Abstract
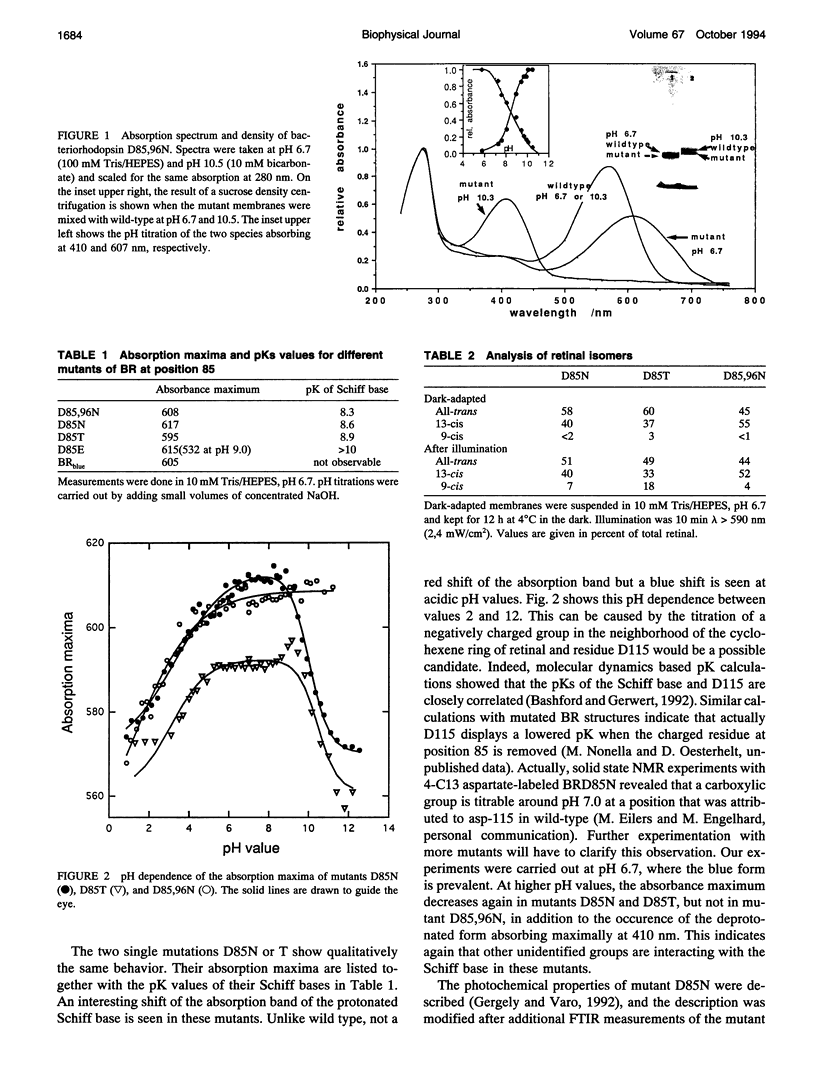
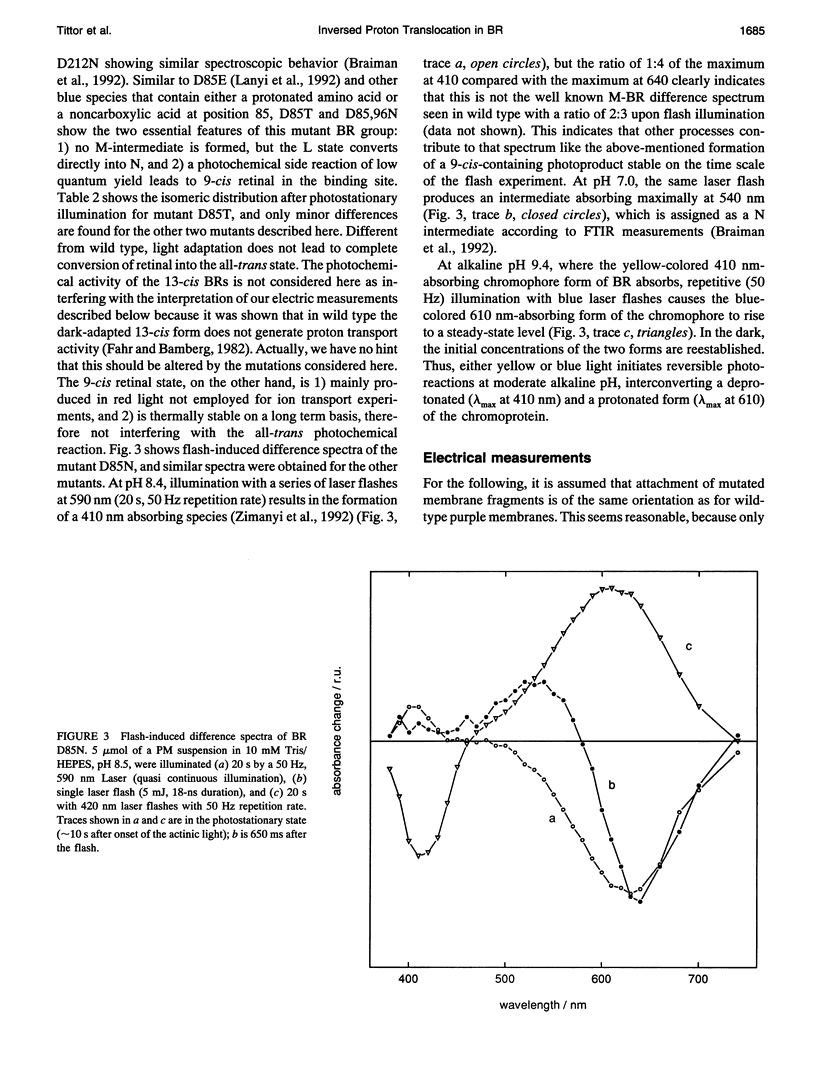
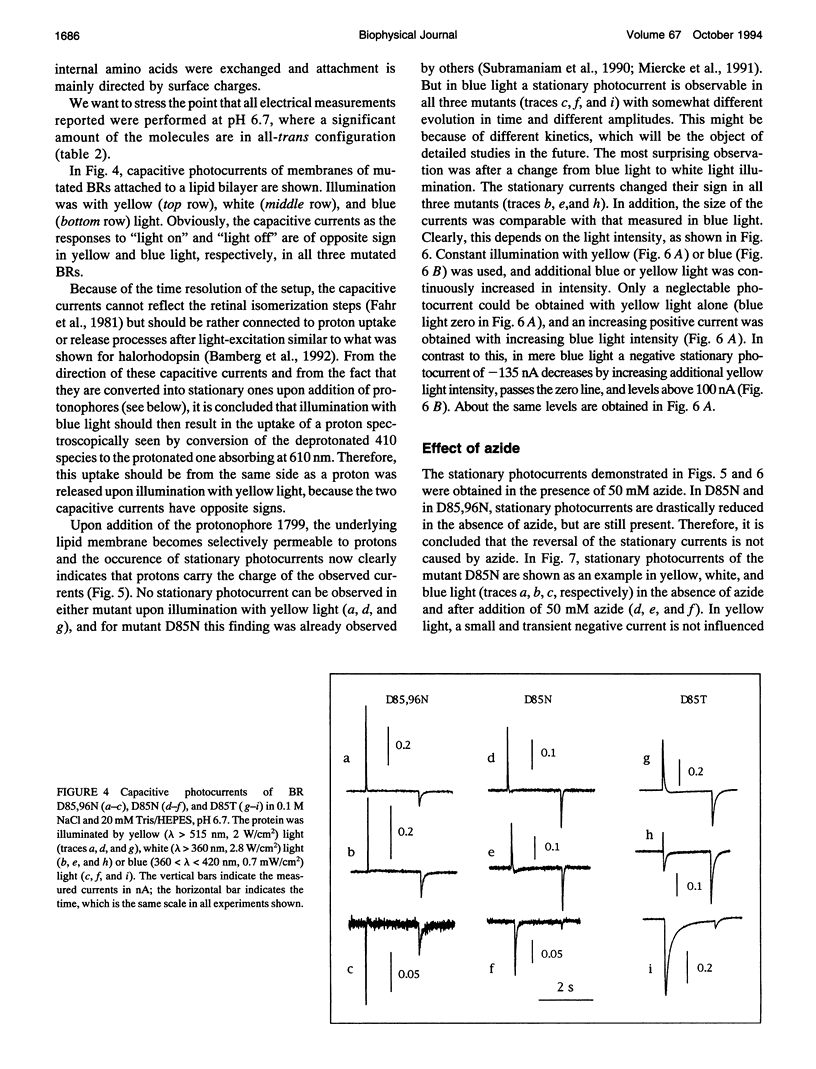
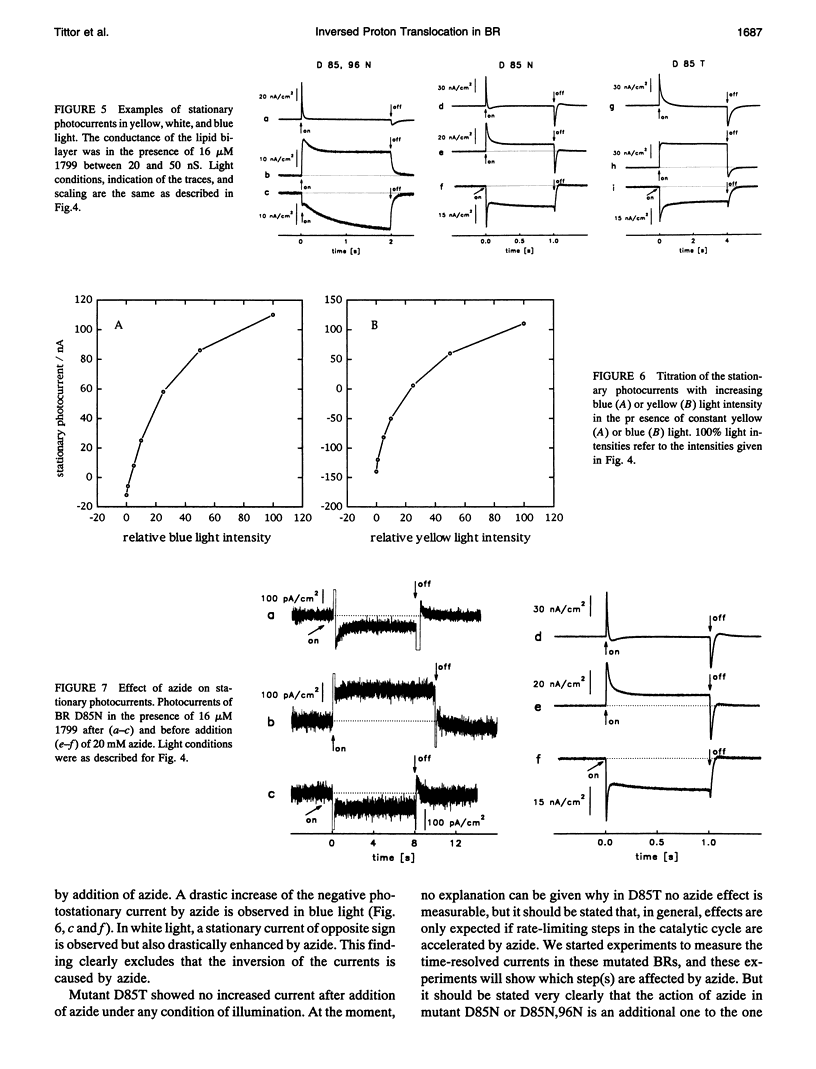
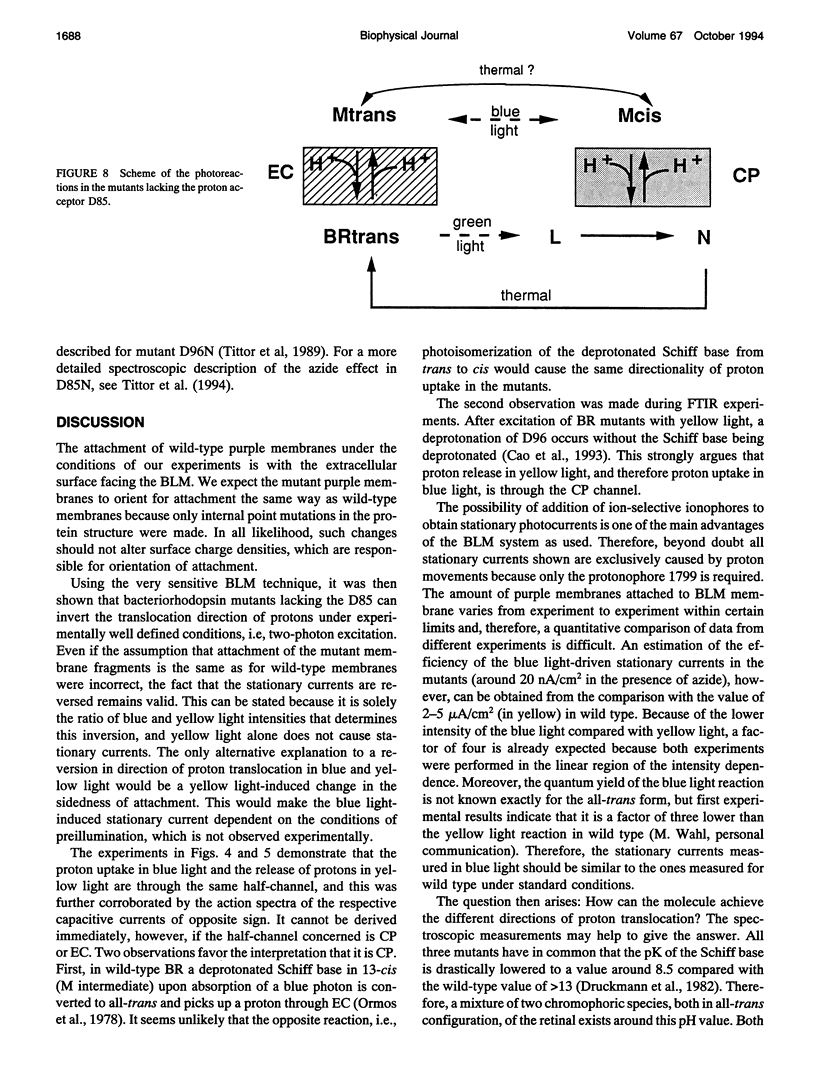
Proton translocation activity of bacteriorhodopsin mutants lacking the proton acceptor Asp-85 was investigated using the black lipid membrane technique. Mutants D85N, D85T, and D85,96N were constructed and homologously expressed in Halobacterium salinarium to yield a membrane fraction with a buoyant density of 1.18 g/cm3, i.e., identical to that of wild-type purple membrane. In all mutants, the absorbance maximum was red-shifted between 27 and 49 nm compared with wild type, and the pKa values of the respective Schiff bases were reduced to between 8.3 and 8.9 compared with the value of > 13 in wild type. Therefore, a mixture of chromophores absorbing at 410 nm (deprotonated form) and around 600 nm (protonated form) exists at physiological pH. In continuous blue light, the deprotonated form generates stationary photocurrents. The currents are enhanced by a factor of up to 50 upon addition of azide in D85N and D85,96N mutants, whereas D85T shows no azide effect. The direction of these currents is the same as in wild type in yellow light. Yellow light alone is not sufficient to generate stationary currents in the mutants, but increasing yellow light intensity in the presence of blue light leads to an inversion of the current. Because all currents are carried by protons, this two-photon process demonstrates an inverted proton translocation by BR mutants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bamberg E., Tittor J., Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Klinger A. L., Doebler R. Fourier transform infrared spectroscopic analysis of altered reaction pathways in site-directed mutants: the D212N mutant of bacteriorhodopsin expressed in Halobacterium halobium. Biophys J. 1992 Apr;62(1):56–58. doi: 10.1016/S0006-3495(92)81777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H. J., Fendler K., Bamberg E., Tittor J., Oesterhelt D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 1989 Jun;8(6):1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Váró G., Klinger A. L., Czajkowsky D. M., Braiman M. S., Needleman R., Lanyi J. K. Proton transfer from Asp-96 to the bacteriorhodopsin Schiff base is caused by a decrease of the pKa of Asp-96 which follows a protein backbone conformational change. Biochemistry. 1993 Mar 2;32(8):1981–1990. doi: 10.1021/bi00059a015. [DOI] [PubMed] [Google Scholar]
- Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C., Doolittle W. F. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989 Jan;35(1):148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Dancsházy Z., Karvaly B. Incorporation of bacteriorhodopsin into a bilayer lipid membrane; a photoelectric-spectroscopic study. FEBS Lett. 1976 Dec 15;72(1):136–138. doi: 10.1016/0014-5793(76)80829-0. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S., Ottolenghi M., Pande A., Pande J., Callender R. H. Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry. 1982 Sep 28;21(20):4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- Ferrando E., Schweiger U., Oesterhelt D. Homologous bacterio-opsin-encoding gene expression via site-specific vector integration. Gene. 1993 Mar 15;125(1):41–47. doi: 10.1016/0378-1119(93)90743-m. [DOI] [PubMed] [Google Scholar]
- Greenhalgh D. A., Subramaniam S., Alexiev U., Otto H., Heyn M. P., Khorana H. G. Effect of introducing different carboxylate-containing side chains at position 85 on chromophore formation and proton transport in bacteriorhodopsin. J Biol Chem. 1992 Dec 25;267(36):25734–25738. [PubMed] [Google Scholar]
- Hegemann P., Oesterbelt D., Steiner M. The photocycle of the chloride pump halorhodopsin. I: Azide-catalyzed deprotonation of the chromophore is a side reaction of photocycle intermediates inactivating the pump. EMBO J. 1985 Sep;4(9):2347–2350. doi: 10.1002/j.1460-2075.1985.tb03937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz M., Drachev L. A., Mogi T., Otto H., Kaulen A. D., Heyn M. P., Skulachev V. P., Khorana H. G. Replacement of aspartic acid-96 by asparagine in bacteriorhodopsin slows both the decay of the M intermediate and the associated proton movement. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2167–2171. doi: 10.1073/pnas.86.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Khorana H. G. Mechanism of light-dependent proton translocation by bacteriorhodopsin. J Bacteriol. 1993 Mar;175(6):1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K. Proton translocation mechanism and energetics in the light-driven pump bacteriorhodopsin. Biochim Biophys Acta. 1993 Dec 7;1183(2):241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K., Tittor J., Váró G., Krippahl G., Oesterhelt D. Influence of the size and protonation state of acidic residue 85 on the absorption spectrum and photoreaction of the bacteriorhodopsin chromophore. Biochim Biophys Acta. 1992 Jan 30;1099(1):102–110. [PubMed] [Google Scholar]
- Marinetti T., Subramaniam S., Mogi T., Marti T., Khorana H. G. Replacement of aspartic residues 85, 96, 115, or 212 affects the quantum yield and kinetics of proton release and uptake by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jan;86(2):529–533. doi: 10.1073/pnas.86.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies R. A., Lin S. W., Ames J. B., Pollard W. T. From femtoseconds to biology: mechanism of bacteriorhodopsin's light-driven proton pump. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- Miercke L. J., Betlach M. C., Mitra A. K., Shand R. F., Fong S. K., Stroud R. M. Wild-type and mutant bacteriorhodopsins D85N, D96N, and R82Q: purification to homogeneity, pH dependence of pumping, and electron diffraction. Biochemistry. 1991 Mar 26;30(12):3088–3098. doi: 10.1021/bi00226a016. [DOI] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Marti T., Chao B. H., Khorana H. G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B. F., Chang M., Duschl A., Lanyi J., Needleman R. An efficient system for the synthesis of bacteriorhodopsin in Halobacterium halobium. Gene. 1990 May 31;90(1):169–172. doi: 10.1016/0378-1119(90)90456-2. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Tittor J., Bamberg E. A unifying concept for ion translocation by retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- Ormos P., Dancsházy Z., Karvaly B. Mechanism of generation and regulation of photopotential by bacteriorhodopsin in bimolecular lipid membrane. Biochim Biophys Acta. 1978 Aug 8;503(2):304–315. doi: 10.1016/0005-2728(78)90190-1. [DOI] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Stern L. J., Engel F., Khorana H. G., Heyn M. P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild K. J. FTIR difference spectroscopy of bacteriorhodopsin: toward a molecular model. J Bioenerg Biomembr. 1992 Apr;24(2):147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Marti T., Khorana H. G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82----Ala and Asp-85----Glu: the blue form is inactive in proton translocation. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittor J., Soell C., Oesterhelt D., Butt H. J., Bamberg E. A defective proton pump, point-mutated bacteriorhodopsin Asp96----Asn is fully reactivated by azide. EMBO J. 1989 Nov;8(11):3477–3482. doi: 10.1002/j.1460-2075.1989.tb08512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G. J., Miercke L. J., Thorgeirsson T. E., Kliger D. S., Betlach M. C., Stroud R. M. Bacteriorhodopsin D85N: three spectroscopic species in equilibrium. Biochemistry. 1993 Feb 9;32(5):1332–1337. doi: 10.1021/bi00056a019. [DOI] [PubMed] [Google Scholar]
- Uhl R., Meyer B., Desel H. A polychromatic flash photolysis apparatus (PFPA). J Biochem Biophys Methods. 1984 Nov;10(1-2):35–48. doi: 10.1016/0165-022x(84)90048-4. [DOI] [PubMed] [Google Scholar]
- Zimányi L., Cao Y., Chang M., Ni B., Needleman R., Lanyi J. K. The two consecutive M substates in the photocycle of bacteriorhodopsin are affected specifically by the D85N and D96N residue replacements. Photochem Photobiol. 1992 Dec;56(6):1049–1055. doi: 10.1111/j.1751-1097.1992.tb09728.x. [DOI] [PubMed] [Google Scholar]



