Abstract
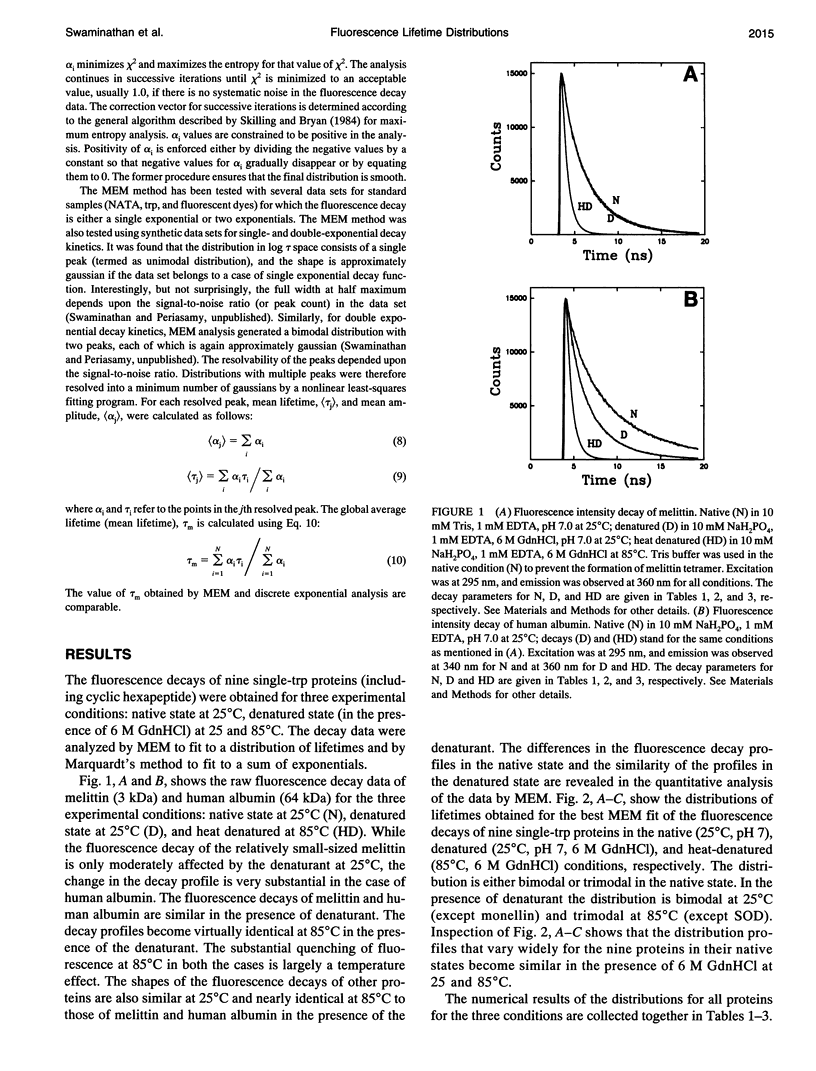
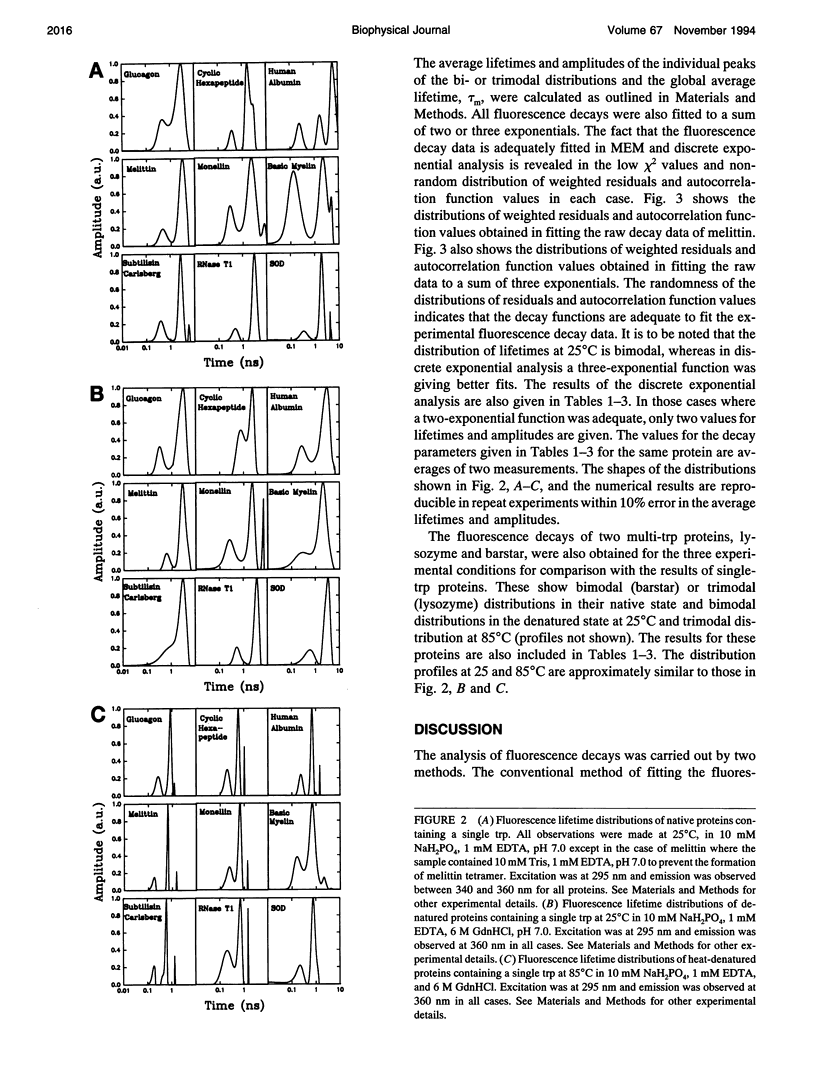
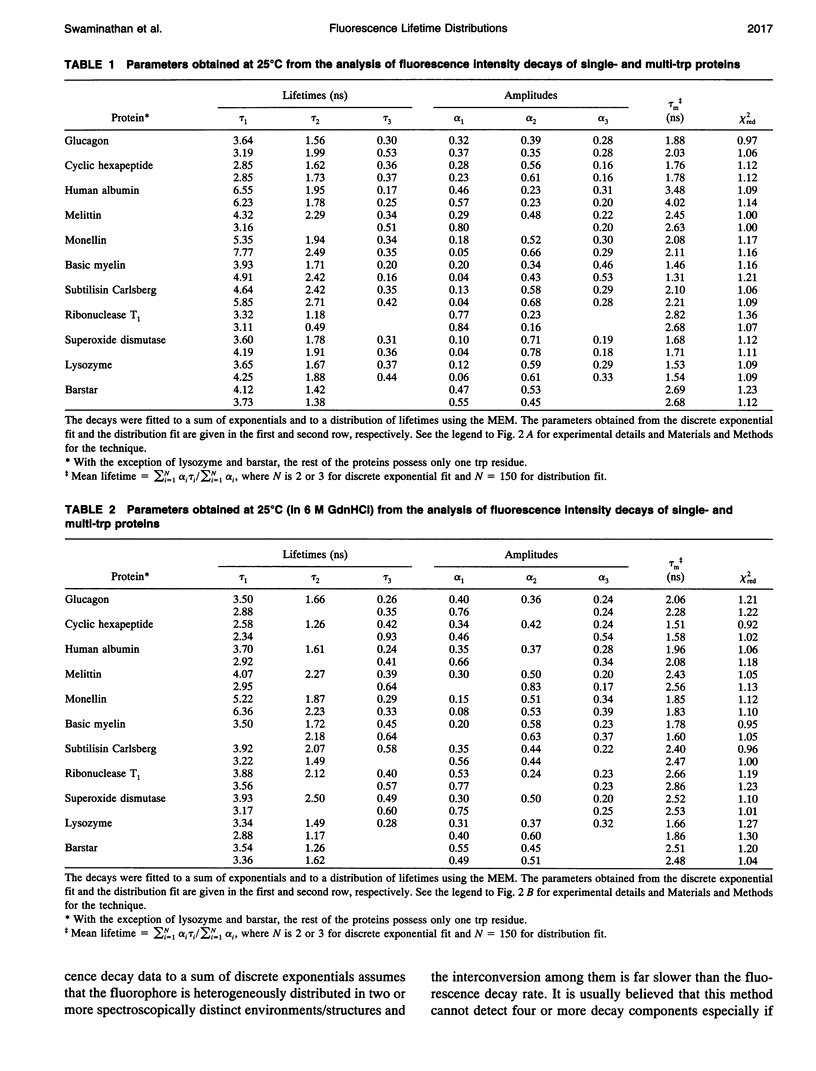
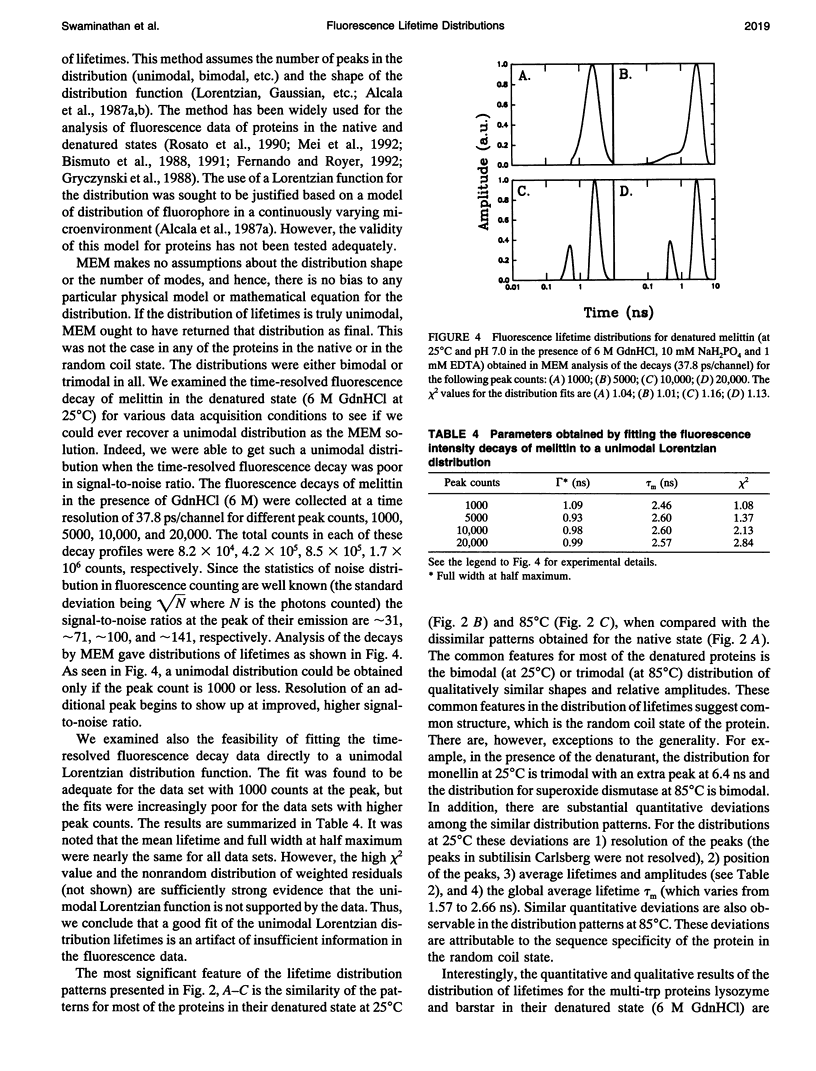
The picosecond time-resolved fluorescence decay data of nine single-tryptophan (trp) proteins and two multi-trp proteins in their native and denatured states were analyzed by the maximum entropy method (MEM). In the denatured state (6 M guanidine hydrochloride) a majority of the single-trp proteins show bimodal (at 25 degrees C) and trimodal (at 85 degrees C) distributions with similar patterns and similar values for average lifetimes. In the native state of the proteins the lifetime distributions were bimodal or trimodal. These results (multimodal distributions) are contradictory to the unimodal Lorentzian distribution of lifetimes reported for some proteins in the native and denatured states. MEM analysis gives a unimodal distribution of lifetimes only when the signal-to-noise ratio is poor in the time-resolved fluorescence decay data. The unimodal distribution model is therefore not realistic for proteins in the native and denatured states. The fluorescence decay components of the bi- or trimodal distribution are associated with the rotamer structures of the indole moiety when the protein is in the random coil state.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcala J. R., Gratton E., Prendergast F. G. Fluorescence lifetime distributions in proteins. Biophys J. 1987 Apr;51(4):597–604. doi: 10.1016/S0006-3495(87)83384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala J. R., Gratton E., Prendergast F. G. Interpretation of fluorescence decays in proteins using continuous lifetime distributions. Biophys J. 1987 Jun;51(6):925–936. doi: 10.1016/S0006-3495(87)83420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1992 May 11;20 (Suppl):2019–2022. doi: 10.1093/nar/20.suppl.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Bismuto E., Gratton E., Irace G. Effect of unfolding on the tryptophanyl fluorescence lifetime distribution in apomyoglobin. Biochemistry. 1988 Mar 22;27(6):2132–2136. doi: 10.1021/bi00406a047. [DOI] [PubMed] [Google Scholar]
- Bismuto E., Sirangelo I., Irace G. Conformational dynamics of unfolded peptides as a function of chain length: a frequency domain fluorescence approach. Arch Biochem Biophys. 1991 Nov 15;291(1):38–42. doi: 10.1016/0003-9861(91)90102-o. [DOI] [PubMed] [Google Scholar]
- Chen R. F., Knutson J. R., Ziffer H., Porter D. Fluorescence of tryptophan dipeptides: correlations with the rotamer model. Biochemistry. 1991 May 28;30(21):5184–5195. doi: 10.1021/bi00235a011. [DOI] [PubMed] [Google Scholar]
- Eftink M. R. Fluorescence techniques for studying protein structure. Methods Biochem Anal. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- Fernando T., Royer C. A. Unfolding of trp repressor studied using fluorescence spectroscopic techniques. Biochemistry. 1992 Jul 28;31(29):6683–6691. doi: 10.1021/bi00144a007. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal Biochem. 1974 Jun;59(2):583–598. doi: 10.1016/0003-2697(74)90312-1. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. The fluorescence decay of tryptophan residues in native and denatured proteins. Biochim Biophys Acta. 1976 Apr 14;427(2):663–678. doi: 10.1016/0005-2795(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Gryczynski I., Eftink M., Lakowicz J. R. Conformation heterogeneity in proteins as an origin of heterogeneous fluorescence decays, illustrated by native and denatured ribonuclease T1. Biochim Biophys Acta. 1988 Jun 13;954(3):244–252. doi: 10.1016/0167-4838(88)90079-9. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Saenger W. Specific protein-nucleic acid recognition in ribonuclease T1-2'-guanylic acid complex: an X-ray study. Nature. 1982 Sep 2;299(5878):27–31. doi: 10.1038/299027a0. [DOI] [PubMed] [Google Scholar]
- Ito A. S., Castrucci A. M., Hruby V. J., Hadley M. E., Krajcarski D. T., Szabo A. G. Structure-activity correlations of melanotropin peptides in model lipids by tryptophan fluorescence studies. Biochemistry. 1993 Nov 16;32(45):12264–12272. doi: 10.1021/bi00096a041. [DOI] [PubMed] [Google Scholar]
- Livesey A. K., Brochon J. C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys J. 1987 Nov;52(5):693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei G., Rosato N., Silva N., Jr, Rusch R., Gratton E., Savini I., Finazzi-Agrò A. Denaturation of human Cu/Zn superoxide dismutase by guanidine hydrochloride: a dynamic fluorescence study. Biochemistry. 1992 Aug 18;31(32):7224–7230. doi: 10.1021/bi00147a003. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992 Sep 11;257(5076):1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Rosato N., Gratton E., Mei G., Finazzi-Agrò A. Fluorescence lifetime distributions in human superoxide dismutase. Effect of temperature and denaturation. Biophys J. 1990 Oct;58(4):817–822. doi: 10.1016/S0006-3495(90)82427-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. B., Wyssbrod H. R., Porter R. A., Schwartz G. P., Michaels C. A., Laws W. R. Correlation of tryptophan fluorescence intensity decay parameters with 1H NMR-determined rotamer conformations: [tryptophan2]oxytocin. Biochemistry. 1992 Feb 18;31(6):1585–1594. doi: 10.1021/bi00121a002. [DOI] [PubMed] [Google Scholar]
- Vincent M., Brochon J. C., Merola F., Jordi W., Gallay J. Nanosecond dynamics of horse heart apocytochrome c in aqueous solution as studied by time-resolved fluorescence of the single tryptophan residue (Trp-59). Biochemistry. 1988 Nov 29;27(24):8752–8761. doi: 10.1021/bi00424a010. [DOI] [PubMed] [Google Scholar]
- WEBER G. Rotational Brownian motion and polarization of the fluorescence of solutions. Adv Protein Chem. 1953;8:415–459. doi: 10.1016/s0065-3233(08)60096-0. [DOI] [PubMed] [Google Scholar]


