Abstract
αvβ8 integrin is highly expressed in the vertebrate CNS, and mice lacking the αv or β8 genes develop cerebral hemorrhage due to defective interactions between blood vessels and αvβ8-expressing neural cells. Although αvβ8 binds many of the same extracellular matrix protein ligands as other integrins, very little is known about the intracellular signal transduction events used by αvβ8 to regulate CNS development. Here we identify Band 4.1B as an intracellular factor that interacts selectively with the β8 cytoplasmic tail. The association with αvβ8 occurs via the band 4.1B C-terminal domain, a region highly conserved among the various Band 4.1 family members. Indeed, we show that β8 integrin interacts directly with the C-terminal domains of several Band 4.1 proteins and colocalizes with them in cultured astrocytes and in the brain. These data identify a previously uncharacterized interaction between an integrin and Band 4.1 family members and suggest an important functional role for αvβ8-Band 4.1 interactions in the development and maintenance of the CNS.
Keywords: central nervous system, extracellular matrix, intracellular signal transduction
The formation of the vertebrate CNS requires the precise regulation of cell adhesion and signaling events, including those involving members of the integrin family of proteins (1-3). Integrins are heterodimeric cell surface receptors consisting of α and β subunits that signal upon binding to extracellular matrix ligands (4). In many cases, integrin signaling involves recruitment of intracellular effectors that contain common 4.1/ezrin/radixin/moesin (FERM) domains (4). The Band 4.1 proteins are members of the FERM domain family and include 4.1B, 4.1G, 4.1N, and 4.1R (5, 6). All Band 4.1 family members are highly expressed in the mammalian CNS; however, rather little is known about their functions during CNS development and maintenance. Functional links between integrins and Band 4.1 proteins have not been analyzed.
Here we have identified Band 4.1B as a protein that interacts selectively with the αvβ8 integrin, which plays an important role in the development of the CNS (7-9). The association between αvβ8 and Band 4.1B is independent of the Band 4.1B FERM domain and rather occurs via the Band 4.1B C-terminal domain (CTD), a region highly conserved among the Band 4.1 family members. Indeed, we show that the CTDs of multiple Band 4.1 proteins interact directly with the β8 integrin cytoplasmic domain (β8cyto) domain and display overlapping expression patterns with αvβ8 in situ. These data suggest mechanisms for αvβ8-mediated adhesion and signaling and implicate the αvβ8-Band 4.1 interactions as important for proper CNS development.
Materials and Methods
Plasmids and Antibodies. A cDNA encoding the 54-aa human β8cyto was inserted in the yeast two-hybrid (Y2H) plasmid pGBKT7 (Clontech) and used to screen a human fetal brain cDNA library (Clontech). The various Band 4.1 CTD constructs were expressed by using the pGADT7 (Clontech) plasmid (see Supporting Text, which is published as supporting information on the PNAS web site). The full length human β8 integrin cDNA was inserted into the pcDNA3.1A-V5/His vector (Invitrogen). The human full length or truncated Band 4.1B cDNAs were inserted into pcDNA3.1A-myc/His or pCMV-Myc (Clontech) plasmids.
The rabbit anti-4.1Β antiserum was generated by using a synthetic peptide sequence of human Band 4.1B, CTTKGISQTNLITTVTPEKKA. The affinity-purified rabbit anti-β8 and goat anti-4.1B polyclonal antibodies have been described (6, 9). The following antibodies were purchased: anti-Nestin (Developmental Studies Hybridoma Bank, Iowa City, IA), smooth muscle α actin (Sigma), anti-αv (Chemicon), anti-paxillin (BD Transduction Laboratories), anti-4.1N (Protein Express, Chiba, Japan), and Alexa-conjugated secondary antibodies (Molecular Probes).
Transfection. The various constructs (Fig. 3) were transiently expressed in COS or 293T cells by using LF2000 lipofectin (Invitrogen). Cells were lysed in 50 mM Tris, pH 7.4/150 mM NaCl/1% Nonidet P-40/2.5 mM EDTA, and a complete protease inhibitor mixture (Roche, Mannheim, Germany). Detergent-soluble lysates were immunoprecipitated with an anti-myc polyclonal antibody (Santa Cruz Biotechnology). Immunoprecipitates were then resolved by SDS/PAGE, and Western blots were probed with anti-V5 or anti-myc monoclonal antibodies (Invitrogen). In some experiments, detergent-insoluble fractions were washed thoroughly with lysis buffer, and the insoluble pellets were resuspended in boiling 2× SDS sample buffer containing protease inhibitors. Detergent-soluble and -insoluble fractions were then resolved by SDS/PAGE. Alternatively, transfected cells were surface-labeled with EZ-Link Sulfo-NHSLC-Biotin (Pierce). Lysates were immunoprecipitated with antiserum, resolved by SDS/PAGE, and immunoblotted with Streptavidin-horseradish peroxidase (Vector Laboratories).
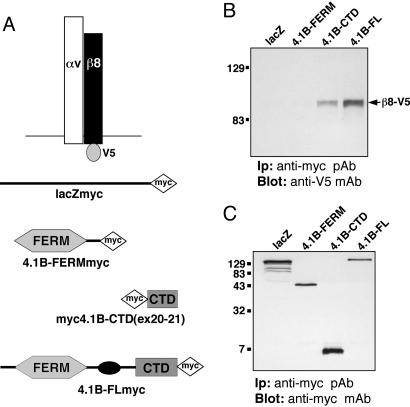
Fig. 3.
αvβ8 integrin and Band 4.1B CTD physically associate in transfected mammalian cells. (A) V5- and myc-tagged constructs used for transfection and coimmunoprecipitation analysis. (B and C) β8-V5 interacts with Band 4.1B via the Band 4.1B CTD encoded by exons 20 and 21. As shown in B, αvβ8-V5 coimmunoprecipitates with full length Band 4.1B-myc or with the CTD region encoded by exons 20 and 21 (CTDex20-21-myc). No interaction is detected between αvβ8-V5 and the Band 4.1B FERM-myc or the lacZ-myc control. (C) Relative amounts of myc-tagged proteins were immunoprecipitated the various transfected cell lysates.
Immunofluorescence. Adult mouse brains were fresh-frozen in Tissue Tek OCT compound (Miles). Midgestation mouse embryos were dissected, fixed with 4% paraformaldehyde for 1 h, cryopreserved in sucrose, and embedded in OCT compound. Alternatively, astrocytes cultured from neonatal mice as described (9) were plated onto coverslips coated with 10 μg/ml LAP-TGFβ1 (Research Diagnostics, Flanders, NJ) or laminin (Sigma) for 90 min. Cells were then fixed, permeabilized, and incubated with primary and fluorescent secondary antibodies.
Results
Primary Amino Acid Sequence Comparisons of β8cyto. Alignment of the cytoplasmic domains of the five β subunits that pair with αv revealed that the β8cyto shares little primary amino acid sequence homology with β1A, β3A, β5, and β6 (Fig. 1A). For example, the juxtamembrane amino acid sequence of β8 shares no primary amino acid sequence homology with that of other β subunits. This region is involved in a “molecular handshake” with the adjacent integrin α subunit (10), which maintains the integrin extracellular domain in an inactive state (4). The α/β cytoplasmic association is disrupted upon interaction between the β subunit and the intracellular protein talin, leading to “inside-out” integrin activation (11). Talin is recruited to the β cytoplasmic tail via an NPXY tetrapeptide motif found in β1A, β2, β3A, β5, and β6. This NPXY sequence is absent from β8 (Fig. 1 A). These results suggest that αvβ8 integrin activation and signal transduction may occur via mechanisms distinct from those used by other integrins.
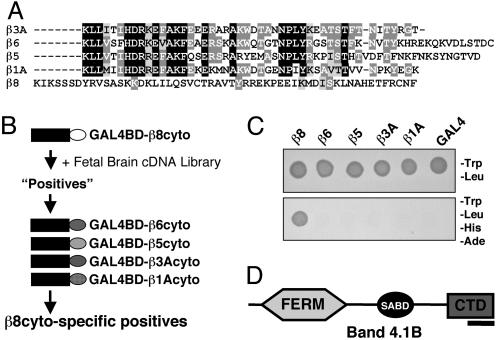
Fig. 1.
The β8 integrin cytoplasmic tail interacts with Band 4.1B. (A) Amino acid sequence alignment of the cytoplasmic domains of β integrin subunits that pair with αv integrin. Identical amino acids are highlighted in black or dark gray and conserved amino acids in light gray. β8cyto lacks primary amino acid sequence homology with other β subunits and does not contain the NPXY motif or the conserved juxtamembrane sequence. (B) A Y2H screen to identify proteins that interact selectively with β8cyto. The β8cyto sequence (white oval) was used to screen a human fetal brain cDNA library. Positive interactors were then screened by using the cytoplasmic tails of other integrins. (C) One example of a β8cyto-specific interacting positive, as determined by growth on selective media. (D) The β8cyto-specific positive shown in C encodes Band 4.1B. The portion of Band 4.1B that interacts with β8cyto is within the C-terminal portion (indicated by black bar).
Band 4.1B Selectively Interacts with β8 Integrin. We performed a Y2H screen to identify factors that interact selectively with β8cyto (Fig. 1B). A single clone identified from the Y2H library encoded Band 4.1B (Fig. 1 C and D), a member of the Band 4.1 family of proteins (12, 13). Mammalian Band 4.1 proteins contain three distinct functional domains: an N-terminal FERM domain, a central spectrin/actin-binding domain, and a CTD (6, 14, 15). The Band 4.1B cDNA identified from the Y2H screen encoded the last 63 amino acids of the CTD (Figs. 1D and 2A). The CTD amino acid sequences are highly conserved among the mammalian 4.1 family members (15). The last 63 amino acids of the Band 4.1B CTD, encoded exactly by exons 20 and 21, are >75% identical with the other mammalian 4.1 family members (Fig. 2 A). This region also contains ≈60% amino acid sequence homology with a worm Band 4.1 homolog and the fly Band 4.1 protein, coracle (14, 16). We postulated that the CTDs from the other 4.1 family members might also interact with β8cyto. Indeed, in the Y2H system, we detected an interaction with the isolated CTDs from 4.1G, 4.1N, and 4.1R (Fig. 2C).
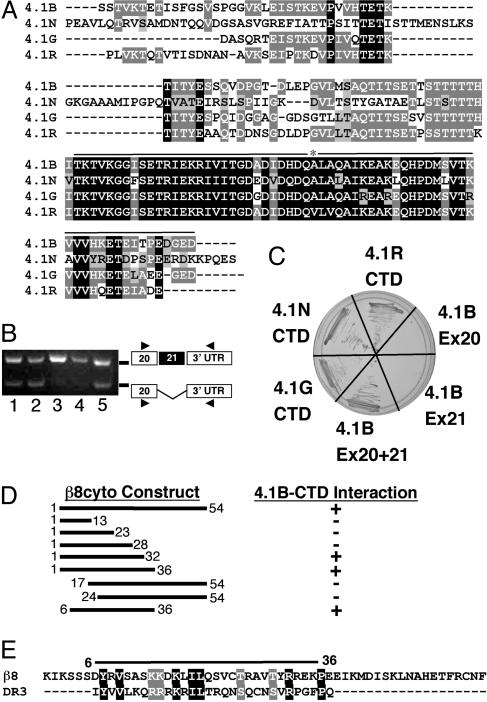
Fig. 2.
The β8 integrin cytoplasmic tail interacts with the region of the Band 4.1B CTD encoded by exons 20 and 21. (A) Alignment of CTD amino acid sequences of human Bands 4.1B, 4.1N, 4.1G, and 4.1R. Identical amino acids are highlighted in black or dark gray and conserved amino acids in light gray. Exons 20 and 21 (black line) encode the portion of Band 4.1B identified by the Y2H screen; asterisk marks boundary between exons 20 and 21. (B) Band 4.1B exon 21 is alternatively spliced. The presence or absence of Band 4.1B exon 21 was determined by using RNA isolated from wild-type mouse astrocytes (lane 1), αv integrin-null astrocytes (lane 2), adult mouse heart (lane 3), adult mouse brain (lane 4), or embryonic day (E)11.5 mouse embryo (lane 5). Exon 21 is included in the adult heart and brain samples but is absent in a portion of the transcripts from astrocytes and E11.5 embryos. (C) The CTDs from Band 4.1G, 4.1N, 4.1R, or 4.1B interact with β8cyto; however, the Band 4.1B segments encoded by individual exons 20 or 21 do not interact with β8cyto. (D) Deletion analysis identifies a 31-aa portion of the β8cyto as necessary for interaction with the Band 4.1B CTD in the Y2H system. (E) The 31-aa sequence of the β8cyto portion that interacts with the 4.1B CTD shares sequence homology with the second intracellular loop of Dopamine Receptor 3 (DR3), which also binds to multiple 4.1 CTDs.
Using RNAs isolated from cultured primary astrocytes, embryonic heads, and adult brain and heart, we showed that Band 4.1B exon 21 is differentially spliced in a cell- and tissue-specific manner (Fig. 2B). With this knowledge, we hypothesized that alternative splicing of exon 21 could affect the 4.1B CTD interaction with β8cyto. Using the Y2H system, no interaction was detected with the Band 4.1B protein fragments encoded by individual exons 20 and 21 (Fig. 2C). These data show that a 63-aa portion of the Band 4.1B CTD, encoded by exons 20 and 21, interacts specifically with the cytoplasmic sequence of β8 integrin, and that this interaction can be regulated by alternative splicing of Band 4.1B exon 21.
We generated various N- and C-terminal truncations of β8cyto (Fig. 2D) and tested these constructs in the Y2H system for interactions with the Band 4.1B CTD. We identified a 31-aa portion of the β8cyto as necessary for interaction with Band 4.1B CTD (Fig. 2D). This region of β8cyto contains significant sequence homology with the second cytoplasmic loop of Dopamine Receptor 3 (DR3-D2L) (Fig. 2E), which also binds to multiple Band 4.1 CTDs (17). Thus, this 31-aa region of β8cyto contains conserved sequences that may serve as Band 4.1 CTD interaction motifs.
αvβ8 and the Band 4.1B CTD Interact in Mammalian Cells. We verified the Y2H data by testing for an interaction between Band 4.1B and αvβ8 integrin using mammalian cells (COS) expressing various epitope-tagged constructs (Fig. 3A). COS cells express detectable levels of endogenous αvβ8 protein but do not express endogenous Band 4.1B proteins (Fig. 6, which is published as supporting information on the PNAS web site). αvβ8 coimmunoprecipitated with full length Band 4.1B or a fragment of the Band 4.1B CTD encoded by exons 20 and 21 (Fig. 3B). V5-tagged β8 also showed partial colocalization with myc-tagged full length Band 4.1B or the isolated CTD in transiently transfected COS cells (Fig. 7, which is published as supporting information on the PNAS web site). β8 integrin also showed partial colocalization with the isolated Band 4.1B FERM domain (Fig. 7); however, coimmunoprecipitation revealed no physical association between αvβ8 and the Band 4.1B FERM domain or with β-galactosidase (Fig. 3B). We reproducibly detected more detergent-soluble αvβ8 protein coimmunoprecipitating with full length 4.1B in comparison with the isolated 4.1B CTD fragment (Fig. 3B). This was not due to a lower expression level of the myc-tagged Band 4.1B CTD protein (Fig. 3C) or to significant differences in detergent solubility for β8 integrin, the isolated CTD fragment, and full length Band 4.1B (Fig. 6). Previous papers report (17-19) that overexpression of isolated Band 4.1 CTDs reduced the level of associated cell surface receptors. However, the levels of cell surface-expressed αvβ8 are not altered upon coexpression with Band 4.1B CTD or other Band 4.1B constructs (Fig. 6). Thus, we believe the differential interaction with αvβ8 can be explained by the fact that the CTD in the context of the entire 4.1B protein is important for a more efficient association with αvβ8.
αvβ8 and Band 4.1B Are Expressed in Primary Astrocytes and Colocalize at LAP-TGFβ1 Adhesion Sites. Astrocytes cultured from neonatal mouse brain expressed several cell surface integrins, including collagen, laminin, and fibronectin receptors (Fig. 4A), as well as αvβ5 and αvβ8 integrins (Fig. 4B) (20). In contrast, astrocytes cultured from αv-null neonatal mice did not express cell surface αvβ5 or αvβ8 (Fig. 4B). Cultured astrocytes also expressed a 126-kDa Band 4.1B immunoreactive band (Fig. 4C); this molecular weight coincides with the size for Band 4.1B protein described in previous reports (12, 13). The relative amount of Band 4.1B does not change in αv-/- astrocytes or neonatal brains (Fig. 4C). A similar immunoreactive band at 126 kDa was detected in the adult mouse brain. Additionally, protein bands were detected at ≈112 and 115 kDa (Fig. 4C). Because Band 4.1B mRNA is alternatively spliced, these proteins likely represent adult-specific isoforms of Band 4.1B.
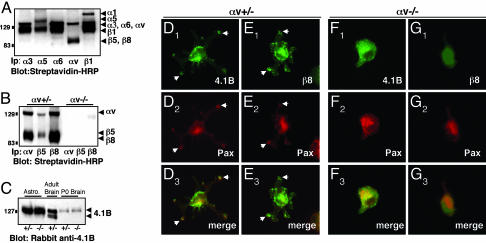
Fig. 4.
αvβ8 integrin and Band 4.1B localize to adhesion sites when astrocytes are plated on the αvβ8 ligand, LAP-TGFβ1. (A-C) Primary astrocytes express endogenous αvβ8 and Band 4.1B. (A) Astrocytes express multiple cell surface integrins as shown by immunoprecipitation with different anti-integrin antibodies. (B) Astrocytes cultured from αv+/- neonates, express αvβ5 and αvβ8, whereas αvβ5 and αvβ8 are not expressed at the cell surface of αv-/- astrocytes. (C) Protein lysates were prepared from αv+/- or αv-/- astrocytes or from neonatal brains and immunoblotted with anti-4.1B antibody. Similar levels of 4.1B protein are present in αv+/- and αv-/- samples. Adult brain lysates were immunoblotted with anti-4.1B. (D-G) Primary astrocytes cultured from neonatal mice; either αv+/- (D and E) or αv-/- (F and G) were plated on LAP-TGFβ1. Cells were immunostained with anti-4.1B or -β8 antibodies in combination with anti-paxillin to visualize adhesion sites. αv+/- astrocytes (D and E) extend multipolar processes when plated on LAP-TGFβ1. Band 4.1B (D1) and β8 integrin (E1) colocalize with paxillin (D2 and E2) in contact sites (arrows in D1-3 and E1-3). αv-/- astrocytes that do adhere to LAP-TGFβ1 do not spread and form contact sites (F1-3 and G1-3).
We colocalized Band 4.1B and αvβ8 in cultured astrocytes plated on LAP-TGFβ1. Band 4.1B (Fig. 4D) and αvβ8 (Fig. 4E) protein were present in adhesion sites in some astrocytes, as shown by colocalization with paxillin (Fig. 4 D and E). αv-/- astrocytes adhered poorly to LAP-TGFβ1, and most cells detached during the immunostaining procedure. αv-/- astrocytes that did adhere, however, did not spread and form contact sites (Fig. 4 F and G). This was not due to a general adhesion defect, because αv-/- astrocytes plated on laminin attached, spread, and formed cell contact sites similar to αv+/- astrocytes (Fig. 8, which is published as supporting information on the PNAS web site). Band 4.1B and β8 integrin (Fig. 9, which is published as supporting information on the PNAS web site) localization revealed a diffuse pattern on laminin, with no localization to contact sites.
We determined whether expression of full length Band 4.1B or the isolated CTD or FERM domain affected cell adhesion to LAP-TGFβ1. We detected no quantitatively significant differences in the adhesion of transiently transfected COS cells to LAP-TGFβ1 (Fig. 9). Similarly, overexpression of full length Band 4.1B or the isolated CTD fragment in COS cells did not result in significant changes in cell spreading on LAP-TGFβ1 (Fig. 7). In contrast, COS or 293T cells overexpressing the isolated Band 4.1B FERM domain appeared more spread on LAP (Fig. 7), as well as LM (data not shown).
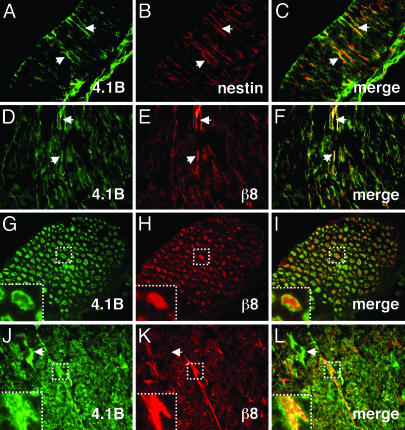
αvβ8 Integrin and Band 4.1 Proteins Colocalize in the Brain. We analyzed the expression patterns of Band 4.1B and β8 integrin in the embryonic and adult mouse brain. We previously showed that αvβ8 integrin is expressed on embryonic neuroepithelial and radial glial processes (9). Here, we demonstrate a similar embryonic glial expression pattern for Band 4.1B, as shown by colocalization with nestin (Fig. 5 A-C). Additionally, we showed that αvβ8 and Band 4.1B are expressed on some axons in the embryonic hindbrain (Fig. 5 D-F).
Fig. 5.
αvβ8 and Band 4.1B colocalize in neural cells in the embryonic and postnatal central nervous system. (A-C) Band 4.1B is expressed on embryonic neuroepithelial processes (arrows in A), as shown by colocalization with nestin (arrows in B and C). (D-F) Band 4.1B (D) and β8 integrin (E) colocalize (F) on neuronal cell bodies and axons in the embryonic hindbrain. Arrows (D-F) show axonal coexpression. (G-I) Band 4.1B (G) and β8 (H) are expressed on axons and colocalize (I) at axonal membranes in the adult brainstem. Areas in dashed boxes highlight Band 4.1B and β8 colocalization (G-I Insets) at higher magnification. (J-L) Band 4.1B (J) and β8 integrin (K) colocalize (L) to axons in the adult cerebellum. Band 4.1B is also expressed in neuronal cell bodies (arrows in J and L) where β8 integrin is absent (arrow in K). For all colocalization experiments, Band 4.1B was detected with a goat polyclonal antibody (D, G, and J), and β8 was detected with a rabbit polyclonal antibody (E, H, and K).
In the postnatal brain, αvβ8 integrin and Band 4.1B colocalized at the plasma membrane on white-matter axons, particularly in the cerebellum, brainstem, and spinal cord (Fig. 5 G-L). We also detected Band 4.1B protein expression on neuronal cell bodies in the cerebellum (Fig. 5J). However, we did not observe a similar expression pattern for αvβ8 (Fig. 5K), suggesting that Band 4.1B may be involved in other neural cell processes.
We also observed a blood vessel-associated expression pattern for αvβ8 protein throughout the adult brain (Fig. 10, which is published as supporting information on the PNAS web site). αvβ8 protein colocalized with smooth muscle α-actin, which is expressed primarily by vascular smooth muscle cells (Fig. 10). Glial endfeet are closely juxtaposed to most blood vessels in the CNS. Indeed, while this manuscript was under review, immunoelectron microscopy localized αvβ8 to perivascular glial endfeet (21). In the adult brain, Band 4.1N, but not Band 4.1B, was also expressed in a perivascular pattern (Fig. 10). These data suggest that αvβ8 adhesion and/or signaling likely involve interactions with distinct Band 4.1 family members and bolster the Y2H results (Fig. 2), which showed that the β8 cytoplasmic tail interacts with multiple Band 4.1 protein CTDs.
Discussion
A central conclusion of this work is that the highly conserved CTD of Band 4.1B and those of other members of the Band 4.1 family selectively interact with αvβ8 but not with other αv-containing integrins. How might Band 4.1 proteins be involved in αvβ8-mediated function? One possibility is that interaction with Band 4.1 may affect the activation state of αvβ8, thus altering the affinity or avidity for extracellular matrix (ECM) ligands. This function would be similar to the role of talin during inside-out activation of other integrins. Using transient cotransfection analyses, we have not detected significant contributions of full length Band 4.1B or isolated CTD or FERM domains on ECM adhesion. However, it remains to be determined whether Band 4.1B may play subtle roles in regulating the activation state of αvβ8. Second, Band 4.1 interaction with αvβ8 may also be involved in outside-in integrin signaling. Indeed, we show that astrocyte adhesion to LAP-TGFβ1 leads to localization of αvβ8 and Band 4.1B at adhesion sites. Thus, αvβ8 adhesion appears to induce an association with Band 4.1 proteins, which may in turn affect other signaling cascades. Thus, Band 4.1 proteins may serve as a signaling nexus for αvβ8 integrin and probably other cell surface and intracellular proteins.
The functional importance of the Band 4.1 C terminus is highlighted by studies of coracle, the Drosophila Band 4.1 protein. Coracle plays an important role in the formation of epithelial septate junctions (22, 23). Many coracle mutant alleles encode truncated proteins that contain the N-terminal FERM domain but lack large portions of the C terminus, including the CTD. Because the CTDs of coracle and vertebrate Band 4.1 proteins are ≈60% identical, it is interesting to speculate that Band 4.1B may be playing a similar role in the regulation of vertebrate cell-cell or cell-extracellular matrix interactions, likely via interactions with αvβ8 integrin.
Other experimental data support functional links between αvβ8 integrin and members of the Band 4.1 superfamily. For example, the human Band 4.1B gene product is down-regulated in some lung adenocarcinomas (24). Interestingly, αvβ8 integrin protein expression is also reduced in some epithelial-derived human lung tumors (25), suggesting that dysregulation of signaling pathways involving Band 4.1B and αvβ8 may be involved in the progression of lung epithelial tumorigenesis. Last, we have recently generated a mutant mouse model where the αv integrin gene is selectively ablated in the CNS (9). These mice develop neonatal cerebral hemorrhage, leading to spinocerebellar axonal degeneration and a variety of neurological phenotypes. The neurological deficits observed in the αv conditional knockouts are strikingly similar to symptoms observed in human patients with the hereditary disease cerebral cavernous malformation 1 (CCM1). The causative gene associated with CCM1 onset encodes the FERM domain-containing protein Krit1 (26). It will be interesting to determine whether signaling events involving αvβ8 integrin, Band 4.1 proteins, and Krit1 are involved in normal cerebrovascular development, and how dysregulation of these events may lead to the pathogenesis of CCM1.
Supplementary Material
Acknowledgments
We thank Frank Solomon and Adam Lacy-Hulbert for critical reading of the manuscript. We thank Denise Crowley for histology assistance and Drs. Joel-Anne Chassis and John Conboy (Lawrence Berkeley National Laboratory, Berkeley, CA) for providing goat anti-Band 4.1B antiserum. Stephen Nishimura (University of California, San Francisco) provided the human β8 integrin cDNA, and Takahiro Nagase (Kazusa DNA Research Institute, Chiba, Japan) provided the human full-length Band 4.1B cDNA. This work was supported by a grant from the National Heart, Lung, and Blood Institute, National Institutes of Health (P01-HL66105) and by the Howard Hughes Medical Institute. R.O.H. is an Investigator and J.H.M. was an Associate of the Howard Hughes Medical Institute.
Abbreviations: β8cyto, β8 integrin cytoplasmic domain; CTD, C-terminal domain; FERM, 4.1/ezrin/radixin/moesin; LAP, latency-associated peptide; Y2H, yeast two-hybrid.
References
- 1.Thiery, J. P. (2003) Curr. Opin. Genet. Dev. 13, 365-371. [DOI] [PubMed] [Google Scholar]
- 2.Ross, M. E. & Walsh, C. A. (2001) Annu. Rev. Neurosci. 24, 1041-1070. [DOI] [PubMed] [Google Scholar]
- 3.Huber, A. B., Kolodkin, A. L., Ginty, D. D. & Cloutier, J. F. (2003) Annu. Rev. Neurosci. 26, 509-563. [DOI] [PubMed] [Google Scholar]
- 4.Hynes, R. O. (2002) Cell 110, 673-687. [DOI] [PubMed] [Google Scholar]
- 5.Hoover, K. B. & Bryant, P. J. (2000) Curr. Opin. Cell Biol. 12, 229-234. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher, A., Edwards, K. & Fehon, R. G. (2002) Nat. Rev. Mol. Cell. Biol. 3, 586-599. [DOI] [PubMed] [Google Scholar]
- 7.McCarty, J. H., Monahan-Earley, R. A., Brown, L. F., Keller, M., Gerhardt, H., Rubin, K., Shani, M., Dvorak, H. F., Wolburg, H., Bader, B. L., et al. (2002) Mol. Cell. Biol. 22, 7667-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, J., Motejlek, K., Wang, D., Zang, K., Schmidt, A. & Reichardt, L. F. (2002) Development (Cambridge, U.K.) 129, 2891-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty, J. H., Lacy-Hulbert, A., Charest, A., Bronson, R. T., Crowley, D., Housman, D., Savill, J., Roes, J. & Hynes, R. O. (2005) Development (Cambridge, U.K.) 132, 165-176. [DOI] [PubMed] [Google Scholar]
- 10.Vinogradova, O., Velyvis, A., Velyviene, A., Hu, B., Haas, T., Plow, E. & Qin, J. (2002) Cell 110, 587-597. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood, D. A., Yan, B., de Pereda, J. M., Alvarez, B. G., Fujioka, Y., Liddington, R. C. & Ginsberg, M. H. (2002) J. Biol. Chem. 277, 21749-21758. [DOI] [PubMed] [Google Scholar]
- 12.Parra, M., Gascard, P., Walensky, L. D., Gimm, J. A., Blackshaw, S., Chan, N., Takakuwa, Y., Berger, T., Lee, G., Chasis, J. A., et al. (2000) J. Biol. Chem. 275, 3247-3255. [DOI] [PubMed] [Google Scholar]
- 13.Yamakawa, H., Ohara, R., Nakajima, D., Nakayama, M. & Ohara, O. (1999) Brain Res. Mol. Brain Res. 70, 197-209. [DOI] [PubMed] [Google Scholar]
- 14.Scott, C., Phillips, G. W. & Baines, A. J. (2001) Eur. J. Biochem. 268, 3709-3717. [DOI] [PubMed] [Google Scholar]
- 15.Gimm, J. A., An, X., Nunomura, W. & Mohandas, N. (2002) Biochemistry 41, 7275-7282. [DOI] [PubMed] [Google Scholar]
- 16.Fehon, R. G., Dawson, I. A. & Artavanis-Tsakonas, S. (1994) Development (Cambridge, U.K.) 120, 545-557. [DOI] [PubMed] [Google Scholar]
- 17.Binda, A. V., Kabbani, N., Lin, R. & Levenson, R. (2002) Mol. Pharmacol. 62, 507-513. [DOI] [PubMed] [Google Scholar]
- 18.Shen, L., Liang, F., Walensky, L. D. & Huganir, R. L. (2000) J. Neurosci. 20, 7932-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, D., Yan, H., Othman, T., Turner, C. P., Woolf, T. & Rivkees, S. A. (2004) Biochem. J. 377, 51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milner, R., Huang, X., Wu, J., Nishimura, S., Pytela, R., Sheppard, D., ffrench-Constant, C. (1999) J. Cell Sci. 112, 4271-4279. [DOI] [PubMed] [Google Scholar]
- 21.Cambier, S., Gline, S., Mu, D., Collins, R., Araya, J., Dolganov, G., Einheber, S., Boudreau, N. & Nishimura, S. L. (2005) Am. J. Pathol. 166, 1883-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgartner, S., Littleton, J. T., Broadie, K., Bhat, M. A., Harbecke, R., Lengyel, J. A., Chiquet-Ehrismann, R., Prokop, A. & Bellen, H. J. (1996) Cell 87, 1059-1068. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, R. S., Ward, R. E., Schweizer, L. & Fehon, R. G. (1998) Mol. Biol. Cell 9, 3505-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran, Y. K., Bogler, O., Gorse, K. M., Wieland, I., Green, M. R. & Newsham, I. F. (1999) Cancer Res. 59, 35-43. [PubMed] [Google Scholar]
- 25.Cambier, S., Mu, D. Z., O'Connell, D., Boylen, K., Travis, W., Liu, W. H., Broaddus, V. C. & Nishimura, S. L. (2000) Cancer Res. 60, 7084-7093. [PubMed] [Google Scholar]
- 26.Marchuk, D. A., Srinivasan, S., Squire, T. L. & Zawistowski, J. S. (2003) Hum. Mol. Genet. 12 Spec. No. 1, R97-112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.