Abstract
Hereditary coproporphyria is an autosomal dominant disorder resulting from the half-normal activity of coproporphyrinogen oxidase (CPO), a mitochondrial enzyme catalyzing the antepenultimate step in heme biosynthesis. The mechanism by which CPO catalyzes oxidative decarboxylation, in an extraordinary metal- and cofactor-independent manner, is poorly understood. Here, we report the crystal structure of human CPO at 1.58-Å resolution. The structure reveals a previously uncharacterized tertiary topology comprising an unusually flat seven-stranded β-sheet sandwiched by α-helices. In the biologically active dimer (KD = 5 × 10-7 M), one monomer rotates relative to the second by ≈40° to create an intersubunit interface in close proximity to two independent enzymatic sites. The unexpected finding of citrate at the active site allows us to assign Ser-244, His-258, Asn-260, Arg-262, Asp-282, and Arg-332 as residues mediating substrate recognition and decarboxylation. We favor a mechanism in which oxygen serves as the immediate electron acceptor, and a substrate radical or a carbanion with substantial radical character participates in catalysis. Although several mutations in the CPO gene have been described, the molecular basis for how these alterations diminish enzyme activity is unknown. We show that deletion of residues (392-418) encoded by exon six disrupts dimerization. Conversely, harderoporphyria-causing K404E mutation precludes a type I β-turn from retaining the substrate for the second decarboxylation cycle. Together, these findings resolve several questions regarding CPO catalysis and provide insights into hereditary coproporphyria.
Keywords: coproporphyrinogen oxidase, oxidative decarboxylation, mitochondria, x-ray crystallography
The terminal three steps of heme biosynthesis occur within the mitochondria (1, 2). First, coproporphyrinogen III is converted to protoporphyrinogen IX in the intermembrane space (3, 4) by coproporphyrinogen oxidase (CPO) (5, 6). Thus, CPO contains an unusually long (110 residues) N-terminal targeting sequence, required for its import into the mitochondria (7, 8). The substrate for CPO is generated in the cytosol (9) by uroporphyrinogen decarboxylase, and the precise mechanism by which it enters the mitochondria remains to be elucidated. Second, protoporphyrinogen oxidase mediates the six electron oxidation of protoporphyrinogen to protoporphyrin IX. This enzyme is localized to the cytoplasmic side of the inner mitochondrial membrane. Third, ferrochelatase inserts the ferrous iron to generate heme within the matrix of the mitochondria. Hence, the heme biosynthetic pathway is not only partitioned between mitochondria and cytosol, but the last three enzymes are compartmentalized within the mitochondria.
Partial deficiency of CPO leads to hereditary coproporphyria (HCP), an acute hepatic porphyria inherited in an autosomal dominant fashion (10-12). The disease is characterized by abdominal pain, neuropsychiatric symptoms, and/or cutaneous photosensitivity (13). If diagnosed early, HCP can be treated with a high carbohydrate diet and i.v. administration of heme in the form of heme arginate (14). In the majority of heterozygous HCP patients, CPO activity is reduced to ≈50% (15-17), resulting in the excretion of coproporphyrin in urine and stool. In rare homozygous cases, enzyme activity decreases to <10% (18-20). Other factors, including drugs, alcohol, stress, or infection, can precipitate HCP in susceptible individuals (21). Since the cloning of human CPO gene (22, 23), several mutations that diminish enzyme activity have been identified (24).
CPO is an extraordinary enzyme of particular interest to chemistry and medicine. As a homodimer (25), it catalyzes the oxidative decarboxylation of propionic acid side chains of rings A and B of coproporphyrinogen III (refs. 26-29 and Fig. 1A) without using metals (25, 27, 35), reducing agents, thiols, prosthetic groups, organic cofactors, or modified amino acids (36). Whereas the stereochemistry of this reaction has been worked out (31, 32), the molecular oxygen consumption presents an interesting mechanistic puzzle. From a clinical standpoint, a well defined correlation between the genotype and severity of disease is lacking (34). Thus, unlike other hepatic porphyrias, heterozygotes carrying mutations known to cause HCP in homozygotes have the propensity to develop disease.
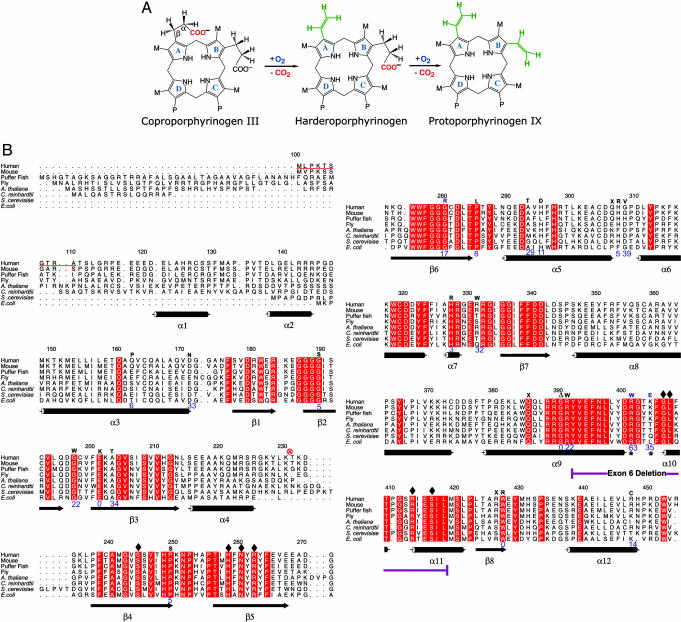
Fig. 1.
CPO chemistry and sequence conservation. (A) Reaction catalyzed by CPO involves both oxidation and decarboxylation (5). CPO sequentially decarboxylates (26, 30) the propionates attached to A and B rings without affecting those on C and D rings. A hydrogen atom from the β-position of the propionate side chain also is removed at each step (31, 32). The chemical identity of the oxidation end product(s) remains to be elucidated. M = CH3 and P = CH2.CH2.COO-.(B) Sequence alignment, secondary structure, and location of HCP-causing mutations in human CPO. The first 110 amino acids are absent in the mature enzyme, for they are part of a mitochondrial targeting signal that is cleaved upon import. In the alignment (generated by using amps and alscript) red represents absolute identity over all sequences present in that part of the alignment. Database of Secondary Structure of Proteins-derived (33) secondary structural assignments are shown directly below the alignment with cylinders indicating α-helices and arrows denoting β-strands. Mutations known to cause HCP are indicated by one letter codes above the human sequence. The enzymatic activity of these variants (24, 34) are shown in blue (% relative to native enzyme). An asterisk denotes residues that affect the second decarboxylation step. Residues that make contact with citrate are indicated by diamonds. ⊗, proteolytic cleavage site.
Here, we describe the high-resolution crystal structure of human CPO and provide molecular mechanisms to explain how disease-causing mutations suppress enzyme activity. We also offer a detailed picture of the active site residues and elaborate on catalytic mechanism(s).
Materials and Methods
Protein Preparation, Crystallization, and Structure Determination. Human CPO was expressed and purified as described (25). Several attempts to crystallize this protein failed because of time-dependent proteolytic cleavage (Fig. 1B). Therefore, we devised a cross-seeding strategy to obtain diffraction-quality crystals, details of which are described in Supporting Text, which is published as supporting information on the PNAS web site. The structure was determined by experimental phasing using selenomethionine-substituted crystals. The crystallographic data statistics are shown in Table 1, which is published as supporting information on the PNAS web site.
Equilibrium Analytical Ultracentrifugation. One hundred twenty microliters of wild-type human CPO was sedimented to equilibrium at two different loading concentrations (A280 of 0.32 and 0.4), five different speeds (18.0, 22.1, 23.4, 26.2, and 28.0 krpm), and at 4°C in a double-sector, epon-filled centerpieces by using an AN60 TI rotor in a Beckman Optima XL-A analytical ultracentrifuge. Scans were taken at 280 nm, once equilibrium was established, by scanning with 20 averages at a 0.001-cm radial step size setting. A detailed account of the methods used in data analysis is provided in Supporting Text.
Results and Discussion
Quality of the Crystal Structure. The electron density for the CPO structure is continuous and well defined. The real space correlation coefficient (37) is excellent, and there are no residues in disallowed regions of the Ramachandran plot. An example electron-density map is shown in Fig. 2A. Some indicators of model quality are detailed in Table 1.
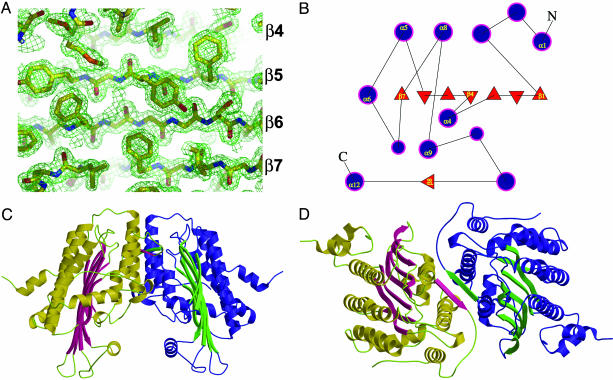
Fig. 2.
Structure of human CPO. (A)2Fo - Fc electron density map (contoured at 1.5 σ) at 1.58 Å with a final model in place. The identity of the β-strands are shown on the right. (B) Topology diagram illustrating the organization of secondary structural elements in human CPO. Filled circles and triangles represent α-helices and β-strands, respectively. (C) Tertiary topology and quaternary structure. (D) Dimer interface.
The Protein Fold. CPO assumes a previously unknown tertiary topology characterized by a large seven-stranded β-sheet that is flanked on both sides by α-helices (Fig. 2B). The up-and-down β-strands are similar to porins, but the β-sheet in CPO is flat (Fig. 2C) and does not form a barrel. The T fold, which was predicted to share structural similarities with CPO (38), also has some resemblance to porins because of the long β-strands, but the β-sheet in the T fold is curved as in porin. Moreover, the T fold has helices on one side of the sheet, but these helices are in a long collapsed hairpin β-α-α-β motif, whereas the connectivity is completely different in CPO.
The flatness of the up-and-down β-sheet in CPO is striking (Fig. 2C). In contrast, seven-stranded β-sheet-containing enzymes usually contain a twisted (TauD; ref. 39) or highly curved (thiol ester dehydrase; ref. 40) β-sheet whose convex or the apolar concave side, respectively, is flanked by helices. Furthermore, in the dehydrase, eight β-bulges contribute effectively toward introducing strong curvature into the sheet. There are only three β-bulges in CPO. The flatness of the CPO sheet is very likely enabled by the abundance of Gly residues found within the β-strands (β2, β3, β4, β6, and β7). In this regard, Richardson and Richardson (41) have noted that high glycine content helps allow either very high or very low curl or twist of β-sheets.
CPO Functions as a Homodimer. The dimensions of the dimer are ≈80 × 60 × 60 Å. The two subunits of the CPO homodimer are related by an ≈40° rotation of one monomer relative to another (Fig. 2D). This rotation is a hinge-like motion about the crystallographic 2-fold axis located roughly parallel to the β-sheet. A dimeric protein the size of CPO (Mr 78,000) is expected to have an accessible surface area (ASA) of ≈28,000 Å2 on the basis of a survey of water-soluble oligomeric proteins (42). Consistent with this prediction, calculations with a 1.4-Å probe reveal that the CPO dimer has an ASA of 28,800 Å2. Roughly 1,300 Å2 per subunit are buried in the dimer interface that is relatively flat (planarity, rms = 2.4 Å) and circular-shaped (length/breadth ratio = 0.92). Twenty-two residues (70% evolutionarily conserved) that comprise this interface hail from five different segments of the polypeptide chain (see Fig. 6, which is published as supporting information on the PNAS web site). Interestingly, β8 is not part of the flat β-sheet, but instead pairs up in an antiparallel fashion, with the corresponding β-strand from the second subunit to generate key contacts at the dimer interface (Fig. 2D). Overall, the interface is made up of 64% nonpolar atoms and 36% polar atoms. Ten intersubunit H bonds also contribute to the dimer stability (Fig. 6). Salt bridges and water-bridged interactions are absent. Taken together, the interacting surface on the CPO monomer is a hydrophobic patch. All of the parameters we have used to describe the CPO interface are in excellent agreement with those found in other homodimers (43, 44). Thus, it is extremely unlikely that CPO will function as a monomer. To quantitatively assess the solution stability of CPO, we have performed analytical ultracentrifugation. The CPO equilibrium distribution fit well to a monomer-dimer-tetramer equilibrium (see Fig. 7 and Table 2, which are published as supporting information on the PNAS web site) with a dissociation constant of 0.5 μM (see Supporting Text)at277K(ΔGdiss of ≈8 kcal·mol-1). Consistent with this finding, yeast (45), human (25), and Escherichia coli (46) proteins are all dimers in solution. Thus, we conclude that the homodimer is the biologically relevant form of CPO.
CPO Structure Lacks a Transition Metal Center. Although it has been established that human CPO is not a metalloprotein (25, 27, 35), a recent work suggests that Mn2+ ion, coordinated tetrahedrally by His residues, participates in catalysis (47). We have screened for metal ions bound to CPO by recording anomalous dispersion effects (48). We subjected both human and bacterial CPO crystals to fluorescence energy scans at the x-ray absorption edge of Cu, Fe, Mn, and Zn. Furthermore, we have grown CPO crystals in the presence of these metal salts and have collected complete anomalous data sets at the maximal f″ values of the absorption edges. Neither the energy scans nor the native anomalous difference Fourier maps provide evidence for bound transition metal ions in CPO. More importantly, the invariant His residues are far apart (14-22 Å) and, therefore, cannot serve as ligands.
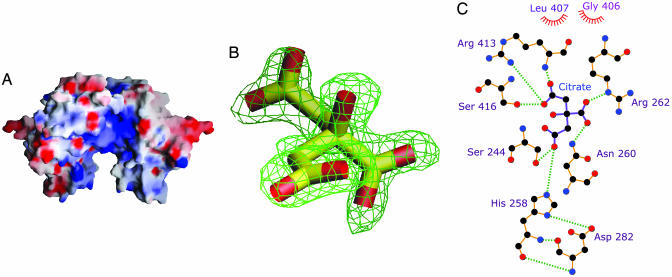
The Active Site. During structure refinement, we serendipitously discovered that an electropositive cleft (Fig. 3A) near the dimer interface had a molecule of citrate (Fig. 3 B and C; see also Fig. 8, which is published as supporting information on the PNAS web site) bound to it. This finding is reminiscent of citrate binding to Src SH2 domain, a fortuitous finding that spearheaded new drug design strategies to treat osteoporosis (51, 52). As in the SH2 case, we too benefited from the use of citrate as a crystallization additive, and it is worth noting that tricarboxylic acid is an elegant mimic of the carboxylate groups in coproporphyrinogen III. In proteins, arginine residues are prime candidates for carboxylate recognition (53), and by comparing >500 unique sequences of CPO (>350 of these sequences are from the environmental samples of the Sargasso Sea; ref. 54), we have identified that Arg-262, Arg-328, Arg-332, and Arg-389 are invariant. In addition, there are no conserved Lys residues. Arg-262 forms a key ionic interaction with citrate and Arg-332 is within striking distance. Other hydrogen-bonded (Gly-411 not shown) and ionic interactions are depicted in Fig. 3C. Thus, where coproporphyrinogen III and harderoporphyringen are concerned, we conclude that Arg-262 mediates substrate recognition. His-258 donates a proton in the form of a hydrogen bond from Nδ1 to O1 of Asp-282 and is likely to function as a diad in a manner common to that of many active-site histidines. The H bond of Nδ1 also positions the imidazole ring rather precisely. Thus, we predict that His-258 is in the correct tautomeric state and optimal orientation for catalysis in the free enzyme. Our assignment of a catalytic role for His-258 is supported by two additional observations: (i)the His-Asp diad configuration results in a basic lone pair of electrons on Nε2 of His-258, and (ii) His258Ala substitution completely abolishes the enzyme activity of mouse CPO (55). Interestingly, the type of residues interacting with citrate and the location of His-Asp/Arg-Ser pairs in CPO is similar to that found in aconitase (56). With the exception of Arg-413 and Ser-416, all of the residues that interact with citrate (Fig. 3C) are strictly conserved and reside in strands β4, β5, and β6. Together they form the decarboxylation corridor, the active site region in charge of substrate recognition and catalysis (Fig. 9, which is published as supporting information on the PNAS web site). The invariant Gly-406 and Leu-407, located in close proximity to a region that affects the second step of CPO catalysis (vide infra), also make nonbonded contacts with citrate. This part of the active site plays an important role in properly orienting the substrate.
Fig. 3.
Active site of human CPO. (A) Electrostatic potential mapped on to the molecular surface. The electropositive active site cleft is readily visible. The blue and red contours represent positive and negative potential (full saturation = 10 kT), respectively (figure was generated by using grasp; ref. 49). (B) Fo - Fc omit electron density of citrate bound at the active site. (C) Schematic illustration of the amino acid residues that make direct contact with the bound citrate. Dashed lines indicate H bonds, and nonbonded contacts are represented by an arc with spokes radiating toward the ligand atoms (figure was generated by using ligplot; ref. 50).
Catalytic Mechanism. CPO catalyzes an unusual metal- and cofactor-independent oxidative decarboxylation. It is well established that CPO abstracts the pro-S hydrogen from the methylene group adjacent to the pyrrole ring (Fig. 1A), leading to the generation of a vinyl group from the remaining three hydrogens and two carbons without rearrangement (31, 32). Such strong stereoselectivity indicates that CPO strictly constrains the orientation of the substrate in the active site, and our structure provides insights into how this result can be achieved (vide supra). However, the precise mechanism for hydrogen abstraction is unknown. Similarities between CPO and urate oxidase have been invoked (38), but the substrate for the latter resembles flavin (57) and, therefore, a well-known redox chemistry is used in catalysis. Because hydride transfer is not an option for CPO, a tyrosyl radical has been proposed in catalysis (36), but substituting conserved Tyr residues (322 and 392) did not affect the activity of the E. coli enzyme (47).
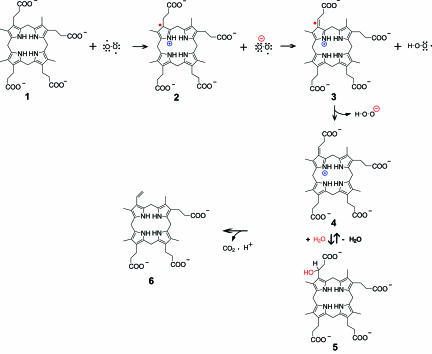
We believe that CPO fits in well with the family of metal- and cofactor-free oxygenases, overlooked by other workers in this field, that capitalize on substrate reactivity to generate radical and/or cabanionic intermediates that react rapidly with molecular oxygen (58). Therefore, we favor a mechanism in which molecular oxygen serves as the immediate electron acceptor (D. Arigoni, personal communication) (Fig. 4). Here, the reaction is likely to proceed in two steps, each of which involve a single electron transfer or radical recombination. In the initial step, oxygen diradical removes an electron from the pyrrole nitrogen, resulting in the generation of a substrate radical cation, 2. Next, superoxide anion reacts with 2 and abstracts the Cβ-H. The “2-center 3-electron” bond (59) shown in 3 provides a rationale for the increased acidity of Cβ-H. A second electron transfer step ensues, resulting in production of the azafulvene cation 4 and H2O2. Now, the decarboxylation can proceed readily in light of the electron sink afforded by the positively charged pyrrole ring of 4. Precedents for this process can be found in pyridoxal-dependent decarboxylases. However, to accommodate Sano's β-hydroxypropionic acid intermediate (60), a Michael addition step 5, analogous to that of enoyl-CoA hydratase (61), is included. We propose that His-258 is localized next to the pyrrole ring A to assist with the formation of substrate cation.
Fig. 4.
Catalytic mechanism. Only the first decarboxylation step is shown; the second step is expected to occur in exactly the same manner after the tricarboxylate intermediate undergoes a 90° counterclockwise rotation at the active site (ref. 26 and D. Arigoni, personal communication).
In our alternate mechanism, a strong base abstracts the Cβ-H proton of coproporphyrinogen III to generate a carbanion intermediate that could react directly with molecular oxygen, through sequential single electron steps, to generate superoxide and a carbon-centered radical. This species would then recombine to yield the hydroperoxide adduct (Cβ-OOH) and can be eliminated with the loss of CO2. His-258 or Ser-244 (as the alkoxide) can serve as a base. Indeed, there are precedents for Ser/Thr functioning as a base in aconitase (56) and urate oxidase (62). It can be argued that there is no sizeable acidity to Cβ-H (pKa ≈ 35), making proton removal from this site difficult. However, precedents exist for deprotonating an unactivated carbon (63) and also for stabilizing high-energy anionic intermediates through nondelocalization mechanisms (64). The ability of O2 to accept electrons from a singlet carbanion is questionable, for it would result in a high-energy triplet excited state (58). In this regard, we would like to draw a corollary between the oxygenase activities of carbanion-forming enzymes and CPO (65, 66). Spin inversion required for the reaction between a triplet oxygen and a singlet substrate can be facilitated by the formation of a caged radical pair or by activation of the carbanion to the triplet state through geometric distortion (67). In light of their interrupted conjugation, porphyrinogens are well known for adopting an 1,3-alternate conformation characterized by substantial nonplanarity. Furthermore, based on the studies with calixpyrroles (68), anion coordination by the pyrrole nitrogens will generate a cone conformation for the macrocycle. Analogously, H bond interactions between the pyrrole NH groups and a protein carboxylate (69) can provide the geometric distortion required to impart triplet character to a carbanion. We posit that the invariant Asp-400 in CPO is uniquely positioned to help introduce such distortion. Both the free-radical and carbanion mechanisms should yield one equivalent of H2O2 for each decarboxylation step. Although CO2 production has been quantified (30), there is no literature on the stoichiometry of H2O2 generation during CPO catalysis.
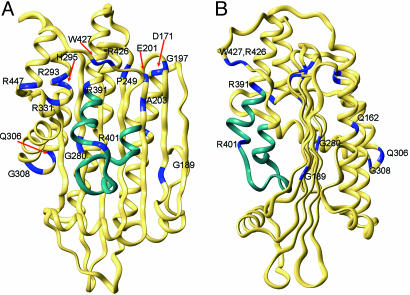
Structural Basis of Disease. There are >20 naturally occurring HCP mutations (Fig. 1B and refs. 24 and 34) and a majority of these mutations lead to substitution of amino acid residues within the structural framework of CPO (Fig. 5). For example, Gln162Pro will disrupt helix α3, and Gly189Ser is expected to perturb strand β2. However, we have found that several mutations distant from the active site can generate dimeric CPO, albeit with little to no activity. Therefore, we will confine our discussion to those mutations for which meaningful insights can be provided solely by inspecting the structure of the native enzyme. First, deletion of the region encoded by exon six, comprising residues 392-418, has been reported in a heterozygous patient, and the resulting protein will be unable to dimerize (Fig. 6). Indeed, we have confirmed this prediction by expressing the variant in E. coli. Trp-427, located on strand β8, makes intersubunit interactions, and, therefore, W427R mutation will also affect dimerization. Second, H327R and R328C will perturb the interaction between helix α7 and the dimerization helix α9 (Fig. 10, which is published as supporting information on the PNAS web site). Third, R331W variant retains sufficient activity to support life in a homozygous setting but can also produce HCP in a heterozygote. Twelve different amino acids are tolerated at position 331 but aromatic residues are not. R331W substitution will abolish the hydrogen bonds between the guanidinium group and the carbonyl of Leu-446 and Arg-447. Interestingly, R447C mutation also results in diminished activity. Finally, K404E causes harderoporphyria, a disease with symptoms unrelated to HCP (34, 70-73). This mutation affects the region that separates helices α9 and α10. Lys-404 is not conserved, and the favored residue at this position is Leu. In the human CPO structure, K404 is part of a type I β-turn. Whereas a positively charged residue at this position is not essential for the second decarboxylation step, introducing a negative charge will produce electrostatic repulsion (or steric hindrance), and the enzyme will lose its ability to hold on to harderoporphyrinogen. From a catalytic standpoint, three key observations illustrate that CPO can function without Lys-404: (i) Individuals who are homozygous for K404E do not suffer from acute attacks diagnostic of HCP, (ii) harderoporphyrinogen is a good substrate for CPO, and (iii) Lys-404 is not evolutionarily conserved. Equally interesting is the R401W mutation that causes atypical HCP in a heterozygous setting. Heterologous expression reveals that this variant, like its K404E counterpart, is defective in the second decarboxylation step. Arg-401 is highly conserved in nearly all CPOs with the exception of Cytophaga hutchinsoni, in which a Lys takes its place. Given the strict conservation of a positive charge at this position, the enzyme most likely prevents the release of the intermediate by an ionic interaction with a propionate group that does not undergo decarboxylation (ring C or D). Thus, the primary role for the 401-405 segment is to deter the premature release of intermediate. It does not participate in substrate recognition or decarboxylation. In sum, our work has revealed the identity of active site residues in CPO and has provided previously uncharacterized insights to understand HCP at the molecular level.
Fig. 5.
Residues and structural regions affected by HCP mutations are shown with two different views of the structure. The polypeptide encoded by exon six is shown in cyan. Regions in blue represent mutation sites.
Note. While this manuscript was under review, the crystal structure of yeast CPO was reported (74). Consistent with the independent crystallographic results reported here for human CPO, the yeast enzyme crystallized as a dimer and has a similar tertiary topology.
Supplementary Material
Acknowledgments
We thank Gerry McDermott (Advanced Light Source, BL 5.0.2) for help with multiwavelength anomalous diffraction data collection; Pierre Nioche for performing the final refinement steps; Sudha Veeraraghavan and Pierre Nioche for figures; Gerard Bricogne for providing buster and Clemens Vonrhein for assistance with using the program; and the Stanford Synchrotron Radiation Laboratories and the Advanced Light Source for generous access to beam time. C.S.R. acknowledges stimulating discussions with Vernon Anderson, Duilio Arigoni, Liisa Holm, Vladimir Kral, Huiying Li, Jane Richardson, Seiyo Sano, John Schloss, Peter Shoolingin-Jordan, Peter Tipton, Sudha Veeraraghavan, and Christopher Walsh and thanks Suzanne Fetzner and Christopher Walsh for sharing their results before publication. This work is supported by the Pew Charitable Trusts through a Pew Scholar Award (to C.S.R.), Robert A. Welch Foundation Grant AU-1574 (to C.S.R.), and Ministry of Education, Youth, and Sports of the Czech Republic Grant 1M 6837805002 (to E.F., M.B., and P.M.). The development of the ultrascan software is supported by National Science Foundation Grant DBI-9974819 (to B.D.).
Author contributions: D.-S.L. and C.S.R. designed research; D.-S.L., B.D., and C.S.R. performed research; D.-S.L., E.F., and P.M. contributed new reagents/analytic tools; D.-S.L., B.D., and C.S.R. analyzed data; and C.S.R. wrote the paper.
This work was presented at the Gordon Conference on Chemistry and Biology of Tetrapyrroles, July 25-30, 2004, Salve Regina University, Newport, RI, and also in part at the Porphyrins and Porphyrias Meeting, September 21-24, 2003, Prague, Czech Republic [Mikula, I., Bodnárová, M., Lee, D.-S., Flachsová, E., Rosipal, R., Zeman, J., Moroz, L., Raman, C. S. & Martásek, P. (2003) Physiol. Res. 52, 18S (abstr.)].
Abbreviations: HCP, hereditary coproporphyria; CPO, coproporphyrinogen oxidase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2AEX).
References
- 1.Sano, S., Inoue, S., Tanabe, Y., Sumiya, C. & Koike, S. (1959) Science 129, 275-276. [DOI] [PubMed] [Google Scholar]
- 2.Dailey, H. A. (2002) Biochem. Soc. Trans. 30, 590-595. [DOI] [PubMed] [Google Scholar]
- 3.Elder, G. H. & Evans, J. O. (1978) Biochem. J. 172, 345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grandchamp, B., Phung, N. & Nordmann, Y. (1978) Biochem. J. 176, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sano, S. & Granick, S. (1961) J. Biol. Chem. 236, 100-107. [PubMed] [Google Scholar]
- 6.Battle, A. M., Del, C., Benson, A. & Rimington, C. (1965) Biochem. J. 97, 731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delfau-Larue, M.-H., Martasek, P. & Grandchamp, B. (1994) Hum. Mol. Genet. 3, 1325-1330. [DOI] [PubMed] [Google Scholar]
- 8.Susa, S., Daimon, M., Ono, H., Li, S., Yoshida, T. & Kato, T. (2002) Tohoku J. Exp. Med. 55, 346-353. [DOI] [PubMed] [Google Scholar]
- 9.Shoolingin-Jordan, P. M. (2003) in The Porphyrin Handbook, eds. Kadish, K. M., Smith, K. M. & Guilard, R. (Elsevier Science, New York), Vol. 12, pp. 33-74. [Google Scholar]
- 10.Berger, H. & Goldberg, A. (1955) Br. Med. J. 2, 85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martasek, P. (1998) Semin. Liver Dis. 18, 25-32. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann, Y. & Puy, H. (2002) Clin. Chim. Acta 325, 17-37. [DOI] [PubMed] [Google Scholar]
- 13.Elder, G. H., Hift, R. J. & Meissner, P. N. (1997) Lancet 349, 1613-1617. [DOI] [PubMed] [Google Scholar]
- 14.Tenhunen, R., Tokola, O. & Linden, I. B. (1987) J. Pharm. Pharmacol. 39, 780-786. [DOI] [PubMed] [Google Scholar]
- 15.Elder, G. H., Evans, J. O., Thomas, N., Cox, R., Brodie, M. J., Moore, M. R., Goldberg, A. & Nicholson, D. C. (1976) Lancet 308, 1217-1219. [Google Scholar]
- 16.Nordmann, Y., Grandchamp, B., Phung, N., de Verneuil, H., Grelier, M. & Noire, J. (1977) Lancet 309, 140. [DOI] [PubMed] [Google Scholar]
- 17.Grandchamp, B. & Nordmann, Y. (1977) Biochem. Biophys. Res. Commun. 74, 1089-1095. [DOI] [PubMed] [Google Scholar]
- 18.Grandchamp, B., Phung, N. & Nordmann, Y. (1977) Lancet 310, 1348-1349. [DOI] [PubMed] [Google Scholar]
- 19.Martasek, P., Nordmann, Y. & Grandchamp, B. (1994) Hum. Mol. Genet. 3, 477-480. [DOI] [PubMed] [Google Scholar]
- 20.Kuhnel, A., Gross, U. & Doss, M. O. (2000) Clin. Biochem. 33, 465-473. [DOI] [PubMed] [Google Scholar]
- 21.Anderson, K. E., Sassa, S., Bishop, D. F. & Desnick, R. J. (1999) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw-Hill, New York), Vol. II, pp. 2961-3062. [Google Scholar]
- 22.Martasek, P., Camadro, J. M., Delfau-Larue, M.-H., Dumas, J.-B., Montagne, J. J., de Verneuil, H., Labbe, P. & Grandchamp, B. (1994) Proc. Natl. Acad. Sci. 91, 3024-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taketani, S., Kohno, H., Furukawa, T., Yoshinaga, T. & Tokunaga, R. (1994) Biochim. Biophys. Acta 1183, 547-549. [DOI] [PubMed] [Google Scholar]
- 24.Rosipal, R., Lamoril, J., Puy, H., Da Silva, V., Gouya, L., De Rooij, F. W. M., Te Velde, K., Nordmann, Y., Martasek, P. & Deybach, J. C. (1999) Hum. Mutat. 13, 44-53. [DOI] [PubMed] [Google Scholar]
- 25.Martasek, P., Camadro, J.-M., Raman, C. S., Lecomte, M. C., Le Caer, J. P., Demeler, B., Grandchamp, B. & Labbe, P. (1997) Cell. Mol. Biol. (Noisy-le-Grand) 43, 47-58. [PubMed] [Google Scholar]
- 26.Elder, G. H., Evans, J. O., Jackson, J. R. & Jackson, A. H. (1978) Biochem. J. 169, 215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshinaga, T. & Sano, S. (1980) J. Biol. Chem. 255, 4722-4726. [PubMed] [Google Scholar]
- 28.Lash, T. D., Mani, U. N., Drinan, M. A., Zhen, C., Hall, T. & Jones, M. A. (1999) J. Org. Chem. 64, 464-477. [Google Scholar]
- 29.Akhtar, M. (2003) in The Porphyrin Handbook, eds. Kadish, K. M., Smith, K. M. & Guilard, R. (Elsevier Science, New York), Vol. 12, pp. 69-86. [Google Scholar]
- 30.Elder, G. H. & Evans, J. O. (1978) Biochem. J. 169, 205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaman, Z., Abboud, M. M. & Akhtar, M. (1972) J. Chem. Soc. Chem. Commun. 1263-1264.
- 32.Battersby, A. R., Baldas, J., Collins, J., Grayson, D. H., James, K. J. & McDonald, E. (1974) J. Chem. Soc. Chem. Commun. 1265-1266.
- 33.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577-2637. [DOI] [PubMed] [Google Scholar]
- 34.Lamoril, J., Puy, H., Whatley, S. D., Martin, C., Woolf, J. R., Da Silva, V., Deybach, J. C. & Elder, G. H. (2001) Am. J. Hum. Genet. 68, 1130-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medlock, A. E. & Dailey, H. A. (1996) J. Biol. Chem. 271, 15765-15770. [DOI] [PubMed] [Google Scholar]
- 36.Yoshinaga, T. & Sano, S. (1980) J. Biol. Chem. 255, 4727-4731. [PubMed] [Google Scholar]
- 37.Reddy, V., Swanson, S. M., Segelke, B., Kantardjieff, K. A., Sacchettini, J. C. & Rupp, B. (2003) Acta Crystallogr. D 59, 2200-2210. [DOI] [PubMed] [Google Scholar]
- 38.Colloc'h, N., Mornon, J. P. & Camadro, J. M. (2002) FEBS Lett. 526, 5-10. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien, J. R., Schuller, D. J., Yang, V. S., Dillard, B. D. & Lanzilotta, W. N. (2003) Biochemistry 42, 5547-5554. [DOI] [PubMed] [Google Scholar]
- 40.Leesong, M., Henderson, B. S., Gillig, J. R., Schwab, J. M. & Smith, J. L. (1996) Structure (London) 4, 253-264. [DOI] [PubMed] [Google Scholar]
- 41.Richardson, J. S. & Richardson, D. C. (1989) in Prediction of Protein Structure and the Principles of Protein Conformation, ed. Fasman, G. D. (Plenum, New York), pp. 1-98.
- 42.Miller, S., Lesk, A. M., Janin, J. & Chothia, C. (1987) Nature 328, 834-837. [DOI] [PubMed] [Google Scholar]
- 43.Jones, S. & Thornton, J. M. (1995) Prog. Biophys. Mol. Biol. 63, 31-65. [DOI] [PubMed] [Google Scholar]
- 44.Valdar, W. S. J. & Thornton, J. M. (2001) Proteins Struct. Funct. Genet. 42, 108-124. [PubMed] [Google Scholar]
- 45.Camadro, J. M., Chambon, H., Jolles, J. & Labbe, P. (1986) Eur. J. Biochem. 156, 579-587. [DOI] [PubMed] [Google Scholar]
- 46.Macieira, S., Martins, B. M. & Huber, R. (2003) FEMS Microbiol. Lett. 226, 31-37. [DOI] [PubMed] [Google Scholar]
- 47.Breckau, D., Mahlitz, E., Sauerwald, A., Layer, G. & Jahn, D. (2003) J. Biol. Chem. 278, 46625-46631. [DOI] [PubMed] [Google Scholar]
- 48.Hendrickson, W. A., Smith, J. L. & Sheriff, S. (1985) Methods Enzymol. 115, 41-55. [DOI] [PubMed] [Google Scholar]
- 49.Nicholls, A., Sharp, K. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 50.Wallace, A. C., Laskowski, R. A. & Thorton, J. M. (1995) Protein Eng. 8, 127-134. [DOI] [PubMed] [Google Scholar]
- 51.Shakespeare, W., Yang, M., Bohacek, R., Cerasoli, F., Stebbins, K., Sundaramoorthi, R., Azimioara, M., Vu, C., Pradeepan, S., Metcalf, C., III, Haraldson, C., Merry, T., et al. (2000) Proc. Natl. Acad. Sci. 97, 9373-9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohacek, R., Dalgarno, D., Hatada, M., Jacobsen, V., Lynch, B., Macek, K., Taylor, M., Metcalf, C., Narula, S., Sawyer, T., et al. (2001) J. Med. Chem. 44, 660-663. [DOI] [PubMed] [Google Scholar]
- 53.Raman, C. S., Martasek, P. & Masters, B. S. S. (2000) in The Porphyrin Handbook, eds. Kadish, K. M., Smith, K. M. & Guilard, R. (Academic, New York), Vol. 4, pp. 293-339. [Google Scholar]
- 54.Venter, J. C., Remington, K., Heidelberg, J. F., Halper, A. L., Rusch, D., Eisen, J. A., Wu, D., Paulsen, I., Nelson, K. E., Nelson, W., Fouts, D. E., Levy, S., et al. (2004) Science 304, 66-74. [DOI] [PubMed] [Google Scholar]
- 55.Kohno, H., Furukawa, T., Tokunaga, R., Taketani, S. & Yoshinaga, T. (1996) Biochim. Biophys. Acta 1292, 156-162. [DOI] [PubMed] [Google Scholar]
- 56.Beinert, H., Kennedy, M. C. & Stout, C. D. (1996) Chem. Rev. 96, 2335-2373. [DOI] [PubMed] [Google Scholar]
- 57.Kahn, K. & Tipton, P. A. (1998) Biochemistry 37, 11651-11659. [DOI] [PubMed] [Google Scholar]
- 58.Fetzner, S. (2002) Appl. Microbiol. Biotechnol. 60, 243-257. [DOI] [PubMed] [Google Scholar]
- 59.Hiberty, P. C., Humbel, S. & Archirel, P. (1994) J. Phys. Chem. 98, 11697-11704. [Google Scholar]
- 60.Sano, S. (1966) J. Biol. Chem. 241, 5276-5283. [PubMed] [Google Scholar]
- 61.Bahnson, B. J., Anderson, V. E. & Petsko, G. A. (2002) Biochemistry 41, 2621-2629. [DOI] [PubMed] [Google Scholar]
- 62.Imhoff, R. D., Power, N. P., Borrok, M. J. & Tipton, P. A. (2003) Biochemistry 42, 4094-4100. [DOI] [PubMed] [Google Scholar]
- 63.Smith, D. M., Buckel, W. & Zipse, H. (2003) Angew. Chem. 42, 1867-1870. [DOI] [PubMed] [Google Scholar]
- 64.Begley, T. P. & Ealick, S. E. (2004) Curr. Opin. Chem. Biol. 8, 508-515. [DOI] [PubMed] [Google Scholar]
- 65.Abell, L. M. & Schloss, J. V. (1991) Biochemistry 30, 7883-7887. [DOI] [PubMed] [Google Scholar]
- 66.Chen, H., Tseng, C. C., Hubbard, B. K. & Walsh, C. T. (2001) Proc. Natl. Acad. Sci. USA 98, 14901-14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andres, J., Safont, V. S. & Tapia, O. (1993) J. Phys. Chem. 97, 7888-7894. [Google Scholar]
- 68.Sessler, J. L. & Gale, P. A. (2000) in The Porphyrin Handbook, eds. Kadish, K. M., Smith, K. M. & Guilard, R. (Academic, New York), Vol. 6, pp. 257-278. [Google Scholar]
- 69.Lovie, G. V., Brownlie, P. D., Lambert, R., Cooper, J. B., Blundell, T. L., Wood, S. P., Warren, M. J., Woodcock, S. C. & Jordan, P. M. (1992) Nature 359, 33-39. [DOI] [PubMed] [Google Scholar]
- 70.Nordmann, Y., Grandchamp, B., de Verneuil, H. & Phung, L. (1983) J. Clin. Invest. 72, 1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doss, M., von Tiepermann, R. & Kopp, W. (1984) Lancet 323, 292. [DOI] [PubMed] [Google Scholar]
- 72.Lamoril, J., Martasek, P., Deybach, J.-C., da Silva, V., Grandchamp, B. & Nordmann, Y. (1995) Human Mol. Genet. 4, 275-278. [DOI] [PubMed] [Google Scholar]
- 73.Lamoril, J., Puy, H., Gouya, L., Rosipal, R., da Silva, V., Grandchamp, B., Foint, T., Bader-Meunier, B., Dommergues, J. P., Deybach, J.-C. & Nordmann, Y. (1998) Blood 91, 1453-1457. [PubMed] [Google Scholar]
- 74.Phillips, J. D., Whitby, F. G., Warby, C. A., Labbe, P., Yang, C., Pflugrath, J. W., Ferrara, J. D., Robinson, H., Kushner, J. P. & Hill, C. P. (2004) J. Biol. Chem. 279, 38960-38968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.