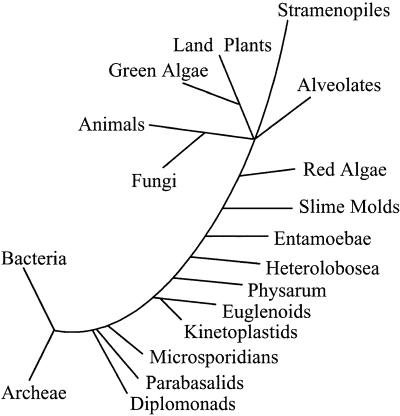
The classification of all living organisms into three major divisions, Archaea (archaebacteria), Bacteria (eubacteria) and Eukarya (eukaryotes), over two decades ago (1) has changed our view of the relationship between nucleated cells (eukaryotes) and those lacking nuclei (archaebacteria and eubacteria). The foundation for this reclassification of living organisms was originally based on molecular phylogenies constructed by using small subunit ribosomal RNAs (rRNAs) (see Fig. 1) and has since been confirmed by analyses of numerous biochemical properties, as well as the entire genome sequence of organisms within each division. Converging molecular comparisons of diverse properties of organisms within and between domains have led to the proposal that the eukaryotic cell is a hybrid arising from a host archaeal cell and eubacterial endosymbionts (2–4).
Figure 1.
Representative tree depicting the three domains of life, Bacteria, Archaea, and Eukaryotes, with emphasis on the eukaryotic branch. The tree is based on small subunit rRNA sequences originally published by Sogin (32) and was redrawn by Dacks and Doolittle (2). This tree has provided enormous intellectual stimuli upon which numerous theories have been built and tested. Construction of eukaryotic phylogenies is a continuous process being steadily fed and modified by the availability of new data in this era of genomics. As discussed by Nixon et al. (11) in this issue of PNAS, the precise nature of deep-branching, divergent lineages and their relationship to other eukaryotes continues to be challenged and debated. [Reproduced with permission from ref. 2 (Copyright 2001, Elsevier Science).]
Eukaryotes are divided into four kingdoms: Animalia, Plantae, Fungi, and Protista. The bulk of research on eukaryotes has been conducted on fungi, animals, and plants, the “crown group” organisms in rRNA trees, whereas relatively little is known about protist biology. Protists were traditionally classified by their morphology into flagellates, ciliates, amoebae, and sporozoa (5). Only a few phyla within these four protist groups have been studied in any detail, compounding our myopic view of eukaryotes. The majority of research conducted on these organisms has focused on a small number of disease-causing organisms, such as the kinetoplastid flagellates (e.g., Trypanosoma spp. and Leishmania spp.) and free-living ciliates (e.g., Tetrahymena). Although limited, these studies have served to underline the importance of casting our nets widely, should we wish to unravel more than a fraction of the biochemical secrets eukaryotic life holds. For example, RNA editing and transsplicing were first discovered in kinetoplastids; similarly, telomeres, cytoskeletal motors, and catalytic RNAs were first uncovered in ciliates.
Giardia lamblia (also known as G. intestinalis) has been the model organism for studies on diplomonads, a group of flagellated protists (6). Diplomonads and their cousins, parabasalids, represented by trichomonads, are thought to be primitive, based on their lack of typical eukaryotic organelles, such as mitochondria, peroxisomes, and nucleoli, their atypical ultrastructure features, and their basal position in rRNA phylogenetic trees (6–8; see Fig. 1). Giardia was initially described by Antoni van Leeuwenhoek in 1681, upon examining his own diarrheal stools with a homemade microscope (9). A few centuries later, the nearly completed genome sequence of the organism, containing approximately 5,800 ORFs, has become available (10). Giardia is a facultative anaerobic, obligate parasite that infects the small intestines of humans and other mammals and is a common cause of diarrhea worldwide (see Fig. 2). The many traits of Giardia that are thought to be atypical for eukaryotic cells (6, 7) have captured the attention of biologists who wish to explore the variation and define the full potential of biochemical processes. The divergence of diplomonads, as well as trichomonads (e.g., parabasalids), at the base of the eukaryotic tree has likewise spurred the interest of evolutionists interested in understanding the evolution of the eukaryotic cell.
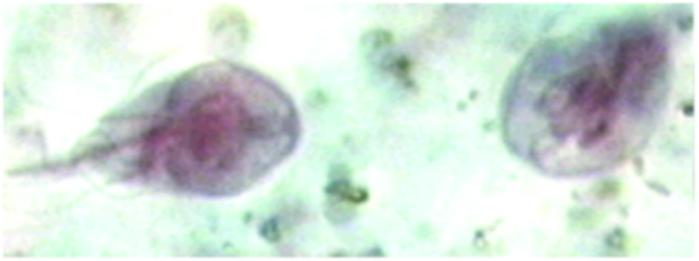
Figure 2.
Giardia lamblia is a facultative anaerobic and obligate parasite. Nixon et al. report the presence of a spliceosomal intron in the genome of Giardia. Before this report, spliceosomal introns had been observed in metazoa, yeast, and, moderately, deep-branching protists (2, 12), but not in the deepest branches of the rRNA-based eukaryotic tree. [Reproduced with permission from the Atlas of Medical Parasitology (AMP), www.cdfound.to.it (Carlo Denegri Foundation).]
Nixon et al., in this issue of PNAS (11), report the presence of a spliceosomal intron in the genome of Giardia. Before this report, spliceosomal introns had been observed in metazoa, yeast, and moderately, deep-branching protists (2, 12), but not in the deepest branches of the rRNA-based eukaryotic tree. These data now leave Parabasalids the only eukaryotic lineage from which a sizeable number of genes have been examined without the detection of introns (13). Interestingly, spliceosomal introns have not been observed in Bacteria and Archaea, although self-splicing introns which do not rely on the spliceosome for removal are found in the former (14). Giardia is, thus, the deepest-branching organism in which spliceosomal introns have been discovered.
In addition to demonstrating the presence of a single, spliceosomal intron in a putative [2Fe-2S] ferredoxin gene in Giardia, Nixon et al. (11) also describe Giardia genes which seem to encode proteins that are conserved in spliceosomes. Spliceosomes are ribonucleoparticles, composed of small nuclear RNAs (snRNAs) and nearly 100 associated proteins, which orchestrate the excision of spliceosomal introns (12, 15, 16). Genes encoding putative SM core peptides, well characterized spliceosomal proteins (12), were identified and phylogenetically analyzed. The six predicted SM peptides examined seem to have arisen by means of multiple duplication and divergence of ancestral archaeal genes, but are similar to protist, yeast, and metazoan homologues, supporting a probable role in splicing. These data imply that SM peptides had assumed their role in RNA splicing before the divergence of Giardia from the main trunk of eukaryotic descent.
Putative Giardia homologues of eukaryotic-specific spliceosomal proteins precursor RNA processing (Prp) 8 and Prp11 and several putative DexH-box RNA helicases (12), which have eubacterial homologues, were also identified. The low-sequence identity of these genes with DexH-box helicases that facilitate RNA arrangements during splicing and Prp proteins precluded the use of phylogenetic analyses to infer their identity and thus solidify their presumed role in splicing. The predicted Prp8 and Prp11 Giardia proteins have only 27% and 30% identity, respectively, with their putative counterparts in Saccharomyces cerevisiae. The low-sequence identity of the putative Prp8 protein is particularly curious, as a predicted Trichomonas Prp8 protein (17) is 48% identical to the yeast homologue, and human and yeast Prp8 proteins are 61% identical, indicating Prp8 to be one of the most conserved spliceosomal proteins. The presence of a putative Prp8 homologue in Trichomonas vaginalis is intriguing and is currently the only data available to suggest that splicing also occurs in this deep-branching protist. Functional analyses will be required to definitively determine whether putative Prp proteins in Giardia or Trichomonas are involved in splicing and, if so, whether the observed sequence divergence of either translates into mechanistic differences in the splicing reaction. The efficiency of splicing may also merit attention in Giardia. The reverse transcription–PCR data which allowed Nixon et al. (11) to identify the sequence of the 35-bp intron examined in their study indicate that less than 50% of RNA derived from the gene has undergone splicing. Whether this result is an artifact of the procedure or indicates the presence of two roughly equal populations of RNA, only one of which gives rise to the predicted [2Fe-2S] ferredoxin, remains to be determined. If two stable RNA populations do exist in vivo, does the abundant unprocessed RNA remain in the nucleus, or is it exported to the cytosol and translated? Is this RNA functional?
The single Giardia intron lacks the 5′ dinucleotide GT that is typically conserved in all known spliceosomal introns.
Introns have now been examined in detail in metazoa and yeast, and elegant in vitro splicing assays have been developed for both systems, which have allowed the essential properties of intron structure in crown group eukaryotes to be defined (12, 15, 16). Two types of introns occur in these organisms, U2-type and U12-type, which are spliced by compositionally distinct spliceosomes (18). The majority of metazoan and yeast introns are the U2-type, characterized by canonical 5′GT and 3′AG dinucleotides that mark the boundaries of the intron and directly interact with spliceosomal components. In yeast, the canonical sequence is extended to 5′GTATGT at the 5′ end of the intron and 5′A/T,T/C,AG at the 3′ end. A strictly conserved internal branch-point sequence, TACTAAC, also is found in yeast introns. The branch point of metazoan introns is marked by conserved sequences, which fit the consensus sequence CTAACT. Although similar 5′, 3′, and branch-point sequences are found in yeast and metazoan introns, yeast introns are unusual as these are generally strictly conserved, relative to metazoan introns where only two-thirds of the nucleotides may match the consensus (16). The single Giardia intron described in the paper in this issue of PNAS conforms to the consensus established for metazoan and yeast U2-type introns, except that it lacks the 5′ dinucleotide GT that is typically conserved in all known spliceosomal introns. Instead, the dinucleotide CT is present at the 5′ splice site of the Giardia intron, followed immediately by the nucleotides ATGT, which perfectly match the consensus typically found immediately downstream of the 5′ splice site (see above). Furthermore, the branch point found in the Giardia intron (AACTAAC) matches the yeast sequence (TACTAAC) in six of seven nucleotides.
A detailed characterization of additional Giardia introns will be necessary to determine whether important functional sequences are strictly conserved, as in yeast, or merely maintained as a consensus. Such studies should also reveal whether the presence of a 5′ CT, rather than GT, is typical in Giardia introns, assuming additional introns will be found. The identification and characterization of Giardia snRNAs, such as U1, U2, U4, U5, and U6, which are known to play critical roles in splicing and are generally conserved, will be essential to determine whether conventional or divergent spliceosomes exist in this protist. In this regard, it is interesting to note that two apparent classes of cis-splicing introns have been found in Euglena—those with conventional structures (19) and that that lack conventional consensus motifs (20). Whether the latter are processed by standard spliceosomes or other, undefined mechanisms is unknown.
Another interesting feature of the single Giardia intron identified is its length of only 35 nucleotides. Although short introns are found in metazoa and are, in fact, the rule in yeast and examined protists, introns less than 50 basepairs are rare. Should very short introns be common in Giardia, spliceosomal components or structure may have diverged to allow their accurate removal. So far, a limited number of introns have been examined in protists from distantly related phyla, including Toxoplasma, Plasmodium (21), Trypanosoma (in which only two cis-spliced introns have been found, both in the same gene, in two different species; ref. 22), and Entamoeba (23). Introns in protists seem to be small (often <100 nucleotides but >50 nucleotides) and scarce, similar to yeast introns. However, given the paucity of current information, caution should be applied when making generalizations about the abundance and size of introns in protists. Unlike yeast, genome projects for most protists are incomplete, and inadequate programs are available to detect introns in these databases, particularly should their sequence not conform to expectations. Recent data on introns in Entamoeba, an anaerobic, amitochondriate, gut parasite which shares many biological properties with Giardia and Trichomonas, indicate that introns are quite common in genes of this protist, contrary to previous belief (ref. 23, and J. Samuelson & B. Loftus, personal communication). Interestingly, an examination of the only published 14 introns in Entamoeba genes (23) reveals a strictly conserved 5′ sequence, 5′GTTTG,T/A, which matches the strongly conserved 5′ sequence 5′GTATGT of yeast introns in five of six positions, Likewise, Entamoeba introns are short, with an average length of 65 nucleotides. Thus, available data on protist introns, although extremely limited, suggest that introns in these organisms may be more structurally similar to their counterparts in yeast than in metazoa. Sequence information that becomes available as protist genome projects are completed should resolve the question of the abundance and size of introns in these genomes and will provide clues regarding the composition of protist spliceosomes, setting the stage for a functional comparison of splicing mechanisms in all eukaryotes by using in vitro assays.
The origin of introns has received tremendous attention from evolutionists, as it has profound implications for the origin of genes per se. The basic question is whether introns were present in the first genes of the common ancestor of all cells (the exon theory of genes or the “intron-early” hypothesis; refs. 24 and 25) and were, in fact, a necessary step in the evolution of complex proteins from much simpler peptides. In this scenario, the lack of spliceosomal introns in extant eubacteria and archaea is explained by evolutionary pressure to streamline their genomes once large, functional proteins had evolved. In contrast, the “intron-late” hypothesis argues that introns have been added to genes in eukaryotes and never existed in homologous genes from eubacteria and archaea (26). Comparison of intron number and position in phylogenetically restricted eukaryotes has provided convincing evidence for intron addition during eukaryotic evolution (27). However, this evidence does not exclude the possible procurement of introns by exon shuffling during the formation of genes. Indeed, there is ample data supporting both the intron-early and intron-late hypotheses, and the question of intron origin remains unanswered (28). Until recently, the lack of introns in particular eukaryotic lineages has been interpreted by some as evidence for the intron-late theory. Although the discovery of an intron in a Giardia gene and the prediction of introns in Trichomonas, based on the strong conservation of a Prp8 gene (17), does not lend direct support for either the intron-early or intron-late theory, these data do strongly predict that introns and a spliceosome sufficient for excision was present in the last common ancestor of extant eukaryotes. Spliceosomes may be a defining feature of eukaryotes, much like the nucleus and endomembrane and cytoskeletal apparati. A clear demonstration that putative Giardia Prp8 and Prp11 are, indeed, homologues of eukaryotic-specific spliceosomal proteins and that putative DexH-box helicases are eubacterial-derived spliceosomal proteins might argue in favor of the intron-late hypothesis, as the presence of a spliceosome composed of components derived from all three domains of life in the progenote seems unlikely. However, apparent catalytic similarities between self-splicing group II introns and spliceosomal introns have led to the suggestion that spliceosomal machinery may have diverged from RNA-based self-splicing (30). This predicts that although spliceosomal introns arose later in evolution, their mechanism of removal essentially evolved from a self-splicing mechanism which could have been present in the progenote.
The evolutionary implications of deep-branching lineages of the eukaryotic tree (Fig. 1) has recently received considerable attention (2, 29, 31). The question has arisen whether these lineages branch deeply because of long-branch attraction, an artifact that results because of a faster rate of evolution of these phyla, which attracts them to distantly related outgroups, usually Archaea. Reinterpretation of eukaryotic phylogenetic trees, using a rate-across-sites correction method, has resulted in a “big-bang hypothesis” for eukaryotic evolution, which posits that all extant eukaryotes are descendants of a sudden radiation that occurred ≈1 billion years ago (31). This hypothesis, which seems to be at odds with the geological and paleontological records (see Nixon et al., ref. 11, for detailed discussion) argues that deep-branching lineages do not reflect an early divergence from the main line of eukaryotic descent but reflect a faster rate of gene evolution. Regardless of whether these lineages are truly ancient or branch deeply in rRNA trees because of a faster rate of evolution, or both, they remain the most divergent eukaryotes examined to date and provide unparalleled opportunities to uncover innovation and conservation of essential eukaryotic mechanisms, such as splicing.
Footnotes
See companion article on page 3701.
References
- 1.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dacks J B, Doolittle W F. Cell. 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle R F. Res Microbiol. 2000;151:85–89. doi: 10.1016/s0923-2508(00)00124-8. [DOI] [PubMed] [Google Scholar]
- 4.Lake J A, Rivera M C. Proc Natl Acad Sci USA. 1994;91:2880–2881. doi: 10.1073/pnas.91.8.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine N D, Corliss J O, Cox F E G, Deroux G, Grain J, Honigberg B M, Leedale G F, Loeblich A R, Lom J, Lynn D, et al. J Protozool. 1980;27:37–58. doi: 10.1111/j.1550-7408.1980.tb04228.x. [DOI] [PubMed] [Google Scholar]
- 6.Gillin F D, Reiner D S, McCaffery J M. Annu Rev Microbiol. 1996;50:679–705. doi: 10.1146/annurev.micro.50.1.679. [DOI] [PubMed] [Google Scholar]
- 7.Adam R D. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benchimol M, Kachar B, de Souza W. Biol Cell. 1993;77:289–295. doi: 10.1016/s0248-4900(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 9.Dobell C. Proc R Soc Med. 1920;13:1–15. [PMC free article] [PubMed] [Google Scholar]
- 10.McArthur A G, Morrison H G, Nixon J E, Passamaneck N Q, Kim U, Hinkle G, Crocker M K, Holder M E, Farr R, Reich C I, et al. FEMS Microbiol Lett. 2000;189:271–273. doi: 10.1111/j.1574-6968.2000.tb09242.x. [DOI] [PubMed] [Google Scholar]
- 11.Nixon J E J, Wang A, Morrison H G, McArthur A G, Sogin M L, Loftus B J, Samuelson J. Proc Natl Acad Sci USA. 2002;99:3701–3705. doi: 10.1073/pnas.042700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staley J P, Guthrie C. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 13.Liston D R, Johnson P J. Parasitol Today. 1998;14:261–265. doi: 10.1016/s0169-4758(98)01264-2. [DOI] [PubMed] [Google Scholar]
- 14.Michel F, Ferat J L. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 15.Sharp P A, Burge C B. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 16.Burge C B, Tuschl T, Sharp P A. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 17.Fast N M, Doolittle F W. Mol Biochem Parasitol. 1999;99:275–278. doi: 10.1016/s0166-6851(99)00017-1. [DOI] [PubMed] [Google Scholar]
- 18.Hall S L, Padgett R A. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 19.Breckenridge D G, Watanabe Y, Greenwood S J, Gray M W, Schnare M N. Proc Natl Acad Sci USA. 1999;96:852–856. doi: 10.1073/pnas.96.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henze K, Badr A, Wettern M, Cerff R, Martin W. Proc Natl Acad Sci USA. 1995;92:9122–9126. doi: 10.1073/pnas.92.20.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahlgen M, Fernandez V, Chen Q, Svard S, Hagblom P. Cell. 1999;96:603–606. doi: 10.1016/s0092-8674(00)80569-3. [DOI] [PubMed] [Google Scholar]
- 22.Mair G, Shi H, Li H, Djikeng A, Aviles H O, Bishop J R, Falcone F H, Gavrilescu C, Montgomery J L, Santori M I, et al. Rna. 2000;6:163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilihoeft U, Campos-Gongora E, Touzni S, Bruchhaus I, Tannich E. Protist. 2001;152:149–156. doi: 10.1078/1434-4610-00053. [DOI] [PubMed] [Google Scholar]
- 24.Doolittle W F. Nature (London) 1978;272:581–582. [Google Scholar]
- 25.Gilbert W. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 26.Palmer J D, Logsdon J M., Jr Curr Opin Genet Dev. 1991;1:470–477. doi: 10.1016/s0959-437x(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 27.Logsdon J M, Jr, Stoltzfus A, Doolittle W F. Curr Biol. 1998;8:R560–R563. doi: 10.1016/s0960-9822(07)00361-2. [DOI] [PubMed] [Google Scholar]
- 28.de Souza S J, Long M, Klein R J, Roy S, Lin S, Gilbert W. Proc Natl Acad Sci USA. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Embley T M, Hirt R P. Curr Opin Genet Dev. 1998;8:624–629. doi: 10.1016/s0959-437x(98)80029-4. [DOI] [PubMed] [Google Scholar]
- 30.Collins C A, Guthrie C. Nat Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 31.Philippe H, Germot A, Moreira D. Curr Opin Genet Dev. 2000;10:596–601. doi: 10.1016/s0959-437x(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 32.Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]