Abstract
The protein defective in hereditary hemochromatosis, called HFE, is similar to MHC class I-type proteins and associates with β2-microglobulin (β2M). Its association with β2M was previously shown to be necessary for its stability, normal intracellular processing, and cell surface expression in transfected COS cells. Here we use stably transfected Chinese hamster ovary cell lines expressing both HFE and β2M or HFE alone to study the effects of β2M on the stability and maturation of the HFE protein and on the role of HFE in transferrin receptor 1 (TfR1)-mediated iron uptake. In agreement with prior studies on other cell lines, we found that overexpression of HFE, without overexpressing β2M, resulted in a decrease in TfR1dependent iron uptake and in lower iron levels in the cells, as evidenced by ferritin and TfR1 levels measured at steady state. However, overexpression of both HFE and β2M had the reverse effect and resulted in an increase in TfR1-dependent iron uptake and increased iron levels in the cells. The HFE-β2M complex did not affect the affinity of TfR1 for transferrin or the internalization rate of transferrin-bound TfR1. Instead, HFE-β2M enhanced the rate of recycling of TfR1 and resulted in an increase in the steady-state level of TfR1 at the cell surface of stably transfected cells. We propose that Chinese hamster ovary cells provide a model to explain the effect of the HFE-β2M complex in duodenal crypt cells, where the HFE-β2M complex appears to facilitate the uptake of transferrin-bound iron to sense the level of body iron stores. Impairment of this process in duodenal crypt cells leads them to be iron poor and to signal the differentiating enterocytes to take up iron excessively after they mature into villus cells in the duodenum of hereditary hemochromatosis patients.
Hereditary hemochromatosis (HH) is an autosomal recessive disorder of iron metabolism, characterized by excessive absorption of dietary iron in the small intestine. The excess iron is stored in the parenchymal cells of major tissues, primarily the liver, pancreas, heart, pituitary, and joints, eventually leading to severe tissue damage (1–3). The pathogenesis of HH is thought to involve a defect in the mechanism controlling iron absorption in the small intestine (4).
The gene defective in hemochromatosis, HFE, encodes an MHC class I-type protein (5). In multiple population studies, 85–90% of HH patients of northern European ancestry were found to be homozygous for the C282Y mutation in HFE. About 5% are compound heterozygotes for C282Y and a second mutation, H63D (5–10). Even earlier, the observation that β2-microglobulin (β2M)-deficient mice develop progressive iron overload similar to that seen in HH patients suggested the involvement of an MHC class I protein in HH (11, 12). Confirmation that mutations in HFE cause HH was provided by observations of mice with targeted disruption of the HFE gene. These HFE knockout mice showed high transferrin saturation and an increase in iron storage in hepatocytes (13–15).
The C282Y mutation in HFE disrupts the disulfide bond in the α3 domain, impairing the normal association of HFE with β2M, and dramatically reduces the cell surface expression of HFE (5, 16, 17). This effect was shown in expression studies of wild-type and mutant HFE in stably transfected mammalian cells (293 cells) (16) and in transiently transfected COS-7 cells (17). Waheed et al. (17) also found that the majority of C282Y mutant protein remains in high molecular weight aggregates, fails to undergo late Golgi processing, and is rapidly degraded. However, 30% of the C282Y mutant protein became endo-H-resistant, suggesting that some of the mutant protein undergoes Golgi processing and is stable, although very little C282Y protein reaches the cell surface in COS cells. With respect to β2M association and cell surface expression, the H63D mutant protein behaved similarly to the wild-type protein (16, 17).
Studies showing similarity between HFE and MHC class I proteins (5, 18), and studies demonstrating the effect of mutations on the synthesis of HFE, β2M association, and cell surface expression (16, 17), suggested a mechanism whereby the C282Y mutation might affect the macromolecular assembly and cell surface expression of HFE. However, these studies do not explain how the HFE protein functions to regulate iron absorption, or how the C282Y mutation might result in the increased iron absorption seen in HH. One link between the HFE-β2M complex and iron homeostasis was provided by the observation that the HFE-β2M complex is physically associated with the transferrin receptor 1 (TfR1) in human placental membranes (19), in crypt enterocytes of duodenum (20), and in cultured human cells (21–23). Furthermore, in vitro biochemical studies on the soluble forms of human HFE-β2M and TfR1 demonstrated pH-dependent associations between TfR1 and HFE-β2M (24–26). Biochemical studies showed that soluble HFE-β2M and Fe-Tf can bind simultaneously to TfR1 to form a quaternary complex (25). The high-affinity binding of HFE with TfR1 suggested that HFE-TfR1 complex formation might be important for the function of HFE protein in iron homeostasis (18–26). This hypothesis was supported by the observation that the transport of HFE to endosomes and the regulation of intracellular iron absorption in some cell lines depend on its association with TfR1 (27).
Several studies on the interaction of HFE with TfR1 in transfected cell lines have attempted to characterize the biological effect of this interaction on iron homeostasis (22, 23, 27–31). Several groups have reported that HeLa cells overexpressing HFE protein showed a decrease in ferritin level, an increase in TfR1, and an increase in iron regulatory protein activity, indicating lower levels of iron stores in the cells (22, 23, 30). In some studies, lower Tf-59Fe uptake was also reported in HeLa cells overexpressing HFE protein (23, 28–30). In all of the above studies on human cell lines, the cells express endogenous levels of human β2M, and only HFE is overexpressed. On the other hand, Montosi et al. (32) found that accumulation of Tf-59Fe was lower in macrophages from HH patients than from control individuals expressing wild-type HFE. Furthermore, the decrease in iron accumulation in macrophages of HH patients, inferred from the ferritin pool, was reversed by 40–60% after transfection with HFE.
The present report shows that the effects of overexpressed HFE in CHO cells are more complicated than those reported on HeLa cells. Overexpression of HFE either enhances or inhibits uptake of Tf-59Fe, depending on whether β2M is also overexpressed in CHO cells. We suggest that the effects of the HFE-β2M complex on CHO cells reflect its normal physiological role in small intestine, where it facilitates sensing of body iron and regulates iron absorption.
Materials and Methods
The antibodies against the C-terminal, 16-aa peptide of HFE, CT16, were the same as described previously (17). Monoclonal mouse anti-human TfR1 antibody was purchased from Zymed. Sheep anti-human ferritin antibody was from The Binding Site (Birmingham, U.K.). Geneticin (G418 sulfate) was from GIBCO. Sulfo-N-hydroxysuccinimide-LC-biotin (in which LC represents hexanoate linker chain) was purchased from Pierce. Rabbit anti-human β2-microglobulin, goat anti-rabbit and anti-sheep IgG-peroxidase, and streptavidin–peroxidase antibodies were from Sigma–Aldrich. Human holo- and apotransferrins and puromycin were from Sigma–Aldrich. 59FeCl3, Na125I, and 35S-translabel were from Amersham Pharmacia.
Cells and Cell Culture.
Chinese hamster ovary (CHO)-K1 cells were transfected with the pCXN vector (33, 34) containing wild-type HFE cDNA together with pCAGGS vector containing human β2M cDNA, as described (17). Electroporation conditions were 25 μF and 1,200 V in a 0.4-cm cuvette by using the Bio-Rad electroporation system. Colonies appeared after 2–3 weeks of selection in G418-containing media. Individual clones were picked, grown to confluency, and analyzed for HFE and β2M protein expression by Western blot by using a mixture of antibodies to HFE and β2M. Stable CHO clones were cultured in MEM supplemented with 10% FCS/200 μg/ml of G418/34.5 μg/ml of proline. TRVb-1 is a CHO cell line deficient in hamster TfR1 that is stably transfected with human TfR1 (35). This clone was a kind gift from T. E. McGraw of Cornell University. TRVb-1 cells were also transfected by electroporation with the pCXN vector containing wild-type HFE cDNA, alone, or in combination with, the pCAGGS vector containing human β2M cDNA and PSV2-PAC for puromycin selection (36), as described for CHO clones. After electroporation, cells were cultured in F-12 medium supplemented with 200 μg/ml of G418 for 2 days. Cells were then fed with increasing concentrations of 10–30 μg/ml of puromycin for 2–3 weeks. Puromycin-resistant colonies were picked and analyzed for the expression of HFE and β2M proteins by Western blot.
Biotinylation of Cell Surface Proteins.
Cells were grown on 35-mm plates for 2 days before biotinylation, washed with ice-cold PBS at 4°C, and incubated with 100 μg/ml of N-hydroxysuccinimide-LC-biotin for 10 min at 4°C. The biotinylation reaction was quenched by removing the medium and incubating the cells with 1% casein in PBS at room temperature for 10 min. The cells were recovered in cold PBS and the cell pellet homogenized in lysis buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM PMSF) containing 1% Nonidet P-40 or 0.2% SDS. The protein concentration was determined by micro-Lowry protein assay (37). The biotinylated proteins were characterized by immunoprecipitation of HFE or TfR1 by using their respective antibodies, and the immune complex was analyzed by SDS/PAGE followed by Western blot.
Metabolic Labeling and Immunoprecipitation.
The cells were metabolically labeled for 30 min with 35S-translabel, 50 μCi (1 Ci = 37 GBq) per 35-mm plate in 1 ml of DME medium without cysteine and methionine and supplemented with 5% heat-inactivated and dialyzed FCS as described (38). In pulse–chase experiments, the cells were labeled for 30 min and chased with cold 10 mM cysteine and 10 mM methionine for the indicated times. The cell pellets were lysed in 1 ml of buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Triton X-100/0.5% deoxycholate/0.1% SDS/1 mM PMSF/1 mM benzamidine). The HFE and TfR1 proteins were immunoprecipitated from the cell lysates by using CT16 and mouse anti-human TfR1 antibodies, respectively, as described (17). The immunoprecipitates were analyzed by SDS/PAGE followed by fluorography.
SDS/PAGE and Western Blot Analysis.
SDS/PAGE was performed under reducing conditions according to Laemmli (39). The polypeptides were transferred electrophoretically to the Immobilon-P membrane. The biotinylated polypeptides on the membrane were visualized by using streptavidin–peroxidase conjugate and chemiluminescent substrate. To detect the TfR1 and ferritin levels, antibodies against TfR1 and ferritin and their secondary antibodies were used.
Ligand Preparation.
Tf-59Fe was prepared by the nitrilotriacetic acid method as described (40). Diferric Tf was labeled with 125I by using iodogen (Pierce) as described (41).
Binding of Diferric 125I-Tf to the Cell Surface Receptors.
The cells were grown on 35-mm plates to confluency, were washed with serum-free MEM, and incubated with the same medium at 37°C for 1 h to remove endogenous Tf. The cells were chilled on ice and incubated with 100 nM 125I-Tf in binding buffer (serum-free MEM containing 100 mM Hepes, pH 7.5/2 mg/ml BSA) at 4°C for 2 h. The unbound ligand was removed, and the cells were washed with ice-cold PBS, lysed in 1% SDS, and counted with scintillation fluid. Nonspecific radioactivity binding was determined by incubation with a 200- to 500-fold excess of nonradioactive Tf-Fe. The protein concentration of the cells from one 35-mm plate was determined. The results of ligand bound per milligram of cell protein at different concentrations of ligand were used to determine the binding constant by the nonlinear saturation curve method or by Scatchard analysis.
Accumulation of Tf-59Fe.
Cells were grown on 35-mm plates to confluency and washed with serum-free MEM medium followed by a 1-h incubation at 37°C to remove endogenous Tf. The cells were then incubated with 100 nM Tf-59Fe in binding buffer at 37°C for different times. At the required times, the cells were incubated with ice-cold buffer (0.15 M glycine⋅HCl, pH 3.0/50 mM NaCl) for 3 min to remove cell surface-bound radioactive Tf-59Fe. The cells were washed with ice-cold PBS, lysed in 1% SDS, and counted with scintillation fluid. The radioactivity accumulated per milligram of cell protein at different times was calculated.
TfR1 Endocytosis.
The endocytosis rate constant for TfR1 internalization was determined by using the In/Sur method (42), as described by McGraw and Maxfield (43). The cells were incubated in duplicate with 25 nM 125I-Tf in binding buffer at 37°C for different times. At the required time, the cells from one plate were washed with ice-cold PBS and counted with 1% SDS for total specific ligand binding. At the same time, the cells from the other plate were first incubated with ice-cold buffer (0.15 M glycine⋅HCl, pH 3.0/50 mM NaCl) for 3 min to remove the cell surface-bound ligand and then washed with ice-cold PBS and counted for internalized ligand. Cell surface-bound ligand was determined by subtracting the internalized radioactivity from the total specific radioactivity bound. The ratios of internalized radioactivity (In) over cell surface-bound radioactivity (Sur) were plotted against time. The slope of the straight line was calculated to represent the internalization rate of TfR1.
Recycling of Endocytosed125I-Tf.
The cells were incubated with 25 nM 125I-Tf in binding buffer at 37°C for 1 h. Cell surface-bound ligand was removed by incubating the cells in ice-cold buffer (0.15 M glycine⋅HCl, pH 3.0/50 mM NaCl) for 3 min and washing with ice-cold PBS. The cells were then chased with binding buffer alone at 37°C for 0–20 min. At the required time, cells were washed with cold PBS, lysed with 1% SDS, and counted in scintillation fluid. The radioactive ligand associated with the cells at zero time chase was used as 100% radioactivity to calculate the radioactive ligand lost during the chase at different times. The slope of the logarithm of percent radioactivity associated at different chase time was calculated and represents the rate of endocytosed 125I-Tf.
Results
Effect of β2M on Stability and Maturation of HFE.
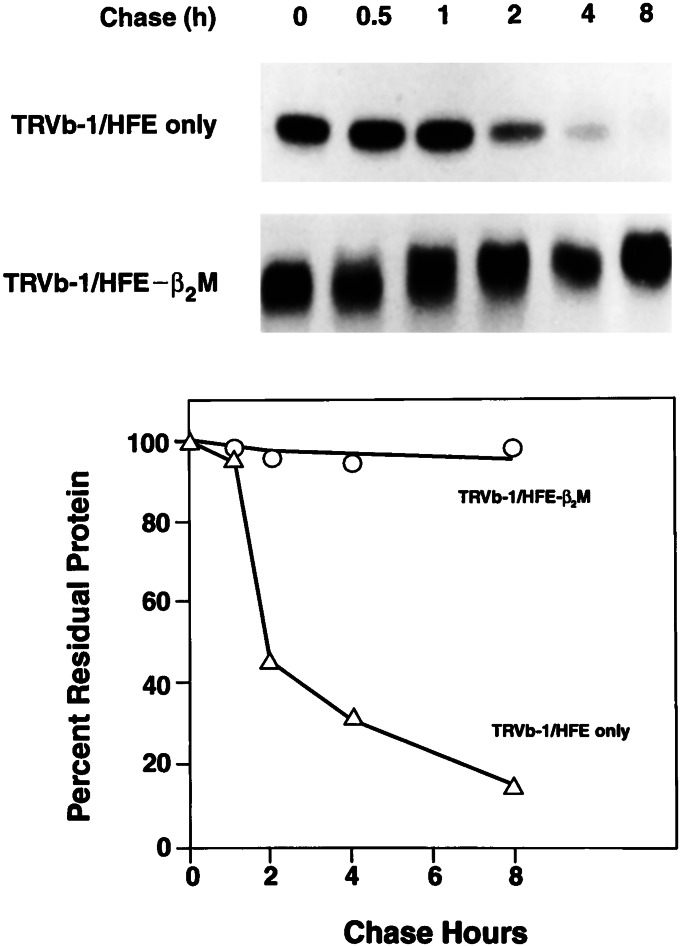
The association of β2M with MHC class I-type molecules is considered necessary for intracellular processing, transport, and cell surface expression of MHC class I proteins (16, 17, 44). The C282Y mutation, the most common mutation causing HH in Caucasians, impairs the ability of HFE to associate with β2M (16, 17). Thus, it was of interest to study the effects of β2M on the fate of HFE by measuring the biosynthesis, maturation, and turnover of HFE in the presence and absence of overexpression of β2M. The results in Fig. 1 show that, in the presence of β2M, the HFE protein is synthesized as a 39- to 41-kDa protein in 30 min, which is matured to a 45- to 52-kDa polypeptide with no apparent turnover within 8 h. When HFE is expressed in the absence of β2M, the HFE protein is synthesized by 30 min, but most of it is degraded rapidly in the first 2 h, then more slowly in the latter phase, with no change in its apparent molecular weight. These results raised the question whether the unprocessed, unstable, and immature HFE protein, produced in the absence of β2M, may have different effects on TfR1-mediated iron uptake than the properly folded, stable, and mature HFE protein made in the presence of β2M.
Figure 1.
Effect of β2M on stability and maturation of HFE protein in TRVb-1 cells. TRVb-1 cells overexpressing HFE only and HFE-β2M were pulse labeled for 30 min with 35S-translabel and chased for 0.5–8 h with nonradioactive methionine and cysteine. The HFE protein was immunoprecipitated and analyzed by SDS/PAGE followed by fluorography. Quantitative analysis of fluorogram is presented (Lower).
Effects of HFE and HFE-β2M on TfR1 Function.
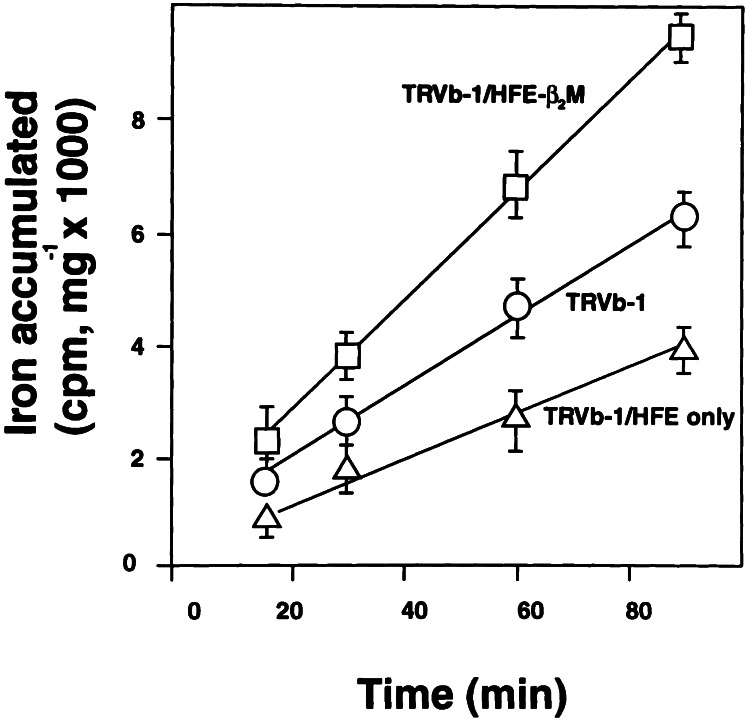
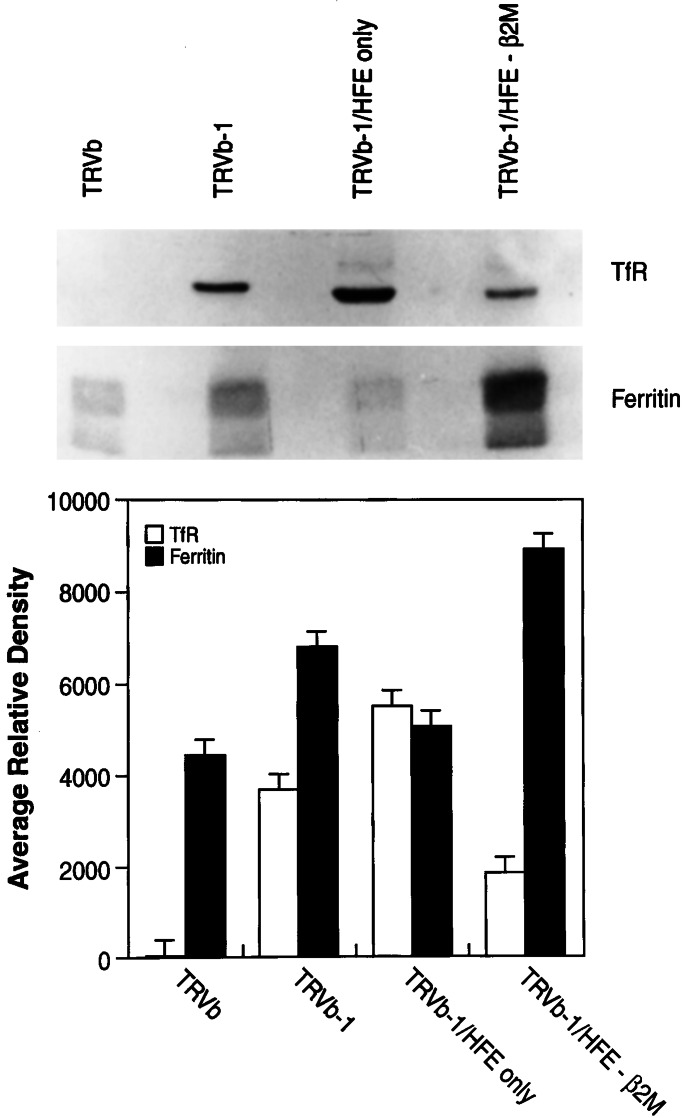
The effect of β2M on the role of HFE in modulating TfR1 function was studied in TRVb-1 cells expressing HFE-β2M or HFE alone. TRVb-1 cells are derivatives of CHO cells that do not express hamster TfR1 but express the human TfR1 as a consequence of stable transfection (35). The results in Fig. 2 show that, in the presence of β2M, HFE enhances TfR1-dependent iron uptake by 50%. On the other hand, in the absence of overexpressed β2M, HFE alone reduces TfR1-dependent iron uptake by nearly 50%. The effects of HFE-β2M and HFE alone on TfR1-dependent iron uptake were further studied by measuring the steady-state levels of ferritin and TfR1 as indicators of the iron status of the transfected cells. The results in Fig. 3 show that TRVb-1 cells expressing HFE-β2M have a 50% decrease in TfR1 and a 30% increase in ferritin content compared with nontransfected TRVb-1 cells, indicating higher iron stores in TRVb-1 cells expressing HFE-β2M. By contrast, TRVb-1 cells expressing HFE alone have a 50% increase in TfR1 and a 40% decrease in ferritin content compared with TRVb-1 cells, suggesting lower iron stores in TRVb-1 cells expressing HFE in the absence of overexpressed β2M. These latter results without β2M are similar to those reported by several groups for HeLa cell lines expressing HFE in the absence of β2M transfection (22, 23, 30).
Figure 2.
Iron accumulation rate in TRVb-1 cells. TRVb-1 cells expressing HFE alone (TRVb-1/HFE only) and HFE-β2M (TRVb-1/HFE-β2M) were incubated with 100 nM Tf-59Fe at 37°C. The cells were washed to remove cell surface-associated radioactivity before intracellular iron radioactivity was determined. The radioactivity accumulated per milligram of cell protein was calculated.
Figure 3.
Effect of HFE and HFE-β2M on iron status of TRVb cell derivatives. (Upper) TRVb cells (CHO cells lacking hamster TfR1); TRVb-1 cells (TRVb cells expressing human TfR1); TRVb-1/HFE only (TRVb-1 cells expressing HFE alone); and TRVb-1/HFE-β2M (TRVb-1 cells expressing HFE and β2M) were grown to confluency. The cell homogenates, equivalent to 30 μg of cell protein, were analyzed by SDS/PAGE followed by Western blot analysis by using TfR1 and ferritin (Ferritin) antibodies (Upper). The polypeptide intensities were quantitated and presented as average relative density (Lower).
Cell Surface Expression of HFE-β2M and Association with Hamster or Human TfR1.
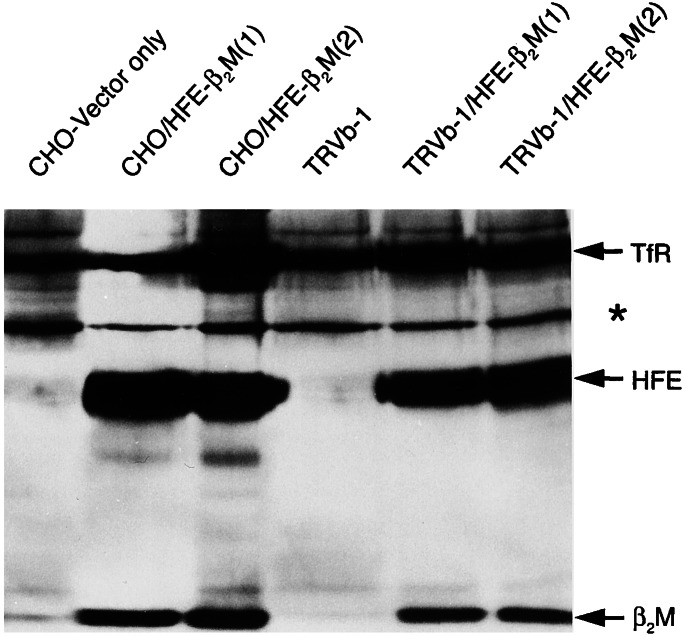
A role for HFE-β2M in TfR1-mediated iron uptake was initially inferred from the observations that HFE-β2M associates with TfR1 in the plasma membrane of human placenta (19), crypt cells (20), stably transfected cells (21–23), and in vitro (18, 24–26), when the truncated secretory forms of the respective proteins are studied. To study the effect of human HFE and β2M expression on TfR1-mediated iron uptake in this system, stable CHO and TRVb-1 clones expressing both HFE and β2M were generated. In the present study, we examined the association at the cell surface of human HFE-β2M with hamster TfR1 in CHO cells and of human HFE-β2M with human TfR1 in TRVb-1 cells. The results in Fig. 4 show that all three members of the TfR1-HFE-β2M ternary complex are immunoprecipitated from the mixture of biotinylated cell surface proteins by monoclonal antibodies against TfR1. Western blot analysis of immunoprecipitates by using streptavidin–peroxidase show all three major polypeptides—TfR1, HFE, and β2M, respectively—both in CHO and TRVb-1 clones expressing human HFE-β2M (see lanes CHO/HFE-β2M or TRVb-1/HFE-β2M). These results suggest that both hamster and human TfR1 associate with human HFE and β2M at the cell surface. Hamster and human TfR1 are immunoprecipitated as single proteins by the anti-TfR1 antibodies in the control cells (see lanes, CHO-vector only or TRVb-1).
Figure 4.
Cell surface expression of HFE-β2M proteins and association with TfR1. The cell surface proteins of CHO and TRVb-1 cells overexpressing HFE-β2M were biotinylated at 4°C. (1) and (2) refer to independent clones. The TfR1-HFE-β2M complex was immunoprecipitated by using TfR1 monoclonal antibodies. The immunoprecipitates were analyzed by SDS/PAGE followed by Western blot by using streptavidin–peroxidase conjugate. The polypeptides for TfR1, HFE, and β2M are marked. A nonspecific polypeptide of 64 kDa was seen in all lanes (*).
The HFE-β2M Complex Enhances TfR1-Mediated Iron Uptake.
Studies of the rate of iron accumulation at 37°C in CHO and TRVb-1 cells expressing HFE-β2M are summarized in Table 1. Tf-59Fe accumulation in CHO and TRVb-1 cells is enhanced by 20 and 70%, respectively, in cells overexpressing both HFE and β2M proteins. These results indicate that the human HFE-β2M complex enhances the function of both hamster and human TfR1 in mediating Tf-59Fe uptake.
Table 1.
Rates of iron accumulation from diferric Tf in CHO and TRVb-1 clones
| Cell types | Fe accumulation rate, cpm/mg/h |
|---|---|
| CHO, vector only | 5,400 ± 100 |
| CHO/HFE-β2M (1) | 6,480 ± 65 |
| CHO/HFE-β2M (2) | 6,265 ± 45 |
| TRVb-1 | 2,133 ± 21 |
| TRVb-1/HFE-β2M (1) | 3,600 ± 36 |
| TRVb-1/HFE-β2M (2) | 3,733 ± 38 |
Effect of HFE-β2M on Tf-Fe-Binding and Endocytotic Properties of TfR1.
To explain the mechanism for an increase in iron accumulation associated with overexpression of HFE-β2M in CHO or TRVb-1 cells, we studied the binding affinities of human Tf to TfR1 at the cell surface and also studied the internalization rate and the rate of recycling of TfR1. The results summarized in Table 2 show the binding constants of transferrin with hamster and human TfR1 (5.94 ± 0.52 nM and 3.29 ± 0.49 nM, respectively). Both independent clones of CHO or TRVb-1 expressing HFE-β2M showed no consistent change in binding affinities for Tf-Fe. Thus, changes in iron uptake cannot be ascribed to a change in the affinity of the TfR1 for human Tf.
Table 2.
Binding constant of diferric Tf at 4°C in CHO and TRVb-1 clones
| Cell types | Binding constant, nM |
|---|---|
| CHO, vector only | 5.94 ± 0.52 |
| CHO/HFE-β2M (1) | 2.18 ± 0.33 |
| CHO/HFE-β2M (2) | 6.21 ± 0.93 |
| TRVb-1 | 3.29 ± 0.49 |
| TRVb-1/HFE-β2M (1) | 5.27 ± 0.79 |
| TRVb-1/HFE-β2M (2) | 3.38 ± 0.51 |
Cell surface binding was measured at 4°C as described (see Materials and Methods). The binding constants are the average of three independent experiments. The values for binding constants were calculated either from the saturation binding curve or by Scatchard analysis of Tf binding results. Where indicated, two independent clones, (1) and (2), were studied.
The results in Table 3 show that the internalization rate of hamster TfR1 is about two times faster than human TfR1. Similar results have been observed by others (43). However, there is no significant effect of HFE-β2M on the rate of internalization of TfR1-bound human Tf-Fe in either CHO or TRVb-1 cells. Finally, we studied the rate of recycling of hamster or human TfR1 in cells expressing HFE-β2M by measuring the rate of exocytosis of endocytosed 125I-Tf. The results in Table 4 show that the rate of recycling of hamster or human TfR1 is enhanced by 20 and 100%, respectively, in CHO and TRVb-1 cells overexpressing human HFE-β2M, compared with their respective controls. The rate of recycling of hamster TfR1 in CHO control cells is about twice that of human TfR1 in control TRVb-1 cells. Similar differences in rates of recycling for hamster and human TfR1 have been reported previously and attributed to a single amino acid difference at position 20 of the cytoplasmic tail of TfR1, which is Cys-20 in the hamster receptor and Tyr-20 in the human receptor (43).
Table 3.
Internalization rate of hamster (CHO) and human (TRVb-1) TfR1
| Cell types | Internalization rate constant, min−1 |
|---|---|
| CHO, vector only | 0.23 ± 0.02 |
| CHO/HFE-β2M (1) | 0.25 ± 0.03 |
| CHO/HFE-β2M (2) | 0.26 ± 0.03 |
| TRVb-1 | 0.12 ± 0.01 |
| TRVb-1/HFE-β2M (1) | 0.10 ± 0.01 |
| TRVb-1/HFE-β2M (2) | 0.09 ± 0.01 |
These results are the average of three separate measurements of the internalization rate constant. The internalization rate for TfR1 was determined by the In/Sur method. TRVb-1 cells express human TfR1, and CHO clones express hamster TfR1. Where indicated, two independent clones, (1) and (2), were analyzed.
Table 4.
Recycling rate of endocytosed 125I-Tf in CHO and TRVb-1 clones
| Cell types | Recycling rate constant, min−1 |
|---|---|
| CHO, vector only | 0.106 ± 0.001 |
| CHO/HFE-β2M (1) | 0.125 ± 0.010 |
| CHO/HFE-β2M (2) | 0.130 ± 0.011 |
| TRVb-1 | 0.044 ± 0.004 |
| TRVb-1/HFE-β2M (1) | 0.092 ± 0.005 |
| TRVb-1/HFE-β2M (2) | 0.104 ± 0.005 |
| TRVb-1/HFE-β2M (3) | 0.100 ± 0.010 |
| TRVb-1/HFE-β2M (4) | 0.096 ± 0.010 |
These results are the average of three separate experiments on the rate of recycling of endocytosed 125I-Tf. Numbers in parentheses identify independent clones.
Effect of Expressing the HFE-β2M Complex on the Number of TfR1 Receptors at the Cell Surface.
Because the binding affinity of Tf to TfR1 and the TfR1 internalization rate are both unaffected, and the recycling rate of 125I-Tf is enhanced, we predicted that the cell surface expression of TfR1 might be increased because of overexpression of HFE-β2M. The steady-state level of TfR1 expression at the cell surface was determined by measuring the binding of human 125I-Tf to the cell surface at 4°C. The results in Fig. 5 show that overexpression of HFE-β2M in CHO and TRVb-1 cells results in a 140 and 180% increase in cell surface binding of transferrin, respectively (Fig. 5B). However, Western blots showed that the total TfR1 content is not increased because of HFE-β2M (Fig. 5A). In fact, overexpression decreases total TfR1 signal (see also Fig. 3). From these results, we infer that overexpression of HFE-β2M increases the cell surface expression of TfR1 without actually decreasing the total cell content of TfR1. The increased cell surface expression of TfR1 (even in the face of decreased total cellular TfR1) may explain the increase in iron accumulation from diferric Tf in cells expressing HFE-β2M.
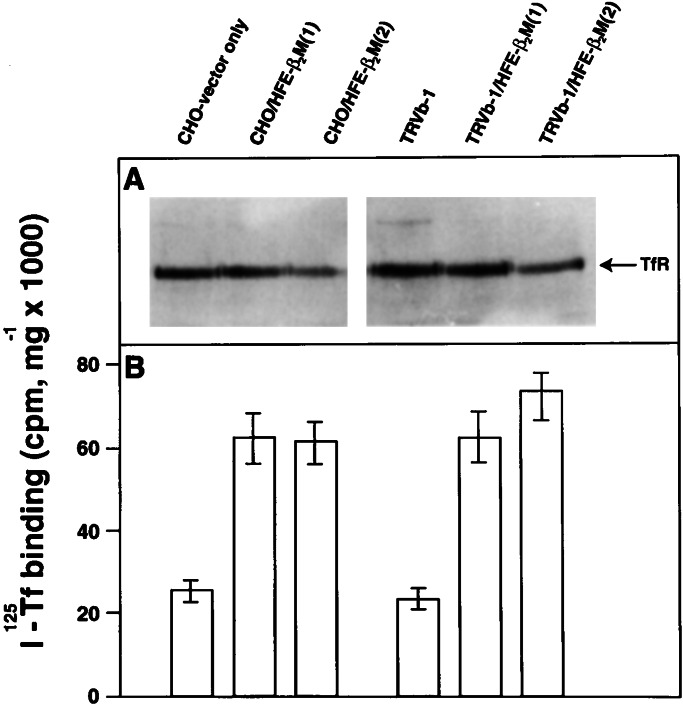
Figure 5.
HFE-β2M enhances the number of TfR1 receptors at the cell surface. CHO and TRVb-1 cells alone and overexpressing HFE-β2M were grown to confluency. (A) The cell homogenates containing 30 μg of cell protein were analyzed by SDS/PAGE followed by Western blot by using TfR1 antibodies. The polypeptide corresponding to total TfR1 (cell surface and intracellular) is marked. The relative intensity of the polypeptide was decreased because of overexpression of HFE-β2M (as in Fig. 3). (B) The cell surface receptor-binding activity was determined by incubating the cells with 100 nM 125I-Tf at 4°C for 1 h. After washing the cells, the cell-associated radioactivity was determined and presented as 125I-Tf binding per milligram of cell protein. The cell surface-binding activity was increased by overexpression of HFE-β2M, despite the lower amount of total cellular TfR1.
Discussion
Although there is overwhelming evidence that mutations in the HFE protein lead to HH (5–10, 13–15, 45, 46), and there is a clear rationale to explain how the C282Y mutation, which interferes with the mutant protein's association with β2M (16, 17), could disrupt the function of the HFE protein, the mechanism by which the C282Y mutation leads to increased absorption of dietary iron has remained a mystery. We previously suggested that the role of the normal HFE protein might be to enhance uptake of Tf-bound Fe by duodenal crypt cells, and that impairment of this function is the source of dysregulation of iron sensing leading to excess absorption of dietary iron (46, 47). However, most prior studies of the effects of HFE on iron uptake by cultured cells have found the reverse effect—namely, impairment of iron uptake by cells overexpressing the normal HFE protein (23, 28–31). The studies reported here address this dilemma.
The present studies agree with most of the prior studies in showing that overexpressed HFE, in the absence of overexpressed β2M, reduces the uptake of Tf-bound Fe (23, 28–30). However, what our studies show, in addition, is that overexpression of wild-type HFE has the opposite effect when β2M is also overexpressed in CHO cells. Whether this would also be true in HeLa cells if β2M were overexpressed remains to be established. At least in this CHO cell culture system, wild-type HFE enhances iron uptake of Tf-bound Fe similar to the role that we have proposed for its function in duodenal crypt cells. Loss of this normal function could explain why duodenal iron status is lower in HH patients than one might predict from their degree of iron overload (48, 49). Macrophages from HH patients show a similarly inappropriate low iron status, despite systemic iron overload (32). Only studies of macrophages from HFE patients have shown an increase in TfR1-mediated uptake of transferrin-bound iron, like that shown here, after transfection with wild-type HFE (32).
The studies presented here suggest that CHO cells (and the CHO-derived cell line TRVb-1, which expresses the human but not the hamster TfR1) are good models for understanding the role of HFE in HH. In the presence of ample amounts of β2M, HFE is normally processed to the mature protein, normally transported to the cell surface, and forms a ternary complex with TfR1, whose function it modulates in a positive manner (enhances iron uptake). In the absence of ample β2M, the HFE fails to undergo normal processing, is unstable, and negatively affects iron uptake. It is in this sense that we propose that CHO cells and TRVb-1 cells may provide a model for duodenal crypt cells. In HH patients, it is postulated that HFE mutations result in the impairment of HFE function in duodenal crypt cells (46, 47). In the mouse models of HH, loss of HFE function results both from the C282Y mutation or knockout mutations in the HFE gene and from deficiency of β2M as in the β2M-knockout mouse (11–15). In fact, the discovery that β2M-deficient mice show an HH phenotype was the initial piece of evidence associating MHC class I-type proteins in iron homeostasis and in HH (11, 12). The CHO and TRVb-1 cells expressing HFE without β2M provide models of the β2M-deficient mouse in which the HFE protein is dysfunctional because β2M is absent.
The mechanism by which normal HFE-β2M enhances iron uptake and recycling of endocytosed Tf in TRVb-1 cells is not yet clear, nor is the mechanism by which the abnormally processed and unstable HFE, which is made in the absence of adequate amounts of β2M, not only fails to enhance iron uptake, but actually depresses iron uptake. Another interesting question is whether mutant HFEs will behave like HFE expressed in the absence of β2M in CHO and TRVb-1 cells. Because HFE knockout and C282Y mutant mice behave phenotypically like β2M-deficient mice, i.e., both have the murine HH phenotype, one would predict that they would (11–15). By analogy with the role of HFE-β2M in modulating uptake of Tf-Fe by CHO and TRVb-1 cells, we suggest: (i) that HFE-β2M normally plays a role in facilitating uptake of Tf-bound iron by duodenal crypt cells and macrophages; (ii) that HFE mutations that impair this function in HH patients contribute to dysregulation of iron absorption, because this uptake process by duodenal crypt cells is key to sensing the level of Tf-bound iron in capillaries surrounding crypt cells of the duodenum where iron absorption is controlled (48, 50, 51); and (iii) this impairment of Tf-bound iron uptake leads the duodenal crypt cells to be iron poor (47–49, 52), even in the face of body iron excess, and to program cells differentiating into mature villus cells to absorb excess iron in HH. Supporting this model, HH patients have increased duodenal expression of the apical iron transporter, divalent metal transporter 1, and the basolateral iron transporter ferroportin 1 (49, 52).
Acknowledgments
We thank Matthew Casey for technical assistance and Tracey Baird and Elizabeth Torno for editorial help. This research was funded by the following National Institutes of Health Grants DK53405, GM34182, and DK40163 (to W.S.S.), HL66225 (to R.E.F.), and DK41816 (to B.R.B.).
Abbreviations
- HH
hereditary hemochromatosis
- β2M
β2-microglobulin
- TfR1
transferrin receptor 1
- CHO
Chinese hamster ovary
References
- 1.Cartwright G E, Edwards C Q, Kravitz K, Skolnick M, Amos D B, Johnson A, Buskjaer L. N Engl J Med. 1979;301:175–179. doi: 10.1056/NEJM197907263010402. [DOI] [PubMed] [Google Scholar]
- 2.Cox T M, Lord D K. Eur J Haematol. 1989;42:113–125. doi: 10.1111/j.1600-0609.1989.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 3.Bacon B R, Tavill A S. In: Hepatology: A Textbook of Liver Disease. Zakim D, Boyer T D, editors. Philadelphia: Saunders; 1996. pp. 1439–1472. [Google Scholar]
- 4.McLaren G D, Nathanson M H, Jacobs A, Trevett D, Thomson W. J Lab Clin Med. 1991;117:390–401. [PubMed] [Google Scholar]
- 5.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 6.Beutler E, Gelbart T, West C, Lee P, Adams M, Blackstone R, Pockros P, Kosty M, Venditti C P, Phatak P D, et al. Blood Cells Mol Dis. 1996;22:187–194. doi: 10.1006/bcmd.1996.0027. [DOI] [PubMed] [Google Scholar]
- 7.Jouanolle A M, Gandon G, Jezequel P, Blayau M, Campion M L, Yaouanq J, Mosser J, Fergelot P, Chauvel B, Bouric P, et al. Nat Genet. 1996;14:251–252. doi: 10.1038/ng1196-251. [DOI] [PubMed] [Google Scholar]
- 8.Jazwinska E C, Cullen L M, Busfield F, Pyper W R, Webb S I, Powell L W, Morris C P, Walsh T P. Nat Genet. 1996;14:249–251. doi: 10.1038/ng1196-249. [DOI] [PubMed] [Google Scholar]
- 9.Carella M, D'Ambrosio L, Totaro A, Grifa A, Valentino M A, Piperno A, Girelli D, Roetto A, Franco B, Gasparini P, et al. Am J Hum Genet. 1997;60:828–832. [PMC free article] [PubMed] [Google Scholar]
- 10.Borot N, Roth M, Malfroy L, Demangel C, Vinel J P, Pascal J P, Coppin H. Immunogenetics. 1997;45:320–324. doi: 10.1007/s002510050211. [DOI] [PubMed] [Google Scholar]
- 11.Santos M, Schilham M W, Rademakers L H, Marx J J, de Sousa M, Clevers H. J Exp Med. 1996;184:1975–1985. doi: 10.1084/jem.184.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg B E, Voland J R. Proc Natl Acad Sci USA. 1996;93:1529–1534. doi: 10.1073/pnas.93.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy J E, Montross L K, Cohen D E, Fleming M D, Andrews N C. Blood. 1999;94:9–11. [PubMed] [Google Scholar]
- 15.Bahram S, Gilfillan S, Kuhn L C, Moret R, Schulze J B, Lebeau A, Schumann K. Proc Natl Acad Sci USA. 1999;96:13312–13317. doi: 10.1073/pnas.96.23.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feder J N, Tsuchihashi Z, Irrinki A, Lee V K, Mapa F A, Morikang E, Prass C E, Starnes S M, Wolff R K, Parkkila S, et al. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 17.Waheed A, Parkkila S, Zhou X Y, Tomatsu S, Tsuchihashi Z, Feder J N, Schatzman R C, Britton R S, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:12384–12389. doi: 10.1073/pnas.94.23.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebron J A, Bennett M J, Vaughn D E, Chirino A J, Snow P M, Mintier G A, Feder J N, Bjorkman P J. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 19.Parkkila S, Waheed A, Britton R S, Bacon B R, Zhou X Y, Tomatsu S, Fleming R E, Sly W S. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waheed A, Parkkila S, Saarnio J, Fleming R E, Zhou X Y, Tomatsu S, Britton R S, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:1579–1584. doi: 10.1073/pnas.96.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feder J N, Penny D M, Irrinki A, Lee V K, Lebron J A, Watson N, Tsuchihashi Z, Sigal E, Bjorkman P J, Schatzman R C. Proc Natl Acad Sci USA. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross C N, Irrinki A, Feder J N, Enns C A. J Biol Chem. 1998;273:22068–22074. doi: 10.1074/jbc.273.34.22068. [DOI] [PubMed] [Google Scholar]
- 23.Corsi B, Levi S, Cozzi A, Corti A, Altimare D, Albertini A, Arosio P. FEBS Lett. 1999;460:149–152. doi: 10.1016/s0014-5793(99)01330-7. [DOI] [PubMed] [Google Scholar]
- 24.Lebron J A, Bjorkman P J. J Mol Biol. 1999;289:1109–1118. doi: 10.1006/jmbi.1999.2842. [DOI] [PubMed] [Google Scholar]
- 25.Lebron J A, West A P, Jr, Bjorkman P J. J Mol Biol. 1999;294:239–245. doi: 10.1006/jmbi.1999.3252. [DOI] [PubMed] [Google Scholar]
- 26.Bennett M J, Lebron J A, Bjorkman P J. Nature (London) 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 27.Ramalingam T S, West A P, Jr, Lebron J A, Nangiana J S, Hogan T H, Enns C A, Bjorkman P J. Nat Cell Biol. 2000;2:953–957. doi: 10.1038/35046611. [DOI] [PubMed] [Google Scholar]
- 28.Salter-Cid L, Brunmark A, Li Y, Leturcq D, Peterson P A, Jackson M R, Yang Y. Proc Natl Acad Sci USA. 1999;96:5434–5439. doi: 10.1073/pnas.96.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy C N, Penny D M, Feder J N, Enns C A. J Biol Chem. 1999;274:9022–9028. doi: 10.1074/jbc.274.13.9022. [DOI] [PubMed] [Google Scholar]
- 30.Riedel H D, Muckenthaler M U, Gehrke S G, Mohr I, Brennan K, Herrmann T, Fitscher B A, Hentze M W, Stremmel W. Blood. 1999;94:3915–3921. [PubMed] [Google Scholar]
- 31.Salter-Cid L, Brunmark A, Peterson P A, Yang Y. Genes Immun. 2000;1:409–417. doi: 10.1038/sj.gene.6363697. [DOI] [PubMed] [Google Scholar]
- 32.Montosi G, Paglia P, Garuti C, Guzman C A, Bastin J M, Colombo M P, Pietrangelo A. Blood. 2000;96:1125–1129. [PubMed] [Google Scholar]
- 33.Tureci O, Sahin U, Vollmar E, Siemer S, Gottert E, Seitz G, Parkkila A K, Shah G N, Grubb J H, Pfreundschuh M, et al. Proc Natl Acad Sci USA. 1998;95:7608–7613. doi: 10.1073/pnas.95.13.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 35.McGraw T E, Greenfield L, Maxfield F R. J Cell Biol. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vara J A, Portela A, Ortin J, Jimenez A. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson G L. Anal Biochem. 1979;100:201–220. doi: 10.1016/0003-2697(79)90222-7. [DOI] [PubMed] [Google Scholar]
- 38.Okuyama T, Waheed A, Kusumoto W, Zhu X L, Sly W S. Arch Biochem Biophys. 1995;320:315–322. doi: 10.1016/0003-9861(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan J, Jordan I, Sturrock A. J Biol Chem. 1991;266:2997–3004. [PubMed] [Google Scholar]
- 41.Parker K C, Strominger J L. Biochemistry. 1983;22:1145–1153. doi: 10.1021/bi00274a024. [DOI] [PubMed] [Google Scholar]
- 42.Wiley H S, Cunningham D D. J Biol Chem. 1982;257:4222–4229. [PubMed] [Google Scholar]
- 43.McGraw T E, Maxfield F R. Cell Regul. 1990;1:369–377. doi: 10.1091/mbc.1.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazaki J, Appella E, Ozato K. Proc Natl Acad Sci USA. 1986;83:757–761. doi: 10.1073/pnas.83.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews N C. Dig Liver Dis. 2000;32:56–61. doi: 10.1016/s1590-8658(00)80045-6. [DOI] [PubMed] [Google Scholar]
- 46.Parkkila S, Niemela O, Britton R S, Fleming R E, Waheed A, Bacon B R, Sly W S. Gastroenterology. 2001;121:1489–1496. doi: 10.1053/gast.2001.29617. [DOI] [PubMed] [Google Scholar]
- 47.Fleming R E, Migas M C, Zhou X, Jiang J, Britton R S, Brunt E M, Tomatsu S, Waheed A, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:3143–3148. doi: 10.1073/pnas.96.6.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietrangelo A, Rocchi E, Casalgrandi G, Rigo G, Ferrari A, Perini M, Ventura E, Cairo G. Gastroenterology. 1992;102:802–809. doi: 10.1016/0016-5085(92)90161-q. [DOI] [PubMed] [Google Scholar]
- 49.Zoller H, Pietrangelo A, Vogel W, Weiss G. Lancet. 1999;353:2120–2123. doi: 10.1016/S0140-6736(98)11179-0. [DOI] [PubMed] [Google Scholar]
- 50.Anderson G J. J Gastroenterol Hepatol. 1996;11:1030–1032. doi: 10.1111/j.1440-1746.1996.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 51.Anderson G J, Powell L W, Halliday J W. Gastroenterology. 1990;98:576–585. doi: 10.1016/0016-5085(90)90276-7. [DOI] [PubMed] [Google Scholar]
- 52.Zoller H, Koch R O, Theurl I, Obrist P, Pietrangelo A, Montosi G, Haile D J, Vogel W, Weiss G. Gastroenterology. 2001;120:1412–1419. doi: 10.1053/gast.2001.24033. [DOI] [PubMed] [Google Scholar]