Abstract
We generated a set of cysteine-to-glycine mutations and screened them to identify a temperature-sensitive allele of the human cytomegalovirus UL122 gene, which encodes the immediate-early 2 transcriptional activating protein. The mutant allele contains a single base pair substitution at amino acid 510. In transcription activation assays, the mutant protein activated the simian virus 40 early and human cytomegalovirus UL112 promoters at 32.5°C but not at 39.5°C. We constructed a mutant virus, BTNtsUL122, in which the wild-type UL122 locus is substituted with the mutant allele. The mutant produced progeny at 32.5°C but not at 39.5°C. Although the mutant virus accumulated immediate-early transcripts and proteins at the nonpermissive temperature, it did not produce any early (UL44 and UL54) and late (UL82) transcripts and it did not replicate its DNA. The mutant's defect at the nonpermissive temperature results, at least in part, from the inability of the temperature-sensitive immediate-early 2 protein to activate early viral promoters, whose products are required for DNA replication and progression into the late phase of the virus growth cycle.
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen. Infection of immunocompetent individuals is generally asymptomatic. However, the virus can cause severe disease in newborns infected in utero and in immunosuppressed individuals, including AIDS and transplant patients (1).
The immediate-early 2 (IE2) protein of HCMV is one of several immediate-early gene products involved in the regulation of viral gene expression during infection. Its transcript is produced from a region of the viral genome known as the major immediate-early locus. The locus is approximately 13 kbp in length and is composed of a strong promoter–enhancer region and a coding region with five exons (2). The IE2 transcript is spliced (3) and contains three exons shared by the immediate-early 1 (IE1) mRNA plus one unique exon, corresponding to the UL122 ORF. The predominant form of the IE2 protein is an 86-kDa nuclear phosphoprotein, which has been shown to transactivate viral and cellular promoters (4–6), often in conjunction with other immediate-early products, such as the IE1 protein (reviewed in ref. 7). IE2 also has been shown to regulate the cell cycle (7–12) and block apoptosis (13, 14).
All of these IE2 functions, however, have been described in transfected cell assays. It has not been possible to analyze the functions of IE2 within the context of a lytic infection for technical reasons. The protein is essential for lytic replication of HCMV (15), and prolonged expression of IE2 in an uninfected cell is cytotoxic (11, 16, 17). Thus, attempts to propagate an IE2 null mutant virus with a complementing cell line have not been successful.
To explore the function of IE2 during infection, we identified an IE2 allele that is temperature-sensitive (ts) for transcriptional activation and constructed an HCMV variant (BTNtsUL122) containing the mutant allele. We show that the mutant as well as the wild-type virus replicates at the permissive temperature of 32.5°C. However, it is severely defective for growth at the nonpermissive temperature of 39.5°C. The mutant is unable to produce progeny virus, replicate its DNA, or generate several early mRNAs at the nonpermissive temperature. Our results are consistent with the view that IE2 performs a master regulatory role during HCMV infection, mediating progression into the early phase of the replication cycle.
Materials and Methods
Plasmids.
The expression vectors pCGN, pCGN-IE2, and pCGN-71 have been described (18). To generate a ts derivative of IE2, exon 5 from pCGN-IE2 was amplified by PCR in two overlapping pieces. One primer contained a single base pair mutation that converted a TGC cysteine codon at amino acid position 510 to a GGC glycine codon (mutation at nucleotide position 169,579 of HCMV; ref. 19). The full-length amplification product was cloned into pGEMT (Promega) and sequenced, and an XbaI-to-BamHI fragment from the clone was used to replace the identical fragment of pCGN-IE2, creating pCGN-C510G.
Temperature sensitivity of the C510G IE2 protein was tested in a transfection assay. U-2OS osteosarcoma cells were subjected to electroporation in growth medium containing 1 μg of pGL3-promoterZ, derived by replacing the luciferase coding region of pGL3-promoter (Promega) with the β-galactosidase coding region, or 2 μg of either pHM142 or pHM214 (20, 21) plus 0.05–2 μg of pCGN-IE2 or pCGN-510. To maintain a constant concentration of DNA in electroporation mixtures, salmon sperm DNA was added when >1 μg of pCGN-IE2 or pCGN-510 was used. Cells were allowed to recover at 37°C for 2 h, and then they were incubated for 24 h at 32.5°C or 39.5°C. Cells were collected in 1 ml of luciferase lysis buffer (0.1 M K2HPO4, pH 7.8/0.2% Triton X-100), and assays were performed by using the Galacto-Light or Galacto-Light β-galactosidase chemiluminescent reporter assay (Tropix).
Viruses.
The mutant virus, BTNtsUL122, was derived from pT-BACwt (15) and contains the IE2 C510G mutation. To introduce the mutation into pT-BACwt, a derivative of the shuttle vector pGS284 (22) was constructed. pGS284–510K contains a UL122 sequence with a kanamycin-resistance expression cassette inserted between BclI and StuI sites (Fig. 1A), and it was used to insert the kanamycin cassette into pT-BACwt by allelic exchange (22). The structure of the resulting clone, BAC IE2-KAN, was verified by sequence analysis, and it was used in another round of allelic exchange with pGS284-C510G to replace the kanamycin cassette with a UL122 sequence including the C510G mutation. Viruses were generated from bacterial artificial chromosomes (BACs) by electroporation of human foreskin fibroblasts (HFFs). Cells were suspended in 260 μl of growth medium containing 10% FCS with 5 μg of BAC DNA and 2 μg of pCGN-71 (23); the electroporation was performed at 260 V and 960 μF in a 0.4-cm cuvette, and the cells were plated into culture dishes, allowed to recover at 37°C for 2 h, and then incubated at 32.5°C for the growth of BTNtsUL122 or 37°C for pT-BACwt-derived virus (BTNwt). It was necessary to cotransfect 5 μg of pCGN-510 with the C510G mutant BAC to produce virus.
Figure 1.
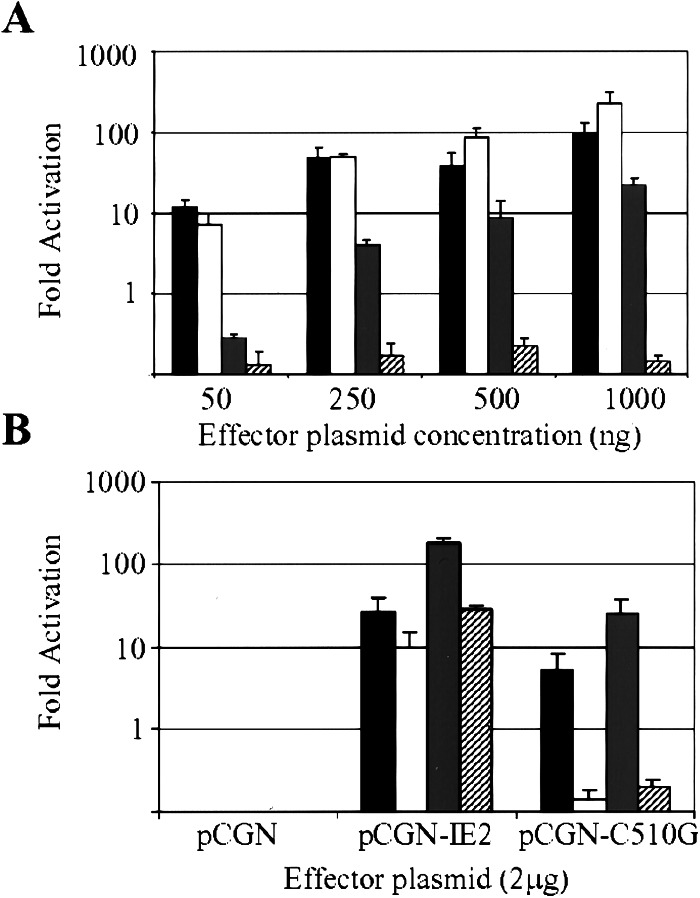
Transcriptional activation by the IE2-C510G ts protein. (A) Activation of the SV40 promoter. U-2OS cells were cotransfected with a reporter, pGL3-promoterZ (1 μg), and indicated amounts of effector, pCGN-IE2 (wt IE2) or pCGN-C510G (ts IE2) plasmids. Cells were incubated at 32.5°C or 39.5°C, and cellular lysates were prepared 24 h later. IE2 activity was measured as a function of the level of β-galactosidase present in each lysate in a β-galactosidase-linked luciferase assay. Relative light units for each sample was divided by the relative light units produced from the cotransfection of the pGL3-promoterZ plasmid and the empty expression vector, pCGN, to calculate a fold-activation. IE2 activation activity is indicated at 32.5°C (wt, solid bars; ts, shaded bars) and at 39.5°C (wt, open bars; ts, striped bars). (B) Activation of the HCMV UL112 promoter. U-2OS cells were cotransfected with 2 μg of pHM142 (solid and open bars) or pHM214 (shaded and striped bars) reporter plasmid plus 2 μg of pCGN-IE2 or pCGN-C510G. Cells then were incubated at 32.5°C (solid and shaded bars) or 39.5°C (open and striped bars). Cell lysates were collected at 24 h after transfection and assayed for luciferase activity. Experiments were performed in triplicate.
Temperature sensitivity of the viruses was tested by monitoring viral growth at 32.5°C or 39.5°C after infection of HFFs at a multiplicity of 0.01 plaque-forming unit (pfu)/cell. Virus yields were determined by plaque assay on HFFs at 32°C.
Analysis of DNA and RNA Accumulation.
For slot-blot assay of viral DNA accumulation, total DNA was prepared from infected cells and used as template in a PCR. The simian virus 40 (SV40) promoter, which controlled expression of a marker gene in both wild-type and mutant viral DNAs, was amplified for 10 rounds, and the product was applied to Hybond N+ (Amersham Pharmacia) and probed with a 32P-labeled DNA fragment corresponding to the SV40 promoter.
To determine which viral immediate-early mRNAs were expressed after infection in the presence of cycloheximide, total RNA was extracted from infected cells by using the TRIzol reagent (GIBCO), subjected to reverse transcription in the presence of [32P]dCTP, and used to probe a HCMV gene array as described previously (24).
To quantitatively measure the levels of viral DNA and mRNA, real-time PCR and reverse transcription–PCR (RT-PCR) were performed by using the iCycler iQ system (Bio-Rad). For amplification of viral DNA sequences, primers designed to amplify the SV40 promoter sequences controlling expression of the marker gene in the viruses were used. For amplification of viral RNA sequences, DNase I-treated total cellular RNA was subjected to RT-PCR. Fluorescence from incorporated SYBR Green (Applied Biosystems) was monitored at the end of each amplification cycle and continuously during the melting curves. pT-BACwt DNA was used to generate standard curves.
Results
A Cysteine-to-Glycine Substitution at Amino Acid 510 Confers a ts Phenotype on the IE2 Protein.
To produce a ts derivative of IE2 that affected the protein's ability to transactivate, cysteines were targeted for mutagenesis, as was done previously to generate a ts VP16 mutant in HSV-1 (25). Eight cysteine-to-glycine mutations were introduced individually, and mutant proteins were tested for their ability to transactivate the SV40 early promoter at 32.5°C and 39.5°C (data not shown). One mutant, C510G, exhibited a ts phenotype; it activated transcription at 32.5°C but not at 39.5°C.
Fig. 1A displays the result of a transcriptional activation assay in which increasing amounts of an effector plasmid encoding wild-type (pCGN-IE2) or mutant (pCGN-C510G) IE2 were cotransfected with a reporter plasmid (pGL3-promoterZ) in which β-galactosidase expression is controlled by the SV40 early promoter. Wild-type IE2 transactivated the SV40 promoter between 100- and 1,000-fold above the basal level of transcription at 32.5 or 39.5°C, and the level of transactivation was directly proportional to the concentration of pCGN-IE2 used in the assay. The mutant IE2 protein transactivated by a factor of >100 at 32.5°C, and the level of transactivation was again proportional to the amount of pCGN-C510G transfected. However, at 39.5°C, the mutant protein did not transactivate the reporter at any concentration of pCGN-C510G tested. Western blot assays demonstrated that wild-type and mutant IE2 proteins accumulated to similar levels after transfection at the two temperatures (data not shown).
The activity of the mutant IE2 protein also was tested on the HCMV UL112 early promoter (Fig. 1B). The reporter plasmid contained either the entire UL112 promoter, with three IE2-binding sites (pHM142), or a minimal UL112 promoter, with only the TATA box domain (pHM214; refs. 20 and 21). Wild-type IE2 transactivated both promoters at both temperatures by a factor of 100–1,000. Although mutant IE2 transactivated the UL112 promoters by a factor of 100 at 32.5°C, it failed to activate transcription at 39.5°C. Thus, the IE2-C510G substitution mutant is able to transactivate an SV40 and HCMV early promoter at 32.5°C, yet it fails to transactivate the same promoters at 39.5°C.
Incorporation of the C510G Mutation into HCMV to Produce BTNtsUL122.
The C510G mutation was introduced into the HCMV genome (Fig. 2A) by using a BAC, pT-BACwt (15). Initially, a kanamycin-resistance cassette was inserted into the UL122 ORF, deleting a 150-bp fragment that included the base pair targeted for mutagenesis. Then, the kanamycin-resistance cassette was replaced with an IE2-specific fragment containing the mutated base pair.
Figure 2.
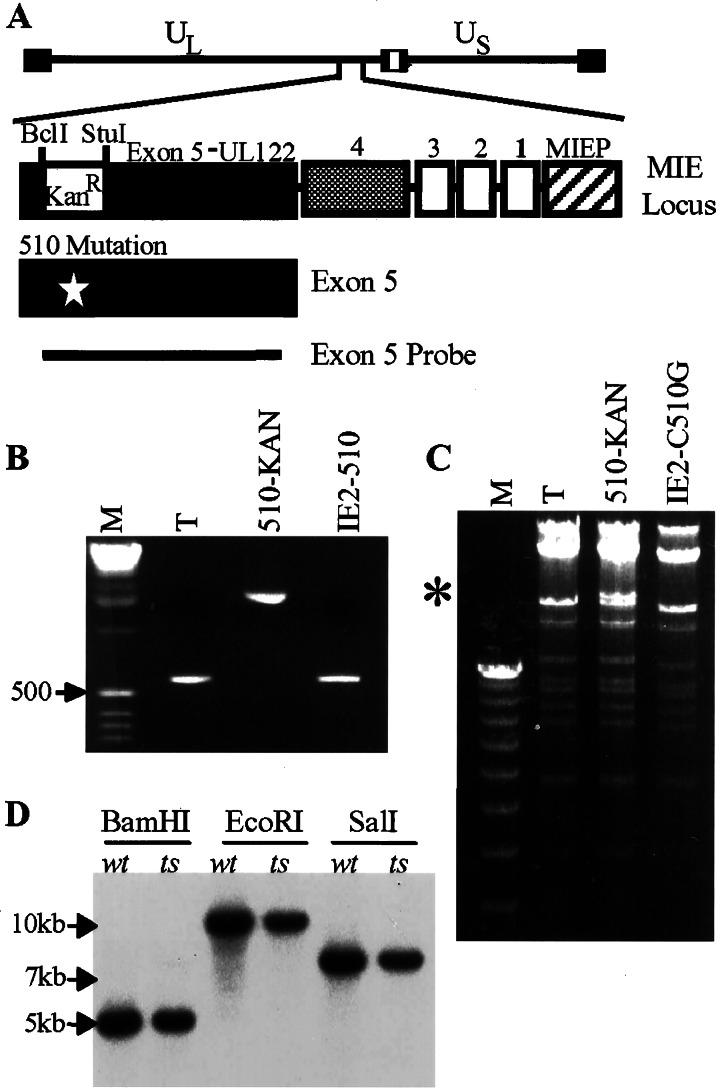
Generation and characterization of BTNtsUL122. (A) Diagram of the major immediate-early (MIE) locus and the approach used to generate the mutant virus. The major immediate-early promoter (MIEP) is shown, and the exons present in MIE mRNAs are numbered. Exon 5 corresponds to the UL122 ORF and is IE2-specific. A kanamycin-resistance (KanR) cassette was inserted into exon 5, deleting a 150-bp segment including the codon for cysteine 510. Cleavage sites (BclI and StuI) flanking the deletion are indicated. The KanR cassette was replaced subsequently with a UL122 sequence that contained the C510G substitution mutation (designated by a star). The exon 5 probe used for Southern blot assays is represented by a line. (B) PCR amplification of a UL122 segment from pT-BACwt (T), KanR insertion BAC (IE2-KAN), and IE2-C510G BAC (IE2-C510G) DNA. M, size markers. (C) BAC DNAs were digested with XbaI, and the resulting fragments were separated by electrophoresis. An asterisk designates the C510G mutation within a segment of the major immediate-early locus substituted with the KanR cassette. M, size markers. (D) Analysis of wild-type (wt) and temperature-sensitive (ts) mutant viral DNA. Infected cell DNAs were digested with indicated restriction enzymes and subjected to Southern blot analysis by using a 32P-labeled probe encompassing a portion of exon 5 as indicated in A. Expected fragment sizes detected by the exon 5 probe are as follows: BamHI, 5.2 kb; EcoRI, 10 kb; SalI, 7.4 kb.
To confirm that the alterations were introduced at the correct location in the BAC clone, a portion of UL122 was amplified by PCR (Fig. 2B). The PCR product spanned the two recombination flanks. As expected, it was 570 bp in length for the wild-type and IE2-C510G BACs, and the addition of the kanamycin-resistance cassette created a product of ≈2 kbp. The wild-type and mutant BACs also were digested with XbaI (Fig. 2C). The major immediate-early locus is contained in a 22.4-kbp fragment that comigrated with a second fragment. The insertion of the kanamycin-resistance cassette increased the size of this band, and removal of the kanamycin cassette, by substitution with a viral sequence containing the C510G mutation, returned the fragment to its original size. The remainder of the mutant BAC genome generated a restriction digest pattern indistinguishable from the wild-type.
Viruses were produced by electroporation of pT-BACwt or pIE2-C510G BAC DNA into HFF cells. It was necessary to include pCGN-510 in the electroporation mixture. This plasmid produces C510G IE2 protein, and the additional mutant protein presumably facilitated the initial phase of infection at 32.5°C within cells receiving mutant BAC DNA. To confirm the genomic structures of the resulting viruses, Southern blot analysis (Fig. 2D) was performed on DNA from cells infected with wild-type (BTNwt) or mutant (BTNtsUL122) virus, probing with a 32P-labeled UL122 fragment. Wild-type and mutant virus DNAs produced fragments of the same size, confirming that the recombination events had occurred correctly. Sequence analysis of the UL122 region from mutant virus DNA verified the presence of the C510G mutation and the absence of any other mutations in the UL122 ORF (data not shown). Immunofluorescent staining demonstrated that IE2 protein accumulated in the nucleus of BTNtsUL122-infected cells at 39.5°C (data not shown).
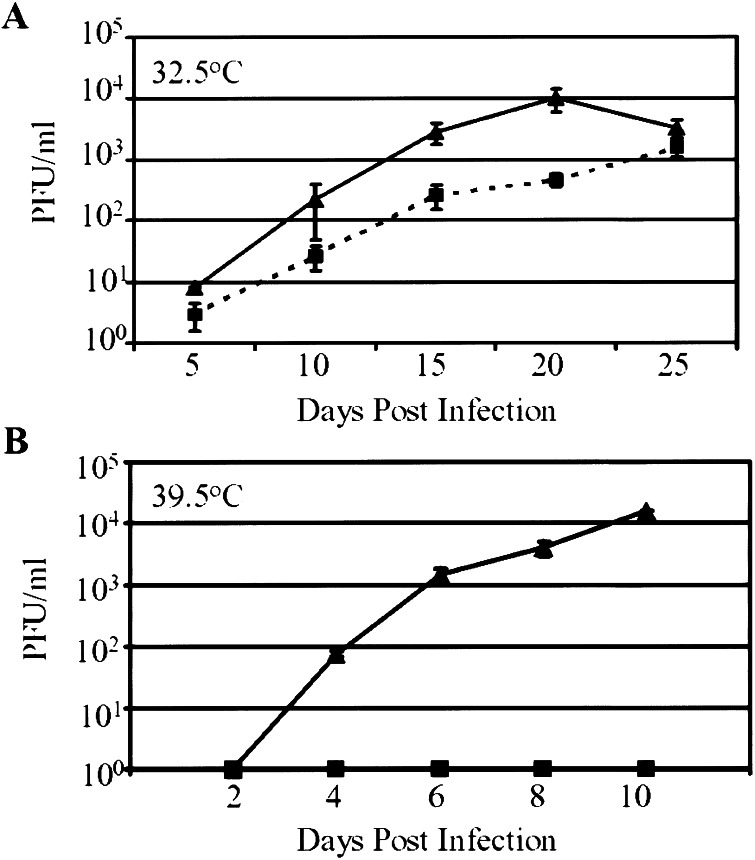
The growth of wild-type and mutant virus was examined at two temperatures. The mutant grew as well as wild-type virus at 32.5°C (Fig. 3A) but failed to produce detectable progeny at 39.5°C (Fig. 3B). In sum, the mutant virus produced from the pIE2-C510G BAC is identical to the parental wild-type virus produced from pT-BACwt, with the exception of the C510G mutation, and it displays a ts growth phenotype.
Figure 3.
Growth kinetics of viruses. HFF cells were infected at a multiplicity of 0.01 pfu/cell with wild-type (▴) or mutant (■) virus. Infected cells were incubated at either 32.5°C (A) or 39.5°C (B). Cultures were harvested at the indicated times after infection, and virus was quantified by plaque assay on HFF cells at 32.5°C. The experiment was performed in triplicate.
BTNtsUL122 Fails to Accumulate Viral DNA at 39.5°C.
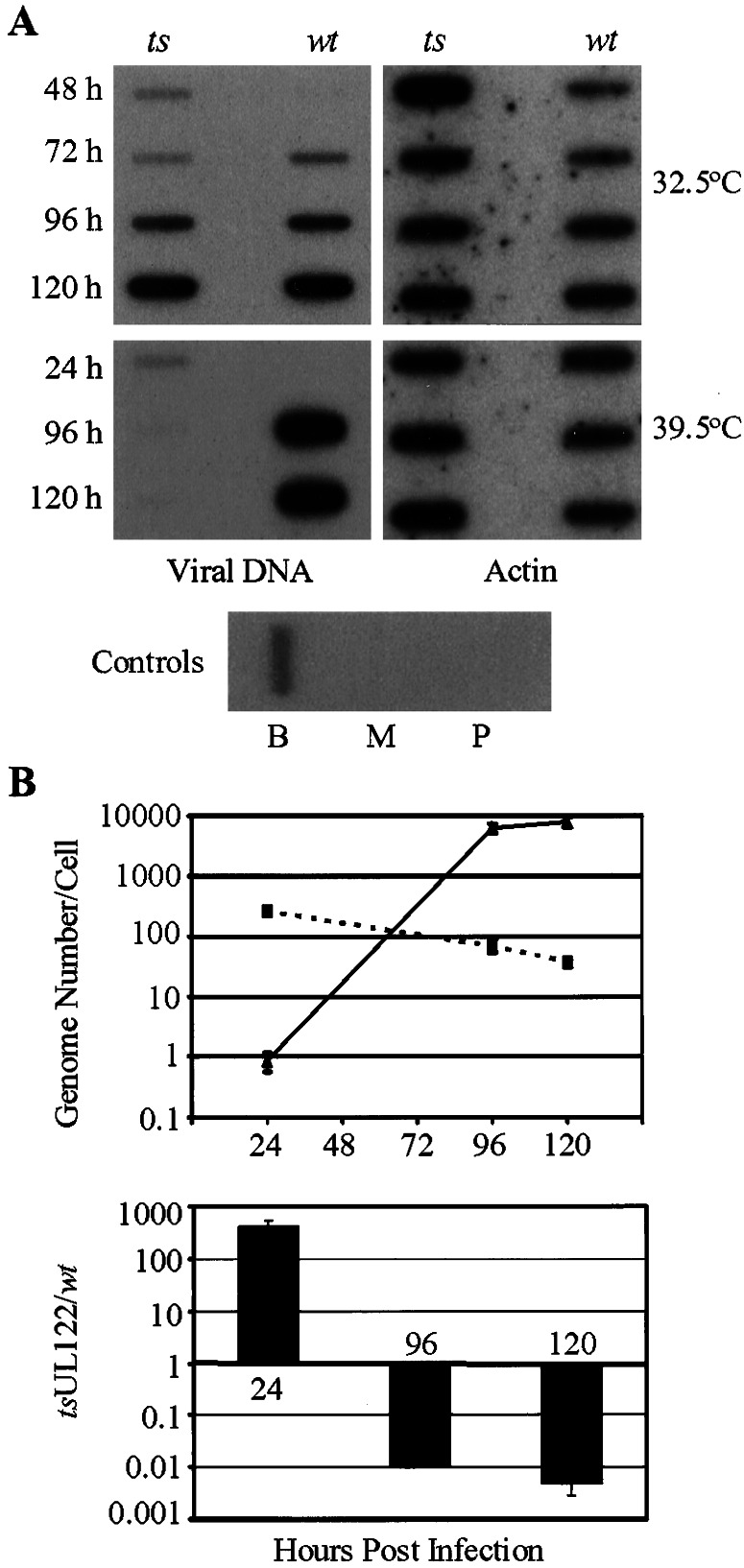
Viral DNA accumulation was examined by slot-blot analysis (Fig. 4A). Cells were infected with wild-type or mutant virus, and total cell DNA was prepared after various growth periods at 39.5°C. Because the infection was performed at a low multiplicity (0.01 pfu/cell), a limitation imposed by the low titer of the mutant virus stock (Fig. 3), a segment of viral DNA was amplified to facilitate detection. The amplification was limited to 10 cycles, which was shown to be within the linear phase of the PCR (data not shown). At 32.5°C, the wild-type and mutant accumulated viral DNA with similar kinetics, producing the same amount of DNA at 120 h, the last time assayed. Although the wild-type virus also produced DNA at 39.5°C, the mutant did not. As a control, the cellular actin gene was assayed, verifying that equivalent concentrations of DNA were used as starting template in all PCRs.
Figure 4.
Accumulation of viral DNA. (A) Slot-blot assay for accumulation of viral DNA. HFF cells were infected with either wild-type (wt) or temperature-sensitive (ts) virus at a multiplicity of 0.01 pfu/per cell and incubated at either 32.5°C or 39.5°C. DNA was extracted at the indicated times after infection. The SV40 early promoter, which controls transcription of the GFP gene in the two viruses, was amplified, and a 32P-labeled fragment of the SV40 promoter was used to detect the amplified viral DNA in a slot-blot assay. The same DNA samples were used as substrates for amplification of the cellular actin gene, which was detected by using a 32P-labeled fragment of the actin gene. Mock-infected cells (M), 0.5 μg of wild-type Towne BAC DNA (T), and a negative PCR control (P) also are shown. (B) Real-time PCR assay to quantify the genome copy number in infected cells at 39.5°C. The SV40 promoter was amplified (40 cycles) from infected-cell DNA. Accumulating PCR products were measured as a function of SYBR green fluorescence. Genome copy number then was calculated based on a standard curve of known BAC copy number. Experiments were performed in triplicate. (Upper) Calculated genome copy number for wild-type (solid line) and mutant virus (dotted line) mutant virus at various times postinfection. (Lower) Fold difference between the mutant genome copy number and the wild-type genome copy number.
At 48 h (32.5°C) and 24 h (39.5°C) after infection, slot-blot analysis detected input mutant but not wild-type virus DNA (Fig. 4A). Because equal numbers of mutant and wild-type pfus were used to infect cells, it seemed likely that the number of particles comprising a pfu was higher for the mutant than the wild-type virus. To test this possibility, real-time PCR was performed to quantify the genome copies present in infected cells (Fig. 4B Upper) and to calculate the particle/pfu ratio for the mutant virus. The HCMV genome copy number was determined by comparison with a standard curve that was generated from PCRs with known starting copy numbers of pT-BACwt DNA. Amplification of the correct PCR product was confirmed by melting-curve analysis and by visualization on an ethidium bromide-stained agarose gel (data not shown). At 39.5°C, wild-type virus-infected cells contained <1 viral genome per cell at 24 h and ≈8,000 genomes per cell at 120 h. In contrast, mutant virus-infected cells contained ≈300 genomes per cell at 24 h and only 40 genomes per cell at 120 h. This experiment confirmed that the mutant virus did not accumulate DNA at the nonpermissive temperature. Further, there are 300 times more viral genomes present in mutant as compared with wild-type infection at 24 h after infection (Fig. 4B Lower). Because viral DNA replication has not begun at this time, this DNA must represent input viral genomes. Consequently, a particle-to-pfu ratio could be estimated based on the number of genomes present at 24 h postinfection, the multiplicity of infection, and by assuming that each virus particle contains one viral genome. One pfu is equivalent to 2.8 × 104 particles for the mutant and 80 particles for the wild-type virus.
BTNtsUL122 Produces IE Viral mRNAs at 39.5°C.
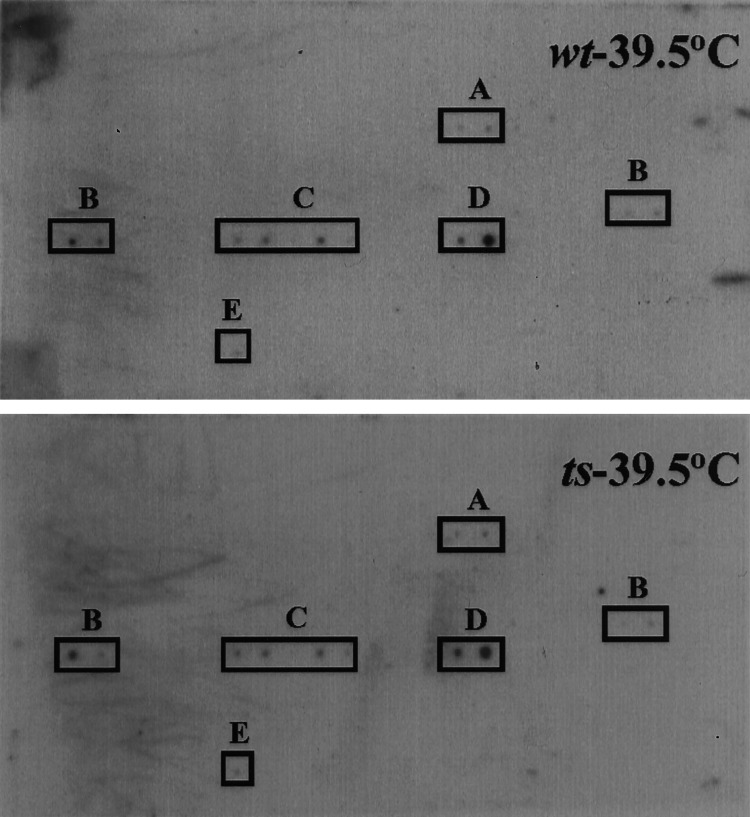
Because the mutant failed to replicate its DNA, we wanted to determine how early in the infectious cycle the block to viral replication occurred at the nonpermissive temperature. Cells were infected in the presence of cycloheximide, which permits the synthesis of only IE mRNAs, to determine whether this class of mRNA was produced by the mutant virus at 39.5°C. RNA was collected at 12 h postinfection and subjected to RT in the presence of [32P]dCTP. The labeled cDNA then was used to probe a gene array (26) that contained the ORFs predicted to be used in the Towne strain of HCMV (Fig. 5). The pattern of immediate-early mRNA expression was identical for the wild-type and mutant viruses. Both viruses expressed UL36 and 37, UL107–110, UL115–119, UL122 (IE2), and UL123 mRNAs. The cellular PLA2 transcript also was present and served as a positive control for hybridization to the array. The mutant is able to enter cells at 39.5°C and express immediate-early genes, yet it fails to initiate viral DNA replication.
Figure 5.
Immediate-early viral mRNA accumulation after infection. HFF cells were infected at a multiplicity of 0.01 pfu/cell with wild-type (wt) or temperature-sensitive (ts) mutant virus in the presence of 100 μg of cycloheximide at 39.5°C. At 12 h postinfection, total RNA was isolated and subjected to RT in the presence of [32P]dCTP, and the labeled cDNA then was used to probe an HCMV array containing 208 predicted HCMV ORFs. Transcripts expressed by the wild-type and mutant viruses are boxed and correspond to: UL36 and UL37 (A), UL107–110 (B), UL115–119 (C), and UL123 and UL122 (D). The cellular PLA2 mRNA (E) was assayed as a control.
BTNtsUL122 Fails to Express Early Viral mRNAs at 39.5°C.
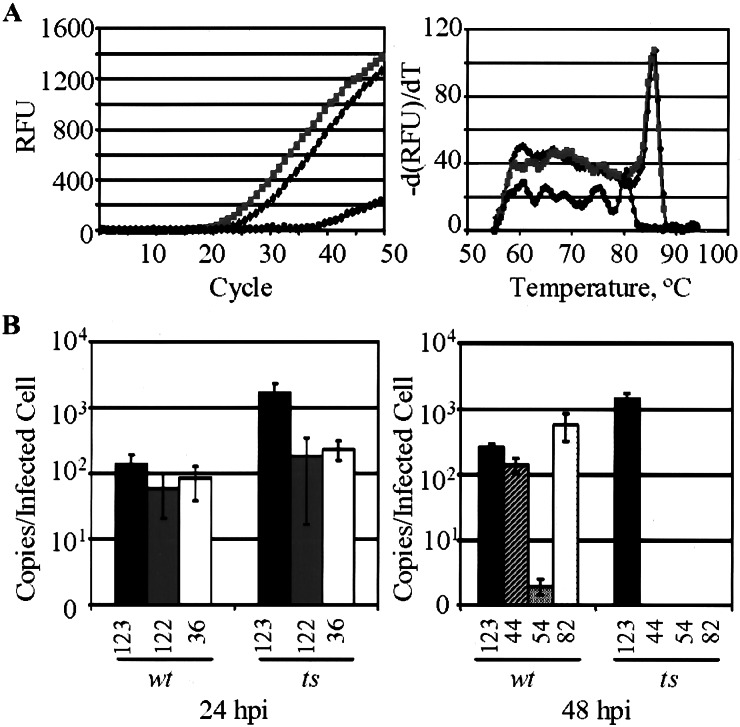
Viral mRNA accumulation was examined by real-time RT-PCR assay to confirm the expression of immediate-early mRNAs and determine whether selected early and late genes are transcribed in cells infected with the mutant virus at 39.5°C. Fig. 6A displays the results of a real-time RT-PCR assay. RNA was prepared at 24 h postinfection or mock infection, and then it was reverse-transcribed and amplified with primers specific to the UL123 ORF. Amplification was monitored for 50 cycles (Fig. 6A Left), and a melting curve of the PCR products (Fig. 6A Right) demonstrated that one specific product was produced in the amplification reaction for both wild-type and mutant samples. The amplification product in the mock-infected sample is not the same as that in infected samples; it is presumably a result of aberrant amplification of primer–dimers during the later cycles.
Figure 6.
Quantitation by real-time RT-PCR analysis of viral mRNA accumulation. HFF cells were infected with wild-type (wt) or temperature-sensitive (ts) mutant virus at a multiplicity of 0.01 pfu/cell. After 24 or 48 h at 39.5°C, total RNA was isolated, and 2-μg portions were subjected to RT and quantitative PCR. (A) Representative experiment. RNA from cells harvested at 24 h postinfection was subjected to RT and amplified in the presence of UL123-specific primers and SYBR green fluorescent dye. (Left) Amplification profile displaying relative fluorescence units (RFU) as a function of cycle number. IE1 transcript is detected in wild-type (■) and mutant (░⃞) infected cells but not mock-infected cells (●). (Right) Melting curve of the PCR products generated for wild-type (■), mutant (░⃞), and mock (●) infections. Fluorescence is plotted as a function of temperature. (B) Expression of viral transcripts after infection with mutant and wild type at 39.5°C. A constant amplicon size (300 bp) was maintained for all transcripts assayed, and all measurements were performed in triplicate. (Left) Expression levels of immediate-early viral transcripts at 24 h postinfection: UL123 (solid bars), UL122 (shaded bars), and UL36 (open bars). (Right) Expression levels at 48 h postinfection for immediate-early (UL123, solid bars; UL122, shaded bars; UL36, open bars), early (UL44, striped bars; UL54, hatched bars), and late (UL82, dotted bars) transcripts.
Fig. 6B shows the levels of various viral transcripts present during wild-type as compared with mutant virus infection at 39.5°C, measured by real-time RT-PCR. Quantities of starting template were calculated from a standard curve generated by amplification of known concentrations of BAC DNA with each primer set. All three immediate-early transcripts tested, UL36, UL122 (IE2), and UL123, were detected at 24 h postinfection with either virus (Fig. 6B Left), consistent with the viral gene array experiment (Fig. 5). The levels of immediate-early transcripts are elevated in mutant relative to wild-type virus-infected cells. This is likely a result of the delivery of more viral genomes in the BTNtsUL122 infection because of its elevated particle-to-pfu ratio (Fig. 4B). UL122 (IE2) and UL123 transcripts are elevated to a greater extent than the UL36 transcript. The mutant IE2 protein may be unable to repress transcription from the major immediate-early promoter at 39.5°C, but we have not yet tested this directly. In contrast to immediate-early transcripts, two early transcripts (UL44 and UL54) and one late transcript (UL82) were not detected at 48 h after infection with the mutant at 39.5°C, even though they accumulated to significant levels after infection with the wild-type virus (Fig. 6B Right). IE2 previously has been shown to be important for the activation of the UL44 and UL54 promoters in transfection-based reporter assays (27–29). IE2 might play a direct role in the activation of the UL82 late promoter, but it remains possible that this promoter fails to activate in the absence of DNA replication.
Discussion
A cysteine-to-glycine substitution mutation at amino acid 510 of the HCMV IE2 protein conferred a ts phenotype to its transcriptional activation function (Fig. 1), and incorporation of the mutant protein into the viral genome produced a conditionally defective virus, BTNtsUL122 (Fig. 3). Although the mutant IE2 protein expressed by BTNtsUL122 accumulated and localized to the nucleus at the nonpermissive temperature, its fluorescent pattern within the nucleus was different from that observed in the wild-type infection (data not shown). Wild-type virus produced IE2 protein that was spread throughout the nucleus with occasional bright spots. The mutant protein was not distributed uniformly. Rather, it congregated toward the center of the nucleus and exhibited a more punctate and granular staining than observed for the wild-type protein. At 39.5°C, the mutant expressed immediate-early mRNAs (Figs. 5 and 6) but failed to accumulate early mRNAs (Fig. 6) or DNA (Fig. 4). Both early mRNAs tested (UL54, DNA polymerase; UL44, processivity factor) encode products required for DNA replication (30, 31), so it is not surprising that the mutant fails to replicate its DNA.
BTNtsUL122 virions were about 300-fold less infectious than BTNwt particles (Fig. 4B). It is possible that the IE2 protein plays a direct role in the assembly of infectious virions, and the C510G mutation interferes with this function. However, we favor the view that the mutant IE2 protein is partially defective in its transcriptional activation function at the permissive temperature, and this partial defect generates a requirement for multiple particles to produce enough protein to initiate an infection. Consistent with this view, 10-fold more pCGN-C510G was needed in transfection experiments to achieve the same level of activation of a reporter gene as that produced by pCGN-IE2 at 32.5°C (Fig. 1A). Further, we were unable to generate infectious virus from BAC DNA containing the C510G IE2 mutation at 32.5°C without inclusion of pCGN-510 in the transfection mixture. Presumably, inclusion of the plasmid increased the level of the partially functional mutant protein to generate sufficient IE2 function to allow BTNtsUL122 to proceed into the early phase of infection.
Several lines of evidence anticipated that IE2 would be required for activation of early genes within infected cells. First, IE2 can activate a wide variety of reporter genes in transfection assays (4–7), including reporters controlled by early HCMV promoters (27–29). Second, Marchini et al. (15) detected immediate-early but not early mRNAs after transfection of cells with BAC DNA from which IE2-specific sequences were deleted. Third, overexpression of the UL84 protein, which binds to IE2 and antagonizes its transcriptional activation function, inhibits expression of early mRNAs in infected cells (32). Finally, a murine cytomegalovirus mutant lacking its ie3 gene failed to express early mRNAs or to replicate viral DNA in noncomplementing cells (33). This gene product appears to be a functional homologue of the HCMV IE2 protein.
The apparently absolute dependence of early gene activation on IE2 is striking. We were unable to detect any UL44 or UL54 RNA after infection with BTNtsUL122 at 39.5°C by using a very sensitive assay—real-time RT-PCR (Fig. 6). Clearly, IE2 function is required for progression from the immediate-early to early phase of infection. Possibly, IE2 functions exclusively as a transcriptional transactivator to mediate expression of early genes. Alternatively, additional functions of the protein, such as cell cycle regulation, are required for progression into the early phase. Work is in progress to further delineate the consequences of the C510G mutation on IE2 function at 39.5°C.
Acknowledgments
We thank J. Flint for thoughtful comments on the manuscript. This work was supported by Grant CA85786 from the National Cancer Institute. W.A.B. received fellowship support from the National Institute of Allergy and Infectious Disease (AI10448).
Abbreviations
- HCMV
human cytomegalovirus
- IE2
immediate-early 2 protein
- ts
temperature-sensitive
- BAC
bacterial artificial chromosome
- HFF
human foreskin fibroblast
- SV40
simian virus 40
- RT-PCR
reverse transcription—PCR
- pfu
plaque-forming unit
References
- 1.Britt B. In: Topley and Wilson's Microbiology and Microbial Infections. Collier L, editor. London: Oxford Univ. Press; 1998. pp. 340–350. [Google Scholar]
- 2.Stenberg R M. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 3.Rawlinson W D, Barrell B G. J Virol. 1993;67:5502–5513. doi: 10.1128/jvi.67.9.5502-5513.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermiston T W, Malone C L, Witte P R, Stinski M F. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staprans S I, Rabert D K, Spector D H. J Virol. 1988;62:3463–3473. doi: 10.1128/jvi.62.9.3463-3473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukac D M, Harel N Y, Tanese N, Alwine J C. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spector D H. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 8.Bonin L R, McDougall J K. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnahan W A, Albrecht T, Thompson E A. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 10.Castillo J P, Yurochko A D, Kowalik T F. J Virol. 2000;74:8028–8037. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy E A, Streblow D N, Nelson J A, Stinski M F. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiebusch L, Hagemeier C. EMBO J. 2001;20:1086–1098. doi: 10.1093/emboj/20.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Shen Y, Shenk T. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac D M, Alwine J C. J Virol. 1999;73:2825–2831. doi: 10.1128/jvi.73.4.2825-2831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchini A, Liu H, Zhu H. J Virol. 2001;75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Zhu H, Shenk T. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mocarski E S, Coucelle C T. In: Fields Virology. 4th Ed. Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 2001. pp. 2629–2674. [Google Scholar]
- 18.Tanaka M, Herr W. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 19.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, III, Kouzarides T, Martignetti J A. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 20.Arlt H, Lang D, Gebert S, Stamminger T. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang D, Gebert S, Arlt H, Stamminger T. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith G A, Enquist L W. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresnahan W A, Shenk T. Proc Natl Acad Sci USA. 2000;97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon A P, Roizman B. J Virol. 1995;69:7658–7667. doi: 10.1128/jvi.69.12.7658-7667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bresnahan W A, Shenk T. Science. 2000;288:2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- 27.Stasiak P C, Mocarski E S. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerry J A, Priddy M A, Stenberg R M. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskenderian A C, Anders D G, Stenberg R M. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pari G S, Anders D G. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pari G S, Kacica M A, Anders D G. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. J Virol. 1997;71:7048–7060. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angulo A, Ghazal P, Messerle M. J Virol. 2000;74:11129–11136. doi: 10.1128/jvi.74.23.11129-11136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]