitrella. Comparison of the protein sequences to Rad51 proteins from other organisms showed that both RAD51 genes originated within the group of plant Rad51 proteins. However, the two proteins form a separate clade in a phylogenetic tree of plant Rad51 proteins. In contrast to RAD51 genes from other multicellular eukaryotes, the Physcomitrella genes are not interrupted by introns. Because introns are a common feature of Physcomitrella genes, the lack of introns in the RAD51 genes is unusual and may indicate the presence of an unusual recombination apparatus in this organism. The presence of duplicated intronless RAD51 genes is unique among eukaryotes. Studies of further members of this lineage are needed to determine whether this feature may be typical of lower plants.
Homologous recombination is essential for chromosomal segregation and participates in DNA replication and repair. The molecular mechanisms of homologous recombination have been studied intensively, mostly in bacteria and yeast. Homology recognition and exchange of homologous DNA strands have a central role in homologous recombination (reviewed in ref. 1). The paradigm of strand exchange proteins, the Escherichia coli RecA protein, is well conserved from bacteria to man, and several structural and functional homologues have been found in eukaryotes (reviewed in ref. 2). The best characterized homologues are the Rad51 and Dmc1 proteins. Although Rad51 seems to be involved in mitotic as well as meiotic recombination, Dmc1 is expressed exclusively in meiosis. The presence of multiple functionally distinct proteins in eukaryotes is likely because of an ancient gene duplication followed by functional divergence early in eukaryotic species evolution (3). In addition to the original RAD51 and DMC1 genes, a series of other genes encoding proteins with low homology to Rad51 have been detected in yeast and other eukaryotes including man and other mammals (reviewed in refs. 2 and 4). The proteins of this group show similarities with Rad51, the most striking of which is that all of them contain Rad51/RecA signatures; however, the low degree of homology [20–30% similarity (4)] to Rad51 distinguishes them from the original Rad51 protein. The group of corresponding genes contains genes that were annotated misleadingly as RAD51, e.g., RAD51B, RAD51C, and RAD51D and genes such as RAD55, RAD57, XRCC2, and XRCC3. These genes do not seem to be functionally redundant with RAD51, because the loss of RAD51 function is lethal in the mouse (5, 6). Although the function of these proteins is poorly understood, it is likely that all of them fulfill particular and specific functions in homologous recombination or DNA-damage repair (7). The fact that loss of Rad51 function in yeast results in a relatively mild phenotype but is lethal in higher eukaryotes suggests that the mammalian gene has acquired additional functions and may be involved in a link to cell cycle control and apoptosis (reviewed in ref. 2). Most eukaryotic RAD51 genes and other genes with homology to RAD51 are interrupted by introns, even those from filamentous fungi such as Neurospora crassa (8), or Penicillium paxilli (EMBL/GenBank/DDBJ databases, accession number AB040404) and the ciliate Tetrahymena thermophila (9). Only some unicellular organisms possess intronless RAD51 genes, and these genes are unique in the genome. These organisms belong to lower eukaryotes such as the yeast Saccharomyces cerevisiae (10) and the yeast Schizosaccharomyces pombe (11) or to a group of ancient eukaryotes that do not have intron-split coding regions (12) such as the unicellular eukaryotic parasite Trypanosoma brucei (13). However, the DMC1 genes of both yeasts do contain introns (14, 15). The role of introns in RAD51, DMC1, and other genes of the RAD51 family is poorly understood. Rad51 and other proteins of this family seem to cooperate, and thus introns may help to increase the diversity of interacting, functional RAD51 proteins by differential splicing of intron-split genes.
In contrast to fungi and animals, there is very little information about RAD51 genes in plants. RAD51 gene sequences are available only from the two dicotyledonous plants Arabidopsis thaliana (16) and tomato (3) and from the monocot maize (17). No RAD51 gene from a lower plant has been described thus far. Bryophytes such as the leafy moss Physcomitrella patens differ from higher, vascular plants such as Arabidopsis in various aspects (18). Most conspicuously, the body plan and development of bryophytes are much simpler. In addition, the gametophyte is the dominant phase in the Physcomitrella life cycle. Recently, Physcomitrella has received considerable attention as a new model organism (18, 19) because of the high efficiency of targeted gene replacement that is unique in the plant kingdom (reviewed in ref. 20). To gain insight into the homologous recombination apparatus of this organism, Physcomitrella RAD51 genes were identified and analyzed, and their gene structure was determined. The Physcomitrella RAD51 genes show an organization highly unusual for a multicellular higher eukaryote. In contrast to the RAD51 genes found in other higher eukaryotes, the Physcomitrella genome contains two highly related genes, neither of which is interrupted by introns. Both genes are fully functional as shown by cDNA cloning and expression analysis. Thus Physcomitrella RAD51 gene organization is unique, and it is the only eukaryotic organism with two intronless RAD51 genes. These facts suggest that the recombination apparatus of Physcomitrella differs from that of other higher eukaryotic organisms.
Materials and Methods
Isolation of Physcomitrella RAD51 Genes.
Genomic DNA prepared from 7-day-old protonema of P. patens (Gransden wild-type strain) as described (21) was used to amplify a 120-bp fragment by PCR with degenerate primers as described (11). The DNA sequence obtained from this PCR product after subcloning was used to design a specific primer (RADL2, GAA TTC AGG ACG GGT AAG AGC CAG A). This primer and a degenerate primer designed from rad51 gene alignments (RADR1, CAN CCT GGT TNG TGA TNA CCA CTG C) amplified a 400-bp fragment from Physcomitrella genomic DNA by using the Roche Expand long-template PCR system (initial denaturation 2 min at 94°C, denaturation 10 sec at 94°C, annealing 30 sec at 56°C, and synthesis 4 min at 68°C for 30 cycles with a time extension of 15 sec followed by a delay of 10 min). The 400-bp product was cloned, and the sequence was verified to encode part of the Rad51 coding region.
To isolate complete genomic clones, a Physcomitrella genomic library (kindly provided by Stavros Bashiardes, University of Leeds, Leeds, U.K.) was screened by PCR for the presence of phages carrying the 400-bp fragment with primers RADRAL (CGA CCT GGT TTG TGA TTA CCA CTG C) and RADBE (TCT GCC ATC TTC TGG CAA CCC CGA A) by using the conditions described above. Sublibraries of 10,000 plaque-forming units were generated from the original genomic library and those diluted 5-fold into sub-sublibraries and screened until a pure λ clone was obtained. The full-length λ insert of this clone was amplified by using primers flanking the cloning sites, and the insert was sequenced by shotgun sequencing to yield the DNA sequence of the genomic region of Pprad51A. This sequence was not identical to that of the 400-bp fragment in the overlapping region. This result suggested the existence of a close homologue.
To isolate this homologue, inverse PCR was performed. MunI-digested Physcomitrella genomic DNA was religated and used as a template in a PCR (initial denaturation 94°C, 2 min; denaturation 92°C, 10 sec; annealing 55°C, 30 sec; and extension 68°C 12 min for 30 cycles) reaction with primers specific for the 400-bp fragment (400 pm-A, CAT GTG ACG CAG AGA GTA TGA C, and 400 pm-F, CGT CAA TTC CAC CTT GCC AAG C). The 4,000-bp PCR fragment generated this way was sequenced by shotgun sequencing and yielded Pprad51B.
The coding regions of both RAD51 genes were amplified from the respective genomic clones by PCR and subcloned into pCR2.1 (Invitrogen) according to the manufacturer's instructions. Plasmid pJA-3 containing the Rad51A coding region was obtained with primers ppa-ATG (CAT ATG GCC ACT GTT AGT GCA G) and ppa-END (GTC GAC TTC GTG TTG AAC GCA TGT C) and plasmid pJB-2 containing the RAD51B gene-coding regions was obtained with primers ppb-ATG (CAT ATG GCC ACT GCC AGT GC) and ppb-END (GTC GAC ACG GCG AAA TTC GCT ACC). These plasmids were sequence-verified.
cDNA clones were isolated from a Physcomitrella cDNA library (kindly provided by S. Bashiardes and A. Cuming, University of Leeds, Leeds, U.K.). The library was made from RNA isolated from 7-day-old protonema tissue grown under 7 days of continuous white light on Physcomitiella standard tissue culture medium (21) medium with ammonium nitrate as the nitrogen source, treated with 10−4 M abscisic acid for 14 h, and constructed in Lambda ZapII vector (Stratagene). The library was screened with radioactively labeled probes obtained from the Rad51A and Rad51B coding regions of pJA-3 and pJB-2. Plaques giving a positive signal were purified further for five rounds of library screening by using standard protocols. Phagemids were converted into plasmids according to the manufacturer's instructions. Plasmids were sequenced from the 5′ and 3′ ends, and the complete inserts of two plasmids that contained the longest cDNA inserts were sequenced fully.
DNA Analysis.
Sequence data were analyzed by using the GCG software package (Genetics Computer Group, Madison, WI), the BLAST program (22), and vector NTISUITE (Informax, Bethesda, MD).
DNA and RNA Hybridization Analysis.
For Southern blots, genomic DNA was isolated from P. patens, digested with restriction enzymes, separated on agarose gels, blotted to Nylon membranes, and hybridized with radioactively labeled probes made from the Rad51A and Rad51B coding regions of pJA-3 and pJB-2 as described (21). The membranes were washed twice at low stringency [2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS at 50°C for 20 min] and then exposed to Kodak Biomax MS film with intensifying screens for 3–4 days.
For Northern blots, 9 μg of poly(A)+ RNA was separated on agarose gels, blotted to Nylon membranes, and hybridized with radioactively labeled probes obtained from fragments containing the entire RAD51A and RAD51B cDNAs as described (23). The membranes were washed twice under high stringency conditions (0.1× SSC/0.1% SDS at 65°C for 30 min) and then exposed to Kodak Biomax MS film with intensifying screens for 3–4 days.
Results
Isolation and Genetic Organization of Physcomitrella RAD51 Genes.
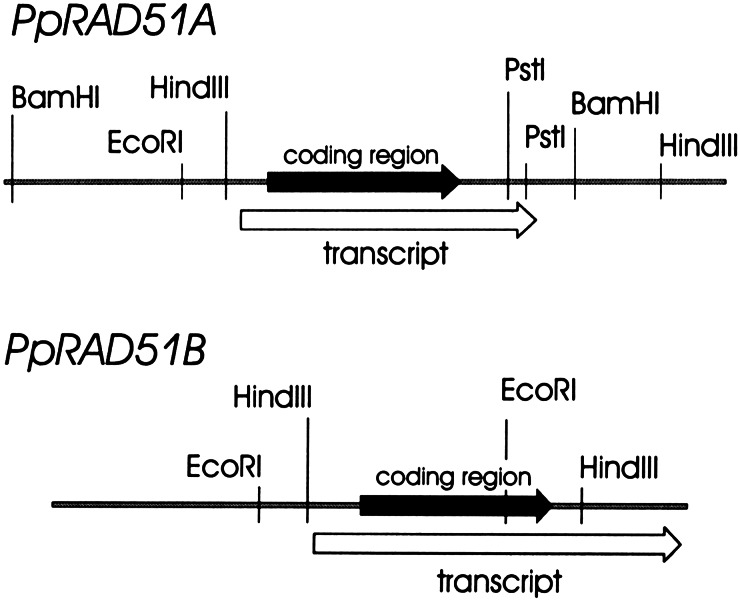
A PCR-based strategy involving several steps (see Materials and Methods) was used to isolate fragments of Physcomitrella genomic DNA that contained RAD51 genes. Surprisingly, two different genomic fragments were obtained that encoded a RAD51 gene. The nucleotide sequence of the two fragments was determined in both strands. Both genomic sequences contained a continuous ORF of 1,029 bp encoding a protein of 342 amino acids. Comparison of the predicted amino acid sequences with the SwissProt and SPTREMBL databases identified both proteins as Rad51 homologues. Both sequences showed highest homology with Rad51 proteins from other organisms, followed by other proteins such as Dmc1 and RecA with weaker homology to RecA-like proteins. Because the proteins were highly homologous but not identical, they were named P. patens Rad51A (PpRad51A) and Rad51B (PpRad51B). To obtain information about the corresponding mRNAs, a Physcomitrella cDNA library was screened with the cloned coding regions. Two sets of cDNA clones were obtained, with one corresponding to the PpRAD51A gene and the other to the PpRAD51B gene. The DNA sequences of longest cDNA clones covered the entire coding regions and were identical to the corresponding genomic DNA sequence over their entire length. Because the amino acid sequences deduced from the genomic sequences matched with Rad51 proteins from other organisms over their full length, and the cDNA sequences were completely identical to the corresponding genomic sequences in their full length, both Physcomitrella RAD51 genes are not interrupted by introns. The deduced gene structures including the location of the coding regions and transcripts are summarized in Fig. 1. The complete sequences of the genomic and cDNA clones are available in the EMBL database under accession numbers AJ344152 and AJ344153, for the genomic sequences, and AJ316537 and AJ316538, for the cDNA sequences.
Figure 1.
Schematic representation of Physcomitrella RAD51 genes. The sequenced genomic regions are shown schematically. The relative positions of the transcripts and coding regions are included. The positions of restriction endonucleases recognition sites used for the Southern blot are shown.
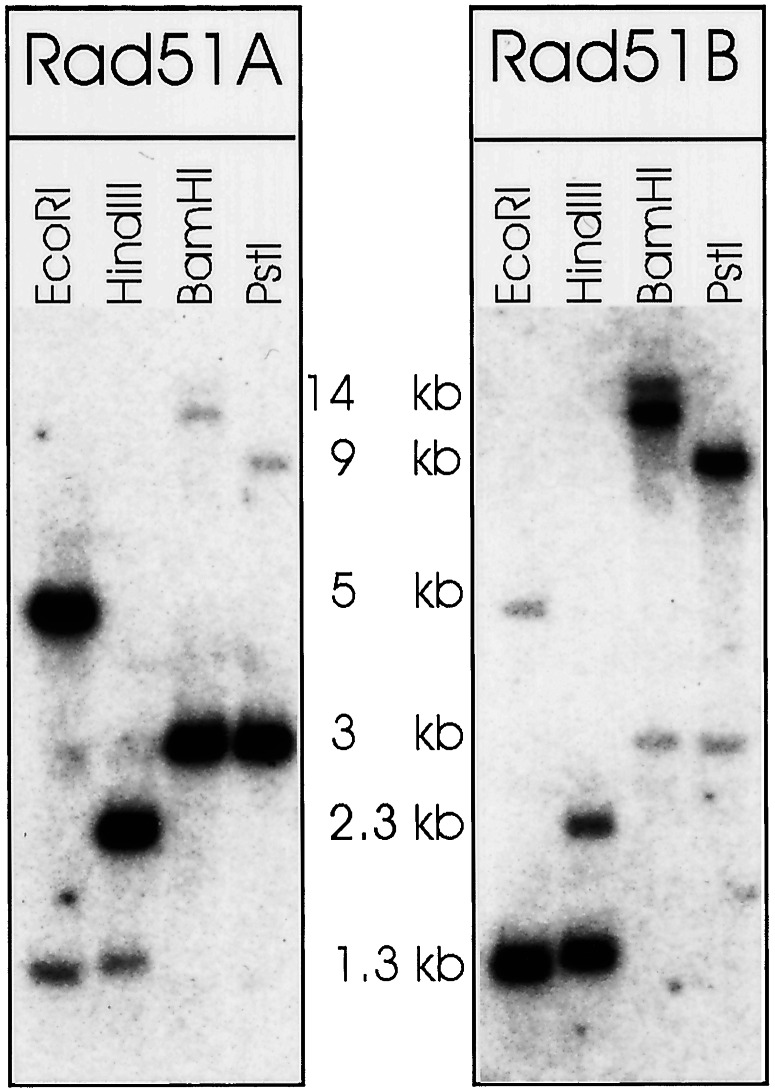
To verify the presence of PpRAD51A and PpRAD51B in the Physcomitrella genome and to search for more RAD51 genes that might be present in the Physcomitrella genome, Southern blotting experiments were performed (Fig. 2). The hybridization pattern observed with both genes was as expected from the genomic DNA sequence and confirmed the presence of both genes. The PpRAD51A-specific probe cross-hybridized weakly with PpRAD51B gene-specific fragments under low-stringency hybridization conditions, and vice versa. Therefore the hybridization conditions allowed detection of DNA sequences in a range of homology comparable to the similarities of the PpRAD51A and PpRAD51B genes. No further additional fragments were detected even after prolonged overexposure of the film. This result indicated that the RAD51A and RAD51B genes are unique in the Physcomitrella genome, and no additional RAD51 genes exist.
Figure 2.
Southern blot analysis of Physcomitrella RAD51 genes. Genomic DNA from P. patens was digested with the restriction endonucleases as indicted and blotted. The membrane was hybridized under low-stringency conditions with RAD51A and RAD51B gene-specific probes, respectively. The figure shows an overexposed film. The approximate sizes of fragments as deduced from a stained DNA size marker (λ DNA digested with PstI) are given.
Gene Structure and Expression.
Both PpRAD51A and PpRAD51B were expressed in vegetative, actively growing protonema tissue (Fig. 3) although at very low level. A single A-gene transcript of 1,550 nucleotides was detected in Northern blots. Two transcripts, one with a size of 1,900 nucleotides and the other with a size of 1,600 nucleotides, were found with the PpRAD51B gene. Both PpRAD51B transcripts were less abundant in protonema tissue than the PpRAD51A transcript. The size of the PpRAD51A gene and the larger PpRAD51B gene transcript corresponded to the length of the cDNA clones, and therefore these clones were close to representing the full-length transcripts predicted from the Northern blot.
Figure 3.
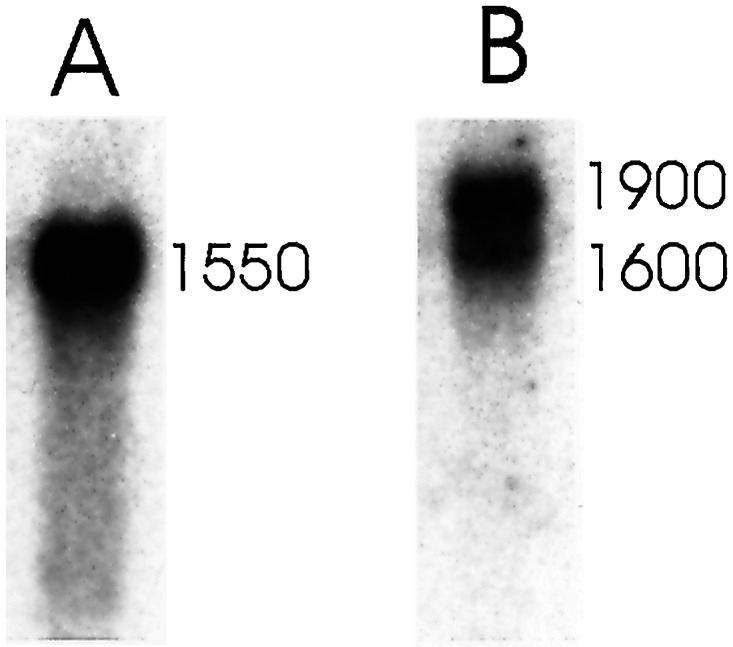
Northern blot analysis of Physcomitrella RAD51 genes. Poly(A)+-selected RNA was prepared from actively growing 7-day-old protonema tissue. The RNA (9 μg per lane) was separated by gel electrophoresis and blotted to a membrane, and the membrane was hybridized under high-stringency conditions with RAD51A (A) and RAD51B (B) gene-specific probes. The approximate sizes of the transcripts as deduced from a stained RNA size marker (0.24–9.5-kb RNA ladder, Life Technologies, Karlsruhe, Germany) are given.
The transcript lengths predicted from the cDNAs were 1,569 bp for PpRAD51A and 1,931 bp for PpRAD51B. Both contained an ORF of 1,029 bp. Well conserved polyadenylation signals were found in the 3′ untranslated region 18 and 27 bp upstream of the poly(A) tail in the PpRAD51A and PpRAD51B transcripts, respectively. Several less well conserved polyadenylation signal sequences were present between the canonical sequence and the end of the coding region in PpRAD51B. The use of one of these cryptic polyadenylation signals is likely to give rise to the second, shorter transcript observed in the Northern blot with the PpRAD51B gene. Both RAD51 genes appeared to have 5′ untranslated regions of normal size (139 bp for PpRAD51A and 212 bp for PpRAD51B), but the 3′ untranslated regions were unusually long (419 bp for PpRAD51A and 690 bp for PpRAD51B). CpG islands indicative of promoters were found in the genomic sequences of both genes upstream of the transcript starts.
The PpRAD51A and PpRAD51B gene nucleotide sequences are 86% identical in the coding region, confirming the remarkable conservation of the two genes. Sequence homology drops to 46 and 58%, respectively, in the 5′ and 3′ untranslated regions and to less than 46% in the 5′ nontranslated region not represented in the cDNA (data not shown). This pattern was reminiscent of the maize and Xenopus RAD51 genes. Both organisms have two highly homologous RAD51 genes. The DNA sequence of the two Xenopus genes is 93% identical in the coding region, and differences in nucleotide sequences were found mostly in the 5′ and 3′ untranslated regions (24). Based on the DNA sequence of the two maize genes (17), these are 84% identical in the coding region and 46 and 47%, respectively, in the 5′ and 3′ untranslated regions.
The Physcomitrella Rad51 Proteins and Their Relationship to Other Rad51 Proteins.
The ORFs of the Pprad51A and Pprad51B DNA genes encode a protein of 342 amino acids that corresponds to a molecular mass of 36.9 kDa for the A gene and 36.8 kDa for the B gene. The size of both proteins was as expected for a Rad51 protein, and both contained the characteristic nucleotide binding domains (Fig. 4), which are conserved between all RecA-like proteins (25). Comparison with other Rad51 proteins confirmed that the Physcomitrella proteins also showed a typical two-domain structure; domain I consisted of 69 amino acids and is rich in positively charged residues, whereas domain II consisted of 269 amino acids that are rich in negatively charged residues. A RecA signature was present between residues 270 and 303.
Figure 4.
Physcomitrella deduced amino acid sequences. The aligned amino acid sequences deduced for the Physcomitrella RAD51A and RAD51B genes are shown in single-letter codes. Blocks of identity are shaded in gray. The Walker A and B motifs are boxed.
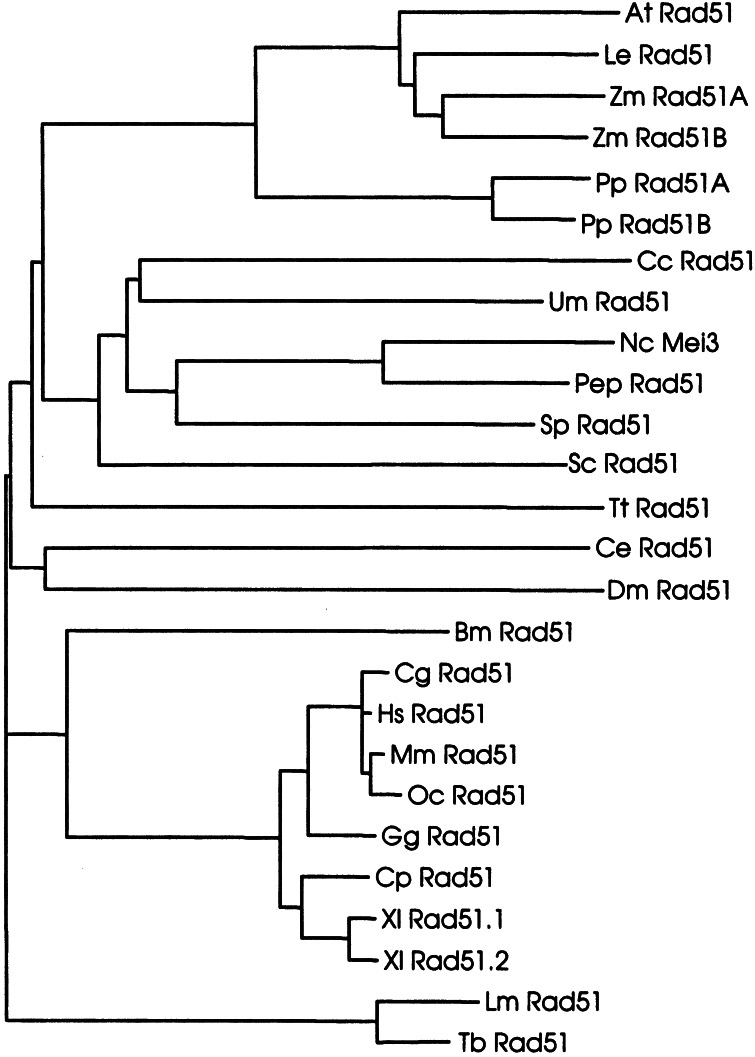
The PpRad51A and PpRad51B proteins (Fig. 4) are 94% identical in amino acid sequence and therefore are highly related. This situation was very similar to that in maize (17) and Xenopus (24), both of which possess two RAD51 genes encoding highly homologous proteins (90 and 98% identity, respectively). To investigate the relationships of the Physcomitrella proteins to Rad51 proteins from other organisms, both proteins were aligned to protein or deduced protein sequences of Rad51 proteins from organisms annotated in the SwissProt and SPTREMBL databases. The results are shown in the form of a phylogenetic tree (Fig. 5). The tree based on the relatedness of Rad51 protein sequences reflected well the relatedness of the respective organisms. The main clades were formed by the vertebrate, fungal, and plant genes, whereas some kinetoplastida, ciliate, and invertebrate genes formed separate branches. The vertebrate genes, the most intensively studied group, separated clearly into mammalian and amphibian genes. The clade of plant genes separated clearly into a group representing the vascular plants containing the genes from Arabidopsis, tomato, and maize and a separate lower land plant group that contained the two Physcomitrella genes. The position of the Physcomitrella Rad51 proteins was in full agreement with the phylogeny of green plants deduced from 18S rDNA sequences (26). Therefore, both Physcomitrella proteins were placed well in accordance with the expected phylogenetic relationships and thus were likely to represent proteins of plant origin forming a clade separate from vascular plants.
Figure 5.
Phylogenetic relationships of Rad51 proteins. The phylogenetic tree shown is based on amino acid sequence distance and was calculated according to the neighbor-joining algorithm (40). The sequences used for the alignment for the organisms and their accession numbers (in brackets after the abbreviations) are: At, A. thaliana (P94102); Bm, Bombix mori (O01679); Cc, Coprinus cinereus (O074569); Ce, Caenorhabditis elegans (O44246); Cg, Cricetulus griseus (P70099); Cp, Cynops pyrrhogaster (Q9W628); Dm, Drosophila melanogaster (Q27297; Q9VAA8); Gg, Gallus gallus (P37383); Hs, Homo sapiens (Q06609); Le, Lycopersicon esculentum (Q40134); Lm, Leishmania major (O61127); Mm, Mus musculus (Q08297); Nc, N. crassa (P87210); Oc, Oryctolagus cuniculus (O77507); Pep, P. paxilli (Q9P956); PpRad51A, P. patens, this work; Pp Rad51B, P. patens, this work; Sc, S. cerevisiae (P25454); Sp, S. pombe (P36601); Tb, T. brucei (O00847); Tt, T. thermophila (O76341); Um, Ustilago maydis (Q99133); Xl rad51.1, Xenopus laevis (Q91918); Xl Rad51.2, X. laevis (Q91917); Zm Rad51A, Zea mays (Q9XED); Zm Rad51B, Z. mays (Q9XED7).
Discussion
The Physcomitrella genome contains two distinct but highly homologous RAD51 genes, PpRAD51A and PpRAD51B. For both, the DNA sequence derived from genomic DNA revealed the presence of an ORF not interrupted by introns. Confirming the absence of introns, the genomic and cDNA sequences were found to be co-linear for both genes. In Southern blots with Physcomitrella genomic DNA, no additional genes were detected at low-stringency hybridization conditions. Moreover, no additional genes were detected by screening of cDNA or genomic libraries. Both genes were expressed actively in Physcomitrella and therefore are functional RAD51 genes.
The presence of two highly homologous, functional RAD51 genes in a genome is unusual. In other organisms studied, RAD51 is present in single copy except for organisms that have a duplicated genome, e.g., X. laevis and maize. These organisms each have two highly homologous RAD51 genes. The Xenopus genes are 98% identical and probably arose in the course of a genome duplication event relatively recently in evolution that gave rise to the pseudotetraploid Xenopus genome (24). The maize genome, probably a fusion of two genomes (27), contains two genes that are 90% identical (17). In addition to the homology in the protein sequences, the genomic DNA sequences of both organisms still carry clear signatures of duplication events. The Physcomitrella Rad51 proteins were 94% identical in their sequence, and the homology extended to the corresponding DNA sequences. The close relatedness of the PpRAD51A and PpRAD51B genes indicated that these genes arose by a recent gene duplication event from a common progenitor. The question arises as to whether this gene duplication reflects a genome duplication event or is restricted to RAD51. There is little information on the Physcomitrella genome, and data available from other genes (28, 29) are too premature to address the question of gene duplication. However, polyploidization in the ancestry of Physcomitrella is discussed (reviewed in ref. 18), and in general polyploidization and subsequent genome duplications are a common phenomenon in plants (30, 31). Genome duplications followed by different degrees of functional diversification, subsequent gene loss, or gene silencing in the ancestry of species can be traced back in many, if not all, plants (32). Moreover, RAD51 genes have been studied intensively in different organisms, and there is a striking correlation between the presence of two RAD51 genes in an organism and the occurrence of genome duplications. From all the organisms studied thus far, only the genomes of Xenopus and maize have two highly homologous RAD51 genes, and these are the two organisms with verified genome duplications in their ancestry. Therefore the similarity of Physcomitrella to Xenopus and maize in their RAD51 gene organization suggests a genome duplication event in the ancestry of Physcomitrella also. Moreover, the close relatedness of the two RAD51 genes suggests a rather recent event in the evolution of the genome of this organism. Although Physcomitrella is haploid, considerable gene redundancy would result from duplication events, and therefore gene analysis in this organism should be treated with caution until the question of genome duplication is resolved.
The Physcomitrella RAD51 genes are free of introns. By contrast, all other multicellular eukaryotes including filamentous fungi such as Neurospora or Penicillium have intron-split RAD51 genes. Organisms with intronless RAD51 genes are unicellular eukaryotes such as budding and fission yeasts, and organisms possessing intronless genomes. Physcomitrella does not belong to the latter group, because introns are found frequently in Physcomitrella genes, and the positions are largely conserved between Physcomitrella and Arabidopsis (28, 29). Introns in RAD51 and other RAD51-like genes may serve important functions. Some RAD51-like genes are differentially spliced, and several splice variants are detected (33–36). Moreover, Rad51 and other Rad51-like proteins (reviewed in refs. 4 and 25) can interact and may form networks of active forms (37). Therefore, differential splicing may be a source of variability for proteins expressed from a single gene and thus may serve to modulate or control Rad51 protein activity. Because differential splicing of RAD51 genes is precluded in Physcomitrella, the presence of two genes may help to increase the variety of Rad51 and Rad51-like proteins that can participate in the networks of proteins active in strand exchange. However, it is possible also that one of the Physcomitrella proteins substitutes for one or more of the other Rad51-like proteins found in other higher eukaryotes. There is a striking correlation between the presence of introns in a close homologue of RAD51, DMC1, and expression during meiosis in yeast and fission yeast. Thus, introns could serve regulatory functions and help to distinguish meiosis-specific functions from general recombination functions. In support of this idea, the presence of introns was found to be correlated with meiosis-specific expression of members of another group of recombination genes in Arabidopsis (38). However, the precise role of introns in RAD51 gene function or regulation remains to be determined.
The bryophytes and other lower plants are not well studied, and no further RAD51 genes from this group of organisms have been described. In the phylogenetic tree presented here, the Physcomitrella Rad51 proteins group together with the other plant proteins but form a separate clade within this group. This result indicates that the Physcomitrella Rad51A and Rad51B proteins and the higher plant proteins have a common origin. However, the phylogenetic data are sparse, and the phylogenetic relationships of this group of organisms are uncertain. Moreover, the Rad51 proteins of several other eukaryotes, especially those of yeast, branch off from the tree relatively early, complicating the exact assignment of phylogenetic relationships. Nevertheless, the agreement of the Rad51-based data with trees based on 18S rDNA sequences (26) strongly suggests that the Physcomitrella genes have evolved in the plant lineage of organisms. Because RAD51 genes of all higher eukaryotes have introns, Physcomitrella must have lost its introns during evolution, either from both RAD51 genes after the gene duplication event or earlier in the ancestral gene. It is difficult to envision how the introns may have been lost. Physcomitrella is an unusual organism in possessing a highly active homologous recombination apparatus in mitotic cells (18–20). It therefore is conceivable that gene conversion events using RAD51 mRNA reverse transcripts as templates caused the loss of introns in the ancestry of Physcomitrella. Another possibility is that reverse transcription of the RAD51 mRNA produced an intronless gene copy that integrated into the genome and the original intron-containing RAD51 genes sequences were subsequently lost. In any case, it will be important to determine whether intronless RAD51 genes are typical for bryophytes and/or other lower land plants or are a rarity specific for Physcomitrella.
Acknowledgments
We thank the crews of the Max-Planck-Institut fuer Zuechtungsforschung and Max-Planck-Institut fuer Molekulare Genetik DNA sequencing facilities, especially Bernd Weisshaar, for their excellent service. We are grateful to Andrew Cuming and Stavros Bashiardes (University of Leeds, Leeds, U.K.) for providing the Physcomitrella genomic and cDNA libraries. Support from European Union Grants BIO4 CT97 2028 and QLK3-CT-2000-00365 is gratefully acknowledged.
Note Added in Proof.
The PpRad51A protein was named PpaRad51.1 and the PpRad51B protein was named PpaRad51.2 by Ayora et al. (40).
Footnotes
References
- 1.Dunderdale H J, West S C. Curr Opin Genet Dev. 1994;4:221–228. doi: 10.1016/s0959-437x(05)80048-6. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, West S C. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 3.Stassen N Y, Logsdon J M, Vora G J, Offenberg H H, Palmer J D, Zolan M E. Curr Genet. 1997;31:144–157. doi: 10.1007/s002940050189. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara A, Ogawa T. Mutat Res. 1999;435:13–21. doi: 10.1016/s0921-8777(99)00033-6. [DOI] [PubMed] [Google Scholar]
- 5.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim D S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takata M, Sasaki M S, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson L H, Takeda S. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatakeyama S, Ishii C, Inoue H. Mol Gen Genet. 1995;249:439–446. doi: 10.1007/BF00287106. [DOI] [PubMed] [Google Scholar]
- 9.Campbell C, Romero D P. Nucleic Acids Res. 1998;26:3165–3172. doi: 10.1093/nar/26.13.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 12.Donelson J E, Gardner M J, El-Sayed N M. Proc Natl Acad Sci USA. 1999;96:2579–2581. doi: 10.1073/pnas.96.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCulloch R, Barry J D. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukushima K, Tanaka Y, Nabeshima K, Yoneki T, Tougan T, Tanaka S, Nojima H. Nucleic Acids Res. 2000;28:2709–2716. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 16.Doutriaux M P, Couteau F, Bergounioux C, White C. Mol Gen Genet. 1998;257:283–291. doi: 10.1007/s004380050649. [DOI] [PubMed] [Google Scholar]
- 17.Franklin A E, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande W Z. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cove D J, Knight C D, Lamparter T. Trends Plant Sci. 1997;2:99–105. [Google Scholar]
- 19.Wood A J, Oliver M J, Cove D J. Bryologist. 2000;103:128–133. [Google Scholar]
- 20.Schaefer D G. Curr Opin Plant Biol. 2001;4:143–150. doi: 10.1016/s1369-5266(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann A H, Codon A C, Ivascu C, Russo V E A, Knight C, Cove D, Schaefer D G, Chakhparonian M, Zryd J P. Mol Gen Genet. 1999;261:92–99. doi: 10.1007/s004380050945. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Markmann-Mulisch U, Reiss B, Mulisch M. Mol Gen Genet. 1999;262:390–399. doi: 10.1007/s004380051098. [DOI] [PubMed] [Google Scholar]
- 24.Maeshima K, Morimatsu K, Shinohara A, Horii T. Gene. 1995;160:195–200. doi: 10.1016/0378-1119(95)00148-y. [DOI] [PubMed] [Google Scholar]
- 25.Thacker J. Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 26.Soltis E D, Soltis P S. Plant Mol Biol. 2000;42:45–75. [PubMed] [Google Scholar]
- 27.Doebley J, Stec A, Wendel J, Edwards M. Proc Natl Acad Sci USA. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Champagne C E M, Ashton N W. New Phytologist. 2001;150:23–36. [Google Scholar]
- 29.Krogan N T, Ashton N W. New Phytol. 2000;147:505–517. doi: 10.1046/j.1469-8137.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- 30.Leitch I J, Bennett M D. Trends Plant Sci. 1997;2:470–476. [Google Scholar]
- 31.Joubes J, Chevalier C. Plant Mol Biol. 2000;43:735–745. doi: 10.1023/a:1006446417196. [DOI] [PubMed] [Google Scholar]
- 32.Wendel J F. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 33.Rinaldo C, Ederle S, Rocco V, Lavolpe A. Mol Gen Genet. 1998;260:289–294. doi: 10.1007/s004380050897. [DOI] [PubMed] [Google Scholar]
- 34.Pittman D L, Weinberg L R, Schimenti J C. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- 35.Cartwright R, Dunn A M, Simpson P J, Tambini C E, Thacker J. Nucleic Acids Res. 1998;26:1653–1659. doi: 10.1093/nar/26.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabata M, Saeki K. Biochem Biophys Res Commun. 1999;257:156–162. doi: 10.1006/bbrc.1999.0413. [DOI] [PubMed] [Google Scholar]
- 37.Dosanjh M K, Collins D W, Fan W F, Lennon G G, Albala J S, Shen Z Y, Schild D. Nucleic Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartung F, Puchta H. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Ayora, S., Piruat, J. I., Luna, R., Reiss, B., Russo, V. E. A., Aguilera, A. & Alonso, J. C. (2002) J. Mol. Biol., in press. [DOI] [PubMed]