Abstract
Interactions between paired homologous genes can lead to changes in gene expression. Such trans-regulatory effects exemplify transvection and are displayed by many genes in Drosophila, in which homologous chromosomes are paired somatically. Transvection involving the yellow cuticle pigmentation gene can occur by at least two mechanisms, one involving the trans-action of enhancers on a paired promoter and a second involving pairing-mediated bypass of a chromatin insulator. A system was developed to evaluate whether the action of the yellow enhancers in trans could be reconstituted outside of the natural near telomeric location of the yellow gene. To this end, transgenic flies were generated that carried a yellow gene modified by the inclusion of strategically placed recognition sites for the Cre and FLP recombinases. Independent action of the recombinases produced a pair of derivative alleles, one enhancerless and the other promoterless, at each transgene location. Transvection between the derivatives was assessed by the degree of interallelic complementation. Complementation was observed at all eight sites tested. These studies demonstrate that yellow transvection can occur at multiple genomic locations and indicate that the Drosophila genome generally is permissive to enhancer action in trans.
Gene expression is controlled by regulatory elements that modulate transcription in appropriate temporal and spatial patterns. In eukaryotes, these control elements often reside in large, complex regions, requiring action over long distances to elicit changes in transcription of a target promoter. In some cases, the control elements of paired homologous genes can interact in trans to alter gene expression. Such trans-regulatory effects illustrate processes known as transvection. Transvection was described first in Drosophila (1), which has homologous chromosomes that are paired somatically. Transvection and related processes have been reported in many organisms besides Drosophila (for example, refs. 2–7, and reviewed in refs. 8–11). These observations suggest that transvection may be generally possible for eukaryotic control regions and that these processes may be important for the normal regulation of some genes. For example, chromosomal pairing is proposed to play a role in gene silencing associated with imprinting and X chromosome inactivation (4, 12).
In Drosophila, interactions between homologous chromosomes can have either a positive or negative effect on transcription. Examples of negative effects include repression of white gene expression by certain alleles of zeste, trans silencing conferred by the insertion of a block of heterochromatin near the brown gene, and pairing-dependent silencing, as exemplified by the effects of Polycomb response elements (reviewed in refs. 8–11, 13, and 14). Examples of positive effects include events at the Ultrabithorax, Abdominal B, decapentaplegic, yellow, and eyes absent genes (1, 15–24).
A useful system for studying the mechanisms involved in positive trans-regulatory effects is the yellow gene, which encodes a protein responsible for the dark pigmentation of larval and adult cuticular structures. Several independent tissue-specific enhancers located 5′ of the transcription start site and within the single intron are required for yellow expression (Fig. 1 and refs. 25 and 26). A large collection of yellow mutations exists that reduce or eliminate pigmentation in some or all cuticle structures. Interestingly, combinations of certain yellow alleles that decrease pigmentation in the same tissue support complementation in a pairing-sensitive fashion such that pigmentation is restored to near wild-type levels (refs. 16, 23, and 27; S.A.O., J. R. Morris, and C.-t.W., unpublished results). Transvection at yellow involves at least two distinct mechanisms (23, 27). In one mode, the wing and body enhancers of one allele trans-activate the yellow promoter present on the paired homologous chromosome. In the second mode, pairing of two alleles alters the effectiveness of an enhancer block conferred by a chromatin insulator. These observations indicate that at least some mechanisms involved in transvection may be related directly to those governing enhancer–promoter interactions.
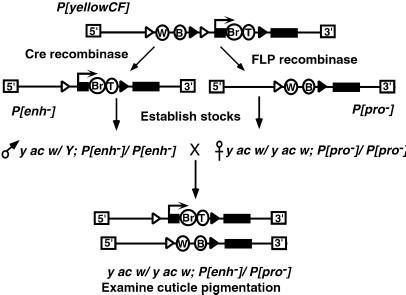
Figure 1.
Strategy for the generation of structurally distinct yellow alleles from a parental transposon. The P[yellowCF] transposon carried a yellow gene with target sites for the Cre (open arrowheads) and FLP recombinases (black arrowheads). The P element ends are shown as boxes labeled 5′ and 3′. Cre recombinase catalyzed excision of the 5′ wing and body enhancers (ovals), producing the P[enh−] derivative. FLP recombinase catalyzed excision of the promoter region (arrow) and intronic (Br, bristle; T, tarsal claw) enhancers, producing the P[pro−] derivative. Males that carried either the P[enh−] or P[pro−] allele were crossed to females that carried either the P[enh−] or P[pro−] allele, and the wing (W) and body (B) cuticle pigmentation of the resulting female progeny was examined to determine if interallelic complementation had occurred.
The features of the yellow gene required for transvection are not understood fully. In this article we address the contribution of chromosomal context in this process. The yellow gene resides ≈110 kb from the X chromosome telomere (28). Because telomeres may play a role in chromosome pairing (29–33), the natural yellow location may be important for establishing the allele proximity necessary for trans-regulatory interactions. To address this issue, we designed a system to test whether the mode of yellow transvection that depends on enhancer action in trans can occur at new sites within the genome. This system evaluated whether the genome was generally permissive to enhancer action in trans and provided insights into the extent of the yellow region required for these processes.
Materials and Methods
Plasmid Construction.
P[yellowCF] was made by cloning a 15.6-kb fragment containing the yellow coding region into the P element transformation vector, Carnegie 4 (34). This fragment included ≈7.5 kb of 5′- and 2.1 kb of 3′-flanking DNA and the recombination target sites for the bacteriophage P1 Cre recombinase (lox P sites) and the yeast FLP recombinase (FRT sites). Direct repeats of lox P sites, located at −2,880 and −900 bp relative to the transcription start site, flanked the yellow wing and body enhancers, whereas direct repeats of FRT sites, located at positions −900 and +1,625 bp relative to the transcription start site, flanked the yellow promoter (Figs. 1 and 2). The lox P and FRT sites were isolated from the p[SFL] plasmid [a gift of Dan Hartl and Mark Siegal, Harvard University (35)]. Restriction mapping confirmed the orientation and position of the recombination target sites in the P[yellowCF].
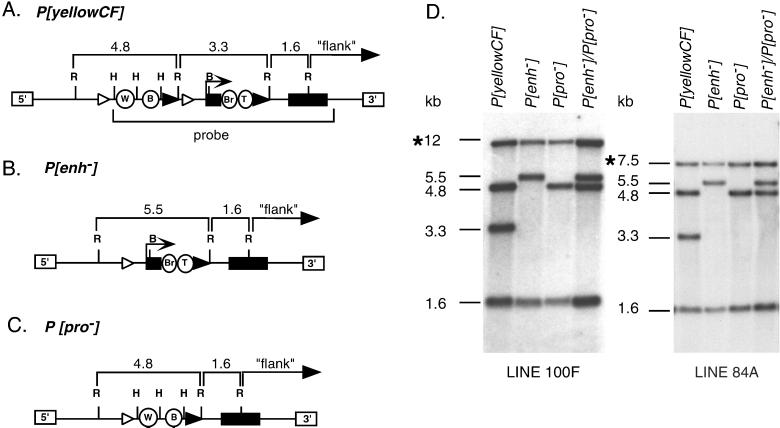
Figure 2.
Southern analysis of P[yellow CF] and derivative lines. The restriction maps of the parental P[yellowCF] (A), P[enh−] (B), and P[pro−] (C) transposons are shown, with the expected sizes of the EcoRI (R) fragments noted above the map. Other enzyme sites shown are HindIII (H) and BamHI (B). W, wing; B, body; Br, bristle; T, tarsal claw. Genomic DNA from two transgenic lines (100F and 84A) was isolated and digested with EcoRI (D). Southern analysis was completed by using a 7.7-kb yellow fragment (marked probe in A). The two lines differ in the size of the flanking band (*), with 100F showing a 12-kb band and 84A showing a 7.5-kb band. The sizes of other bands are shown at the left of each blot.
Germ-Line Transformation.
Germ-line transformation used the y− ac− w1118 host strain that carries a deletion of the natural X-linked yellow gene (25). Unless otherwise noted, all genotypes used in this study carried this y− allele. Embryos of this strain were transformed with 0.5 μg/ml of P[yellowCF] and 0.2 μg/ml of the “wings clipped” helper plasmid pπ 25.7 (36, 37). Transformants were identified by the dark pigmentation of cuticular structures (Table 1). Additional lines were made by mobilization of the P[yellowCF] using a chromosomal source of transposase P[ry+Δ2-3] (99B; ref. 38). Only P[yellowCF] lines in which the P element insertion was homozygous-viable were studied further. Altogether, eight P[yellowCF] homozygous-viable lines with a single P[yellowCF] transposon were studied. The cytological location of each P[yellowCF] transposon was determined by in situ hybridization using the entire yellow gene as a probe (39). Of the lines analyzed, three were obtained by germ-line transformation (92A, 100F, and 102B) and five from mobilization (5B, 18C, 47A, 84A, and 90F).
Table 1.
Pigmentation scores for P[yellowCF] transformants and derivatives
| Insertion site | Cuticle pigmentation score (wing, body)
|
|||
|---|---|---|---|---|
| P[yellowCF] | P[enh−] | P[pro−] | P[enh−]/P[pro−] | |
| 5B | 5, 5 | 2+, 3 | 1, 1 | 4, 4 |
| 18C | 5, 5 | 3, 3+ | 1, 1 | 4, 4 |
| 47A | 5, 5 | 2, 2+ | 1, 1 | 3+, 4 |
| 84A | 5, 5 | 2, 3 | 1, 1 | 4+, 4+ |
| 90F | 5, 5 | 2+, 3 | 1, 1 | 4, 4+ |
| 92A | 5, 5 | 2, 2 | 1, 1 | 4, 4 |
| 100F | 5, 5 | 3, 3 | 1, 1 | 4, 4 |
| 102B | 5, 5 | 3, 3 | 1, 1 | 4+, 4+ |
Pigmentation levels are reported for heterozygous P[yellowCF] (column 2) or derivative females (columns 3 and 4) and for P[enh−]/P[pro−] females (column 5). In all cases, flies carried a deletion of the endogenous yellow gene. Pigmentation scores may vary depending on culture conditions. The scores listed are those most frequently obtained.
Creation of Derivative Alleles.
Enhancerless derivatives of P[yellowCF], referred to as P[enh−], were obtained by crossing females carrying P[yellowCF] to males that contained a constitutively active Cre recombinase transgene (P[w+, Cre], kindly supplied by Dan Hartl and Mark Siegel). Male y− ac− w1118/Y; P[yellowCF]/+; P[w+, Cre]/+ progeny that displayed mutant pigmentation of wing and body tissue were selected. This phenotype indicated that somatic excision of the wing and body enhancers had occurred and suggested that these males contained a deletion event within the germ line. The mutant males were mated to y −ac− w1118/y −ac− w1118; T (2,3)apXa/CyO, MKRS females, in which CyO and MKRS are balancer chromosomes for the second and third chromosomes and carry the dominant Cy and Sb markers, respectively. T (2,3)apXa is a translocation between the second and third chromosomes and carries a dominant apterous mutation (apXa). Progeny with mutant wing and body pigmentation were selected, and homozygous P[enh−] stocks were established.
Promoterless derivatives of P[yellowCF], referred to as P[pro−], were generated by crossing P[yellowCF] females to males that carried a heat shock-inducible source of FLP recombinase (70FLP, kindly supplied by Dan Hartl and Mark Siegel). Embryos aged 15–39 h were subjected to a 1–2-h 37°C heat shock, and adult male progeny with a variegated cuticle phenotype were selected. This phenotype reflected somatic deletion of the yellow promoter from the P[yellowCF] transgene and indicated that these males were likely to have similar germ-line excision events. Variegated males were mated as described above for the P[enh−] derivatives, and homozygous P[pro−] lines were established.
Southern analyses were undertaken to ascertain whether the structures of the P[yellowCF] and derivative alleles were as predicted. Genomic DNA was isolated from adult flies as described by Ashburner (40), digested with EcoRI, separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled random-primed 7.7-kb SalI fragment that encompasses the yellow transcription unit (Fig. 2).
Complementation Tests.
All crosses were conducted at 25°C and 70% humidity on standard corn meal/agar medium. Flies carrying a P[enh−] insertion were crossed to flies carrying a P[pro−] insertion, and the progeny were scored for wing and body pigmentation to determine whether transgenes at ectopic loci could support interallelic complementation. Wing refers to wing blades, and body refers to pigmentation in the abdominal stripes, not the interstripe abdominal cuticle or thoracic cuticle. In general, three females were mated with four to five males, and crosses were transferred every 3 days. Temperature and crowding were monitored carefully, because both affect pigmentation levels. Pigmentation in the wing and body cuticle was scored in 3- to 4-day-old females by using a five-point pigmentation scale (27). A pigmentation score of 1 represents the null or nearly null phenotype, and 5 represents the wild-type or near wild-type state. Pigmentation scores were assigned by comparing progeny to flies obtained from parallel controls. Intermediate scores were determined relative to the pigment levels of y82f29/y1#8 and y2/y1#8 females, which in this study corresponded to scores of 3,3 and 4,4 in the wing and body, respectively. Pigmentation scores were determined in both the Wu and Geyer laboratories and represent an analysis of 20–30 female flies from each of at least three independent crosses. Even under these well defined conditions, pigmentation scores varied slightly between progeny obtained in different crosses. Scores listed in the tables represent the most common phenotype observed. Complementation between derivatives was judged to occur if the pigmentation score of the P[enh−]/P[pro−] females was at least one point higher than that obtained for females heterozygous for the P[enh−] allele.
Results and Discussion
We assessed whether yellow transvection involving enhancer action in trans could occur outside of the natural location of the yellow gene to address the broad question of how generally permissive the genome is to trans-regulatory inputs. Previous studies at yellow demonstrated that enhancer action in trans occurred when the yellow promoter in cis was deleted or compromised (16, 23, 27, 41). By using this information, a method was developed to generate chromosomes bearing structurally distinct yellow alleles at the same ectopic genomic site such that one chromosome carried an allele that lacked its promoter, which presumably would allow its enhancers to activate in trans, whereas the second chromosome carried an allele that lacked its enhancers.
The parental transposon used in our studies was P[yellowCF] (Fig. 1). This transposon was introduced into flies that carried a deletion of the endogenous yellow gene, and eight homozygous-viable transgenic lines were established (Table 1). Six lines had insertions within the euchromatic portion of the chromosomal arms (5B, 18C, 47A, 84A, 90F, and 92A), one line had an insertion close to a telomere (100F), and one line had an insertion on the fourth chromosome (102B), which contains interspersed euchromatic and heterochromatic domains (42). Flies from all lines displayed near wild-type levels of cuticular pigmentation, with scores of 5 in both wing and body (Fig. 3 and Table 1).
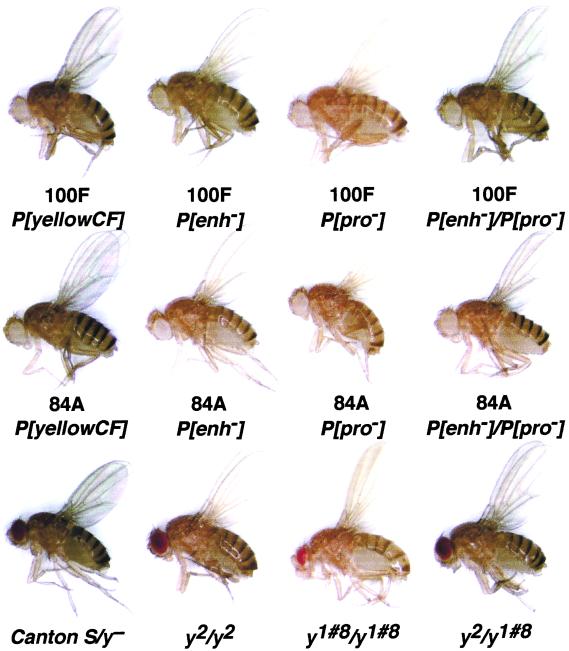
Figure 3.
Body and wing phenotypes of P[yellowCF] flies and derivatives. The top two rows show representative flies from two parental P[yellowCF] transgenic lines (100F and 84A), the corresponding P[enh−]/+ and P[pro−]/+ flies and complementing P[enh−]/P[pro−] flies. The bottom row shows representative flies that have wild-type (Canton S/y−) or mutant (y2/y2 and y1#8/y1#8) wing and body pigmentation and a representative fly that displays interallelic complementation (y2/y1#8).
Southern analysis was undertaken to confirm the structural integrity of the eight P[yellowCF] lines. Southern blots of the DNA samples were hybridized with a yellow fragment that detected only sequences corresponding to the P[yellowCF] transposon, because the endogenous yellow gene was deleted. In each line, four bands of hybridization were observed. Three of the bands were shared among all lines, because they correspond to restriction fragments internal to the P element ends (Fig. 2D). The sizes of these shared bands were as predicted, indicating that each line carried an intact P[yellowCF] transposon. The fourth band was distinctive for each, because it corresponded to DNA sequences from the 3′ end of the P[yellowCF] transposon and a variable amount of flanking genomic DNA (Fig. 2 A and D). The presence of a single variable band confirmed that each P[yellowCF] line studied carried a single insertion of the transposon.
Enhancerless derivatives were obtained by crossing P[yellowCF] flies with flies carrying a Cre recombinase transgene and selecting progeny with reduced cuticular pigmentation (Fig. 1). Heterozygous P[enh−] flies had pigmentation scores ranging from 2 to 3 in the wing and body (Fig. 3 and Table 1). The levels of cuticle pigmentation in P[enh−] flies were compared with those of y82f29 flies, which carry a deletion of the wing and body enhancers of the yellow gene at its natural location (23). We found that heterozygous P[enh−]/+ flies had levels of wing and body pigmentation that were higher than those of y82f29 flies, which had scores of 1,1+ in wing and body, respectively (23). These data suggest that although the major wing and body enhancers were deleted from the P[enh−] allele, low levels of yellow expression still occur in these tissues, presumably because of sequences within the transgene and/or the surrounding chromatin. Studies in other laboratories have indicated that the expression of structurally altered yellow genes can be sensitive to the state of nearby sequences (43, 44). Nonetheless, P[enh−] flies had greatly reduced wing and body cuticle coloration when compared with P[yellowCF] flies, and this reduction allowed distinction of interallelic complementation between derivative alleles.
Promoterless derivatives were obtained by crossing P[yellowCF] flies with flies carrying an FLP recombinase transgene and selecting progeny with reduced cuticular pigmentation (Fig. 1). Pigmentation of all heterozygous P[pro−]/+ flies was reduced to 1 in both the wing and body cuticle (Fig. 3 and Table 1). These scores were indistinguishable from those observed for flies carrying a yellow null allele, y1.
Southern analyses of the enhancerless and promoterless derivative lines were carried out. Only changes in the profile of the bands shared among all P[yellowCF] lines was seen. P[enh−] derivatives had a 5.5-kb band of hybridization that replaced the 4.8- and 3.3-kb bands because of the loss of an EcoRI site in the Cre-induced deletion (Fig. 2B). P[pro−] derivatives retained the 4.8-kb band of hybridization but lost the 3.3-kb band, because the latter fragment resided entirely between the FRT sites (Fig. 2C). These Southern analyses confirmed that the structures of the P[pro−] and P[enh−] derivatives were as predicted. We conclude that the action of the recombinases did not alter the genomic location of any derivative allele, because the size of the unique flanking bands remained unchanged (Fig. 2D).
Flies carrying a P[enh−] allele at one ectopic site were crossed with flies carrying the corresponding P[pro−] allele at the same site, and the level of wing and body pigmentation in the female progeny was assessed to determine whether interallelic complementation could occur (Fig. 1). At all eight genomic sites, P[enh−]/P[pro−] females had levels of wing and body pigmentation that were darker than those of females heterozygous for either the P[enh−] or the P[pro−] allele (Fig. 3 and Table 1). In lines in which the derivative alleles were located on the autosomes, P[enh−]/P[pro−] males also had dark wing and body cuticles, with levels of pigmentation slightly lower than wild-type (data not shown).
Southern analyses of DNA isolated from P[enh−]/P[pro−] flies confirmed that these darkly pigmented flies carried both derivative alleles, because the observed pattern of hybridization was a composite of that seen for P[enh−] and P[pro−] flies (Fig. 2D). Importantly, this pattern did not include the 3.3-kb band, which is diagnostic of the presence of the parental P[yellowCF] transposon, indicating that the observed increase in pigmentation resulted from complementation, not contamination. From these data we conclude that the trans action of the yellow wing and body enhancers on a paired promoter is possible at ectopic locations.
For reference, the pigmentation levels in P[enh−]/P[pro−] females were compared with females carrying complementing yellow alleles at the natural location. These studies used the y2, y1#8 and y1 alleles. The y2 allele is a tissue-specific mutation caused by the insertion of a gypsy retrotransposon between the yellow promoter and the wing and body enhancers. Because gypsy carries a chromatin insulator that blocks enhancer-promoter communication, y2 flies have wing and body pigmentation scores of 1,1+, respectively (45). The y1#8 allele carries a 780-bp deletion of the promoter region and is a null allele. Flies carrying this allele have scores of 1 in the wing and body (46). The y1 allele carries a mutation in the translation start codon, is a null allele, and also gives scores of 1 in both the wing and body (16). Flies carrying both the y2 and y1#8 alleles show complementation, with near wild-type levels of wing and body pigmentation (scores of 4,4; Fig. 3). In contrast, flies carrying the y2 and y1 alleles show no complementation, displaying low levels of wing and body pigmentation (scores of 1+,1+). The level of pigmentation observed in P[enh−]/P[pro−] flies was similar to that observed in y2/y1#8 flies (Fig. 3).
Transvection effects depend on apposition of the interacting genes. In Drosophila, this proximity is most commonly brought about by chromosomal pairing. A variety of genetic tests indicate that loci showing transvection effects differ in their sensitivity to chromosomal rearrangements that are believed to disrupt local pairing (1, 15, 18–21, 47–50). Whereas transvection at some loci can be disrupted by rearrangement break points up to half a chromosome arm away (1, 15, 18), transvection at other loci can occur even in the presence of break points within the interacting regions (19–21). In these cases, it has been proposed that an extensive region of the interacting genes is involved in the capture of regulatory elements (19–21). Furthermore, some homologous sequences, on the order of a few kilobases or less, inserted at different chromosomal locations still retain the ability to alter gene expression, presumably through a pairing-mediated mechanism (reviewed ref. 14). Particularly relevant to our studies is the observation that two transgenes located in different chromosomal positions, one carrying the miniwhite gene and the other carrying the white eye enhancer, supported gene activation (51). Although this activation was observed in only a fraction of the chromosomal sites tested, it indicates that enhancers can activate promoters even when located in nonhomologous genomic sites. Taken together, these data suggest that homologous sequences can have a surprising ability to find each other and pair in the context of a complex genome.
To examine the capacity of yellow sequences to find each other, we asked whether yellow enhancers located at one chromosomal site could activate a yellow promoter located elsewhere. First we assessed whether the y2 allele at its natural location complemented promoterless alleles at five ectopic chromosomal locations (Table 2). Importantly, each allele used in these tests supports transvection when it is able to pair with another yellow allele at a homologous chromosomal site (Fig. 3 and Table 1), implying that any lack of complementation with an allele at a nonhomologous site cannot be attributed to an intrinsic inability of these transgenes to support transvection. In all cases, the y2/y−; P[pro−]/+ flies displayed noncomplementing levels of wing and body pigmentation (Table 2), demonstrating that the ectopic P[pro−] derivatives failed to support transvection with y2. Next we determined whether a P[enh−] allele and a P[pro−] allele from different ectopic sites supported transvection. In these studies, P[pro−] males corresponding to seven ectopic sites were mated to P[enh−] females from these locations. Once again, complementation was not observed (Table 2). These data indicate the importance of allele proximity for yellow transvection.
Table 2.
Complementation scores of P[yellowCF] derivatives inserted at the same or different genomic locations
| Genotype | Cuticle pigmentation score (wing, body)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| y ac w (1B) | P[pro−] 18C | P[pro−] 47A | P[pro−] 84A | P[pro−] 90F | P[pro−] 92A | P[pro−] 100F | P[pro−] 102B | |
| y2 (1B) | 1, 1+ | 1, 1+ | 1, 1+ | ND | 1, 1+ | 1, 1+ | 1, 1+ | ND |
| P[enh−] 18C | 3, 3+ | 4, 4 | 3, 3+ | 3, 3 | 3, 3+ | 3, 3 | 2, 3 | 3, 3 |
| P[enh−] 47A | 2, 2+ | 2+, 2 | 3+, 4 | 2+, 2 | 2+, 2+ | 2, 2 | 2, 2+ | 2, 2 |
| P[enh−] 84A | 2, 3 | 2+, 2+ | 2+, 2+ | 4+, 4+ | 2+, 3 | 2+, 2+ | 2+, 3 | 2+, 2+ |
| P[enh−] 90F | 2+, 3 | 2+, 3 | 2+, 3 | 2+, 2+ | 4, 4+ | 2, 2+ | ND | 2+, 3 |
| P[enh−] 92A | 2, 2 | 2, 2 | 2+, 2+ | 2, 2 | 2, 2 | 4, 4 | 2, 2 | 2, 2 |
| P[enh−] 100F | 3, 3 | 3, 3 | 3, 3+ | ND | 3, 3 | 3, 3 | 4, 4 | 3, 3 |
| P[enh−] 102B | 3, 3 | ND | 3, 3+ | 3, 3 | 3, 3+ | 3, 3 | 3, 3+ | 4+, 4+ |
In these experiments, pigmentation scores are reported for crosses between P[pro−] males and P[enh−] females. Complementing scores are shown in bold. Crosses carried out in the opposite direction gave the same results. ND, not determined.
There are many possible reasons why transvection between nonhomologous sites was not observed. First, yellow regulatory sequences may have difficulty locating each other. In previous circumstances where interactions occurred between nonhomologous sites, alterations of gene expression occurred most frequently when the interacting pair was located on the same chromosome arm (51, 52). In our study, even though three of the tested nonhomologous sites met this criterion (90F, 92A, and 100F), transvection was not seen. Second, if trans-activation of the yellow promoter by the wing and body enhancers requires prolonged and/or intimate interactions between these regulatory elements, then it may be that nonhomologous sites cannot establish the required conditions. Third, the complementation we observed at homologous sites may have depended partially or fully on pairing forces provided by the flanking chromosomal regions. If this is the case, then the lack of sufficiently strong pairing functions within the yellow transgenes and/or the tenacious pairing of the flanking regions may have precluded productive pairing between nonhomologous sites and prevented transvection. Fourth, our pigmentation assay may not be sensitive enough to detect low level complementation between nonhomologous sites. Finally, it is possible that the chromosomal organization within the nucleus may impose constraints on the extent of trans-regulatory interactions exerted between enhancerless and promoterless pairs at nonhomologous sites (53, 54).
In summary, a method was introduced to address whether the Drosophila genome is generally permissive to positive trans-regulatory inputs. Recently, a similar approach addressed whether homologue pairing that influences gene expression could occur in transgenic tobacco (7). In that study, it was found that in certain circumstances, pairing of an intact gene with a homologue lacking its enhancer can increase gene expression, possibly caused by enhancer action in trans. In our studies, we found that yellow transvection between enhancerless and promoterless alleles was reconstituted at eight of eight ectopic sites. These interactions are believed to involve pairing, because interallelic complementation was not observed between yellow alleles at nonhomologous sites. These data indicate that the natural location of the yellow gene is not essential for trans-regulatory interactions and imply that enhancer action in trans may be permitted throughout the genome. Finally, this paper demonstrates that the Cre-FLP system can be used to identify critical sequences that control the trans action of enhancers in vivo, leading to a better understanding of the mechanisms governing enhancer–promoter communication.
Acknowledgments
We thank Mark Siegel and Dan Hartl for P[w+, Cre] and 70FLP fly stocks and the p[SFL] plasmid. We thank Jim Morris, Timothy Parnell, and Robin Roseman for useful discussions, technical assistance, and critical reading of the manuscript. This work was supported by National Institutes of Health Grants GM42539 (to P.K.G.) and GM61936 (to C.-t.W. and P.K.G.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lewis E B. Am Nat. 1954;88:225–239. [Google Scholar]
- 2.Dunaway M, Droge P. Nature (London) 1989;341:657–659. doi: 10.1038/341657a0. [DOI] [PubMed] [Google Scholar]
- 3.Aramayo R, Metzenberg R L. Cell. 1996;86:103–113. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 4.LaSalle J M, Lalande M. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 5.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duvillie B, Bucchini D, Tang T, Jami J, Paldi A. Genomics. 1998;47:52–57. doi: 10.1006/geno.1997.5070. [DOI] [PubMed] [Google Scholar]
- 7.Matzke M, Mette M F, Jakowitsch J, Kanno T, Moscone E A, van Der Winden J, Matzke A J. Genetics. 2001;158:451–461. doi: 10.1093/genetics/158.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S, Comai L. Cell. 1998;93:329–332. doi: 10.1016/s0092-8674(00)81161-7. [DOI] [PubMed] [Google Scholar]
- 9.Pirrotta V. Biochim Biophys Acta. 1999;1424:M1–M8. doi: 10.1016/s0304-419x(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 10.Wu C-t, Morris J R. Curr Opin Genet Dev. 1999;9:237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 11.Kennison J A, Southworth J W. Adv Genet. 2002;46:399–420. doi: 10.1016/s0065-2660(02)46014-2. [DOI] [PubMed] [Google Scholar]
- 12.Marahrens Y. Genes Dev. 1999;13:2624–2632. doi: 10.1101/gad.13.20.2624. [DOI] [PubMed] [Google Scholar]
- 13.Pirrotta V. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 14.Kassis J A. Adv Genet. 2002;46:421–438. doi: 10.1016/s0065-2660(02)46015-4. [DOI] [PubMed] [Google Scholar]
- 15.Gelbart W M. Proc Natl Acad Sci USA. 1982;79:2636–2640. doi: 10.1073/pnas.79.8.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer P K, Green M M, Corces V G. EMBO J. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Laborda A, Gonzalez-Reyes A, Morata G. EMBO J. 1992;11:3645–3652. doi: 10.1002/j.1460-2075.1992.tb05449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiserson W M, Bonini N M, Benzer S. Genetics. 1994;138:1171–1179. doi: 10.1093/genetics/138.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson J E, Sakonju S. Genetics. 1995;139:835–848. doi: 10.1093/genetics/139.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopmann R, Duncan D, Duncan I. Genetics. 1995;139:815–833. doi: 10.1093/genetics/139.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sipos L, Mihaly J, Karch F, Schedl P, Gausz J, Gyurkovics H. Genetics. 1998;149:1031–1050. doi: 10.1093/genetics/149.2.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casares F, Bender W, Merriam J, Sanchez-Herrero E. Genetics. 1997;145:123–137. doi: 10.1093/genetics/145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris J R, Chen J-L, Geyer P K, Wu C-t. Proc Natl Acad Sci USA. 1998;95:10740–10745. doi: 10.1073/pnas.95.18.10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerman J E, Bui Q T, Liu H, Bonini N M. Genetics. 2000;154:237–246. doi: 10.1093/genetics/154.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geyer P K, Corces V G. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 26.Martin M, Meng Y B, Chia W. Mol Gen Genet. 1989;218:118–126. doi: 10.1007/BF00330574. [DOI] [PubMed] [Google Scholar]
- 27.Morris J R, Chen J, Filandrinos S T, Dunn R C, Fisk R, Geyer P K, Wu C-t. Genetics. 1999;151:633–651. doi: 10.1093/genetics/151.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 29.Cockell M, Gasser S M. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 30.Rockmill B, Roeder G S. Genes Dev. 1998;12:2574–2586. doi: 10.1101/gad.12.16.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung J C, Marshall W F, Dernburg A, Agard D A, Sedat J W. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donaldson K M, Karpen G H. Genetics. 1997;145:325–337. doi: 10.1093/genetics/145.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker M Y, Hawley R S. Chromosoma. 2000;109:3–9. doi: 10.1007/s004120050407. [DOI] [PubMed] [Google Scholar]
- 34.Rubin G M, Spradling A C. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegal M L, Hartl D L. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 37.Karess R E, Rubin G M. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 38.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim J K. Drosophila Inf Serv. 1993;72:73–76. [Google Scholar]
- 40.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 41.Morris J R, Geyer P K, Wu C-t. Genes Dev. 1999;13:253–258. doi: 10.1101/gad.13.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun F L, Cuaycong M H, Craig C A, Wallrath L L, Locke J, Elgin S C. Proc Natl Acad Sci USA. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgiev P, Tikhomirova T, Yelagin V, Belenkaya T, Gracheva E, Parshikov A, Evgen'ev M B, Samarina O P, Corces V G. Genetics. 1997;146:583–594. doi: 10.1093/genetics/146.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikhailovsky S, Belenkaya T, Georgiev P. Chromosoma. 1999;108:114–120. doi: 10.1007/s004120050358. [DOI] [PubMed] [Google Scholar]
- 45.Corces V G, Geyer P K. Trends Genet. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- 46.Geyer P K, Richardson K L, Corces V G, Green M M. Proc Natl Acad Sci USA. 1988;85:6455–6459. doi: 10.1073/pnas.85.17.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jack J W, Judd B H. Proc Natl Acad Sci USA. 1979;76:1368–1372. doi: 10.1073/pnas.76.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smolik-Utlaut S M, Gelbart W M. Genetics. 1987;116:285–298. doi: 10.1093/genetics/116.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golic M M, Golic K G. Genetics. 1996;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gubb D, Roote J, Trenear J, Coulson D, Ashburner M. Genetics. 1997;146:919–937. doi: 10.1093/genetics/146.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigrist C J, Pirrotta V. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henikoff S. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 54.Marshall W F, Sedat J W. Results Probl Cell Differ. 1999;25:283–301. doi: 10.1007/978-3-540-69111-2_14. [DOI] [PubMed] [Google Scholar]