Abstract
Mutations in oncogenes and tumor suppressor genes are critical in the development of cancer. A major pathway for the formation of mutations is the replication of unrepaired DNA lesions. To better understand the mechanism of translesion replication (TLR) in mammals, a quantitative assay for TLR in cultured cells was developed. The assay is based on the transient transfection of cultured cells with a gapped plasmid, carrying a site-specific lesion in the gap region. Filling in of the gap by TLR is assayed in a subsequent bioassay, by the ability of the plasmid extracted from the cells, to transform an Escherichia coli indicator strain. Using this method it was found that TLR through a synthetic abasic site in the adenocarcinoma H1299, the osteogenic sarcoma Saos-2, the prostate carcinoma PC3, and the hepatoma Hep3B cell lines occurred with efficiencies of 92 ± 6%, 32 ± 2%, 72 ± 4%, and 26 ± 3%, respectively. DNA sequence analysis showed that 85% of the bypass events in H1299 cells involved insertion of dAMP opposite the synthetic abasic site. Addition of aphidicolin, an inhibitor of DNA polymerases α, δ, and ɛ, caused a 4.4-fold inhibition of bypass. Analysis of two XP-V cell lines, defective in DNA polymerase η, showed bypass of 89%, indicating that polymerase η is not essential for bypass of abasic sites. These results suggest that in human cells bypass of abasic sites does not require the bypass-specific DNA polymerase η, but it does require at least one of the replicative DNA polymerases, α, δ, or ɛ. The quantitative TLR assay is expected to be useful in the molecular analysis of lesion bypass in a large variety of cultured mammalian cells.
The formation of mutations is a key event in several important biological phenomena, most notably cancer and evolution. A major mechanism for the formation of mutations is the replication of DNA lesions that have escaped DNA repair (1). For many years the molecular mechanism of this process, termed translesion replication (TLR), translesion synthesis, or lesion bypass, was not clear. Recently it was discovered that the main enzyme responsible for carrying out lesion bypass is a novel type of DNA polymerase, specialized for replicating across damaged sites in DNA. Such DNA polymerases were found in organisms ranging from Escherichia coli (DNA polymerase V; refs. 2 and 3) to humans (e.g., DNA polymerase η; refs. 4 and 5). These novel DNA polymerases comprise the new Y superfamily of DNA polymerases (6). Remarkably, humans contain four members of the Y family: DNA polymerases η (4, 5), ι (7, 8), and κ (9–12) and hREV1, which has dCMP transferase activity (13, 14). In addition, humans are likely to have an additional polymerase involved in TLR, polymerase ζ, product of the hREV3 and hREV7 genes (refs. 15 and 16; for review see refs. 17–23).
Despite the importance of TLR mechanisms in understanding the role of mutagenesis in cancer, and despite the progress made with the discovery of Y family DNA polymerases, little is known about the mechanism of lesion bypass in mammalian cells. This is partly because of the scarcity of suitable assays at the cellular level. Here we describe the development of a quantitative TLR assay for cells in culture, and its use in the analysis of bypass through an abasic site.
Materials and Methods
Materials.
Restriction nucleases, T4 DNA ligase, and T4 polynucleotide kinase were from New England Biolabs. RPMI 1640 culture medium, Dulbecco's PBS without calcium chloride and magnesium chloride, MEM with Earle's salts and essential and nonessential amino acid, FBS, and aphidicolin were from Sigma. DMEM and a mixture of penicillin and streptomycin for cell culture were from GIBCO/BRL.
DNA.
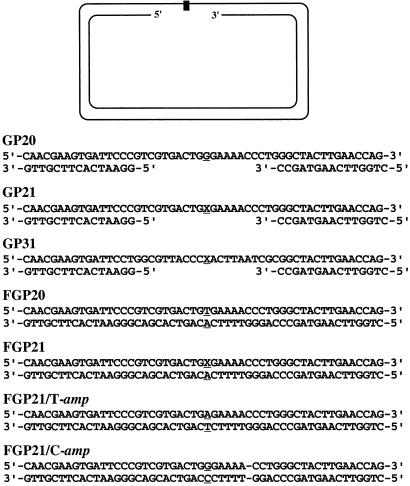
All oligonucleotides were synthesized and purified by the Synthesis Unit of the Biological Services Department of the Weizmann Institute of Science. Oligonucleotides containing an abasic site analog were synthesized similarly by using dSpacer CE phosphoramidite (Glen Research, Sterling, VA) as a building block (24). The construction of the gap-lesion plasmids GP21 and GP31 and the gapped plasmid GP20 (without the lesion; Fig. 1) has been described (25–27). Plasmids FGP21 and FGP20 are similar to GP21 and GP20, respectively, except that they are fully double-stranded. They were prepared similarly to GP21 and GP20, except that the insert oligonucleotides contained no gaps (Fig. 1). FGP21/T-amp and FGP21/C-amp are ampicillin-resistant, kanamycin-sensitive derivatives of FGP21/T and FGP21/C, respectively, intact plasmid isolates that were obtained in TLR experiments with GP21. FGP21/T contains a T opposite the location corresponding to the abasic site, whereas FGP21/C contains a C in this site. In addition, FGP21/C contains a specific marker, namely a deletion of a GC base pair six bases downstream to the site in which the abasic site was originally located (Fig. 1). Each of these plasmids was digested with HindIII and XhoI, to delete a 520-bp fragment from the kan gene, and then the plasmids were ligated to a 945-bp fragment carrying PCR-generated termini with XhoI and HindIII sites, and the bla gene from plasmid pUC18, conferring ampicillin resistance. Plasmid pSA26 (3,356 bp long; cmR) is the ligation product of the EcoRV–HincII fragment (2,712 bp long) from plasmid pACYC184 and the HincII–BglI fragment (644 bp long) from plasmid pOC2 (28).
Figure 1.
Structure of the gap-lesion plasmids and related constructs. (Upper) A schematic picture of a gap-lesion plasmid. The various constructs differ in the sequence in the vicinity of the lesion, which is shown (Lower) for each plasmid. The black rectangle (Upper) and X (Lower) represent the synthetic abasic site.
Cell Cultures.
The human cell lines used in this study were H1299, derived from a large cell lung carcinoma (29); Saos-2, an osteosarcoma-derived cell line (30); PC3, a prostate carcinoma-derived cell line (31); Hep3B, a hepatoma cell line (32); and GM03617 (XP30RO) (33) and GM03618 (XP6DU) (34) fibroblast cell lines derived from XP-V patients. H1299 cells were cultured in RPMI 1640 medium, whereas GM03618 and GM03617 cell lines were cultured in MEM, with Earle's salts and essential and nonessential amino acids. Cells from the other cell lines were maintained in DMEM. Each medium was supplemented with 10% FBS (except for GM03618 cells, which were supplemented with 20% FBS), 10 units/ml of penicillin, and 10 μg/ml of streptomycin. The cells were incubated at 37°C in a 5% CO2 atmosphere. The cell lines were obtained from M. Oren, Weizmann Institute of Science, except for the XP-V cell lines GM03617 and GM03618 that were purchased from Coriell Cell Repositories, Camden, NJ.
In Vivo TLR Assay.
The TLR assay involves the following steps: (i) Transfection of the human cells with a plasmid mixture containing the gap-lesion plasmid (GP21 or GP31; kanR), the internal control plasmid pSA26 (cmR), and the carrier plasmid pUC18. (ii) Extraction of the plasmids from the human cells. (iii) Transformation of an E. coli indicator strain with the plasmid mixture. (iv) Deduction of the extent of lesion bypass repair from the number of transformants. Both the gapped plasmid and plasmid pSA26 were purified to a high degree by using quantitative agarose gel electrophoresis. The protocol was as follows. Human cells were cotransfected with a DNA mixture containing 200 ng of gapped plasmid, 200 ng of the internal control plasmid pSA26, and 10 μg of the carrier plasmid pUC18, using the calcium phosphate method (35). The cells were incubated in their appropriate medium for 20 h at 37°C or 32°C in 5% CO2 atmosphere. At the end of incubation the cells were collected and their plasmid content was extracted by using a plasmid purification kit based on alkaline lysis (Promega). For further purification the DNA was ethanol-precipitated and redissolved in 20 μl of 10 mM Tris⋅HCl, 1 mM EDTA, pH 7.5. Next, 15 μl of the DNA solution obtained was added to 150 μl of E. coli JM109recA competent cells. After a 1-min incubation on ice the mixture was transferred to a cold 0.2-cm electroporation cell (Bio-Rad), and one pulse of 12.5 kV/cm with a time constant of 3.5–6 ms was applied. The cells were then incubated in 1 ml SOC medium at 37°C for 1 h. Transformants were detected by plating the cells in parallel on LB-agar plates containing kanamycin (50 μg/ml) or chloramphenicol (30 μg/ml). For each transfection, the relative normalized amount of recovered plasmids was calculated by dividing the number of GP21* or GP20* transformants (number of colonies on kanamycin plates; asterisks refer to the indicated plasmid after recovery from mammalian cells) by the number of corresponding transformants of the internal standard plasmid pSA26* (number of colonies on chloramphenicol plates). The percentage of lesion bypass repair was calculated by dividing the relative normalized amount of GP21* (gapped plasmid with lesion) by the relative normalized amount of GP20* (gapped plasmid without lesion). When desired, plasmids were extracted from kanR colonies, and the sequence opposite the lesion was determined by automated DNA sequencing analysis in the Biological Services Department of the Weizmann Institute of Science.
Results
Outline of the TLR Assay.
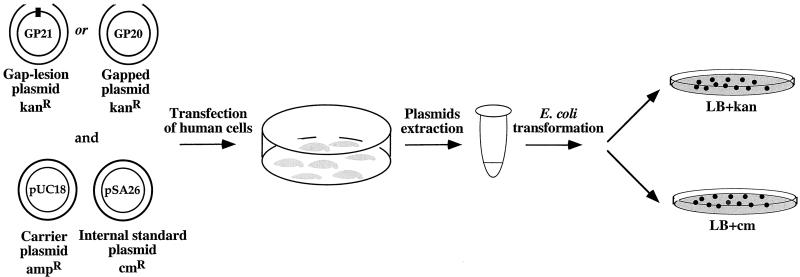
The assay for the quantitative measurement of the extent and specificity of the bypass of a particular DNA lesion in vivo is based on the transient transfection of cultured cells with a gapped plasmid, carrying a site-specific lesion in a single-stranded DNA region (Fig. 1). Such a construct cannot be repaired by recombinational repair in the absence of a homologous DNA copy, nor by excision repair in the absence of a complementary DNA strand. Therefore the only known mechanism by which the plasmid can be repaired in the cells is gap filling by means of TLR. The experimental outline included transfection of cultured cells with the gap-lesion plasmid, followed by an incubation period to allow TLR. The analysis of TLR was done in a subsequent step, by isolation of the plasmid from the mammalian cells, and introducing it into an E. coli indicator strain (Fig. 2). The efficiency of TLR in the mammalian cells is deduced from the number of E. coli transformants. The plasmids used contained neither a mammalian nor a viral origin of replication, and therefore could not replicate in mammalian cells. This choice was made to allow a direct and quantitative correlation between the amount of plasmid DNA rescued from the cells and the amount of plasmid molecules that were repaired by TLR.
Figure 2.
Outline of the quantitative TLR assay in human cells. See text for detail.
For quantitative measurement of lesion bypass, the gapped plasmid (kanR) was cotransfected with two additional plasmids: (i) an intact plasmid, which served as an internal standard (cmR), and (ii) a carrier plasmid (ampR), in high amounts, to allow optimal transfection efficiency. To normalize for the recovery of gapped plasmid from the cells in the absence of a lesion, pairs of transfections were performed in parallel: One with the gap-lesion plasmid and another with a similar gapped plasmid without a lesion. Each transfection mixture contained also the internal standard plasmid and the carrier plasmid.
Control Experiments for the TLR Assay.
A critical requirement for the reliability of the TLR assay is that gap-lesion plasmids that were not repaired in the mammalian cells do not survive in the indicator E. coli strain. This situation was achieved by using the alkaline lysis procedure for extracting the plasmids from the cells. In this procedure gapped or nicked plasmids are denatured, whereas covalently closed plasmids (i.e., plasmids in which the gap was filled in completely) remain intact. To estimate the effectiveness of the alkaline lysis method in selectively reducing the transforming ability of the gapped plasmids, the following experiment was performed. H1299 cells were transfected with the control plasmid pSA26 and the carrier plasmid pUC18, but without the gapped plasmid. The plasmid content of the cells was extracted, and before the alkaline denaturation step, the gapped plasmids GP20 or GP21 were added. The plasmid mixture was then used to transform the E. coli indicator strain. As can be seen in Table 1 (Exp. 1), the ratio of kanR colonies (originating from gapped plasmids GP20 or GP21) to cmR colonies (originating from the covalently closed plasmid pSA26) was very low (5.7 × 10−5 to 1.02 × 10−3), indicating that most of the gapped plasmids did not survive the extraction procedure. When GP20 and GP21 that did not undergo any treatment were used to transform the E. coli strain, they yielded transformation ratios of 0.80 and 0.0048, respectively (Table 1, Exp. 2). This finding indicates that the efficiency of transformation by alkali-treated gapped plasmids was 500- to 1,000-fold lower than gapped plasmids that were not denatured. Notice that GP21 transformed the E. coli strain with a much lower efficiency than GP20, because the indicator strain is recA-defective and therefore unable to bypass abasic sites. When the plasmids were not previously passed through human cells, lesion bypass reached an average of 0.6% (Table 1), consistent with previous results (25, 36). These results show that plasmids that did not undergo gap filling in the mammalian cells are extremely poor in transforming the indicator strain, and their contribution to the observed number of transformants in bypass experiments (see below) is negligible.
Table 1.
Control experiments for the TLR assay
| Exp. | Plasmid examined | Treatment | Transformants
|
Transformants ratio KanR/CmR | Bypass levels | |
|---|---|---|---|---|---|---|
| KanR | CmR | |||||
| 1 | GP21 | Alkali | 2 | 35,000 | 5.7E-05 | |
| GP20 | Alkali | 21 | 20,500 | 1.02E-03 | NA | |
| 2 | GP21 | — | 26 | 5,420 | 0.0048 | |
| GP20 | — | 4200 | 5,250 | 0.80 | 0.60 | |
| 3 | FGP21 | — | 4320 | 4,450 | 0.97 | |
| FGP20 | — | 5280 | 5,400 | 0.98 | NA | |
| 4 | FGP20 | — | 390 | 403 | 0.96 | NA |
In experiments 2 and 3 the plasmid mixtures containing the indicated plasmid (kanR), the internal control plasmid pSA26 (cmR), and the carrier plasmid pUC18 were diluted and directly introduced to the E. coli strain. In experiment 1 H1299 cells were transfected with the control and carrier plasmids, but without the gapped plasmid. Plasmid content of the cells was extracted, and before the alkaline denaturation step, the gapped plasmids GP20 or GP21 were added. In experiment 4 H1299 cells were transfected with a mixture of plasmid FGP20 (kanR) and the control and the carrier plasmids. Plasmid content was extracted and used to transform the E. coli strain. Bypass levels, when applicable, were calculated by dividing the KanR/CmR ratio obtained for GP21 by that obtained for GP20. The average number of transformants obtained was adjusted to 100 μl of transformation mixture. When the number of transformants was high, counting was performed on the appropriately diluted plates; when the number of transformants was low, a larger volume of transformed cells was plated. NA, not applicable.
TLR does not eliminate the abasic site from DNA. On the other hand, the ability of the filled-in plasmid to transform the E. coli depends on the elimination of the abasic site by base excision repair. To test the ability of the TLR products to transform the indicator strain, a plasmid similar to GP21 was constructed, except that it was fully double-stranded. This construct, termed FGP21, mimics the TLR product. In addition, a control plasmid without the lesion, termed FPG20, was constructed. It was found that both constructs transformed the indicator strain with similar normalized efficiencies (Table 1, Exp. 3), indicating that the abasic site was quantitatively repaired in E. coli. This finding implies that gapped plasmid molecules that were filled in in the mammalian cell were capable of transforming the E. coli indicator strain, regardless of the removal of the abasic site in the mammalian cell. The control experiments suggest that the E. coli transformants faithfully represent the number of gap-plasmids filled in by TLR.
Highly Efficient Bypass of a Synthetic Abasic Site in the Human Lung Adenocarcinoma Cell Line H1299.
The assay was used to determine the extent of lesion bypass in the human lung adenocarcinoma H1299 cell line. The experiment was repeated six times, and the percentage of lesion bypass was calculated for each experiment. As can be seen in Table 2 the extent of lesion bypass was very high, with an average efficiency of 92%. There was a considerable variation in the absolute numbers of transformants obtained in different experiments (columns 3 and 4 in Table 2), reflecting differences in transfection, extraction, and transformation. However, the final lesion bypass level had a remarkable reproducibility: The coefficient of variance in the six experiments was only 6.5%. This result indicates that having the appropriate control and internal standard plasmids indeed enables a reliable determination of the extent of lesion bypass in mammalian cells.
Table 2.
Extent of lesion bypass in H1299 cells
| Exp. no | Gapped plasmid | Transformants
|
KanR/CmR | Bypass level, % | |
|---|---|---|---|---|---|
| KanR | CmR | ||||
| 1 | GP21 | 110 | 795 | 0.138 | |
| GP20 | 55 | 400 | 0.138 | 100 | |
| 2 | GP21 | 200 | 1,860 | 0.108 | |
| GP20 | 185 | 1,650 | 0.112 | 96 | |
| 3 | GP21 | 80 | 1,135 | 0.070 | |
| GP20 | 70 | 860 | 0.081 | 86 | |
| 4 | GP21 | 150 | 1,160 | 0.129 | |
| GP20 | 265 | 1,905 | 0.139 | 93 | |
| 5 | GP21 | 185 | 1,825 | 0.101 | |
| GP20 | 220 | 1,845 | 0.119 | 85 | |
| 6 | GP21 | 50 | 380 | 0.132 | |
| GP20 | 80 | 540 | 0.148 | 89 | |
| Average 92 ± 6 | |||||
The plasmids mixture containing the indicated gapped plasmid (kanR), the internal control plasmid pSA26 (cmR), and the carrier plasmid pUC18 was passed through H1299 cells, after which the DNA was extracted and introduced into the E. coli indicator strain. Bypass levels were calculated by dividing the kanR/cmR ratio obtained for GP21 by that obtained for GP20. The average number of transformants obtained was adjusted to 500 μl of transformation mixture. When the number of transformants was high, counting was performed on the appropriately diluted plates; when the number of transformants was low, a larger volume of transformed cells was plated.
Table 2 shows that the number of transformants of the control plasmid (cmR) is an order of magnitude higher than the number of transformants of the gapped plasmid (kanR). Because the plasmid FGP20 (the mimic of the fully repaired GP20) and pSA26 showed similar survival throughout the assay steps (Table 1, Exp. 4), the lower amount of GP20 or GP21 transformants is probably caused by the lower survival of gapped-plasmids in the mammalian cells.
How fast does TLR occur? To answer this question DNA was extracted from the cells at different time points, and the extent of lesion bypass was determined. It was found that bypass reached a level of 60% by 8 h after transfection and continued to rise up to 94% after 26 h.
Bypass of the Abasic Site in Different Cell Lines.
The high extent of bypass obtained in the H1299 cell line and the desire to examine the applicability of the method to different cell lines prompted us to assay lesion bypass in three other human cell lines. These included the osteogenic sarcoma cell line Saos-2, the prostate carcinoma cell line PC3, and the hepatoma cell line Hep3B. As can be seen in Table 3, substantial bypass was obtained in each cell line examined, although there was a considerable difference in bypass: 32% in Saos-2 cells, 72% in PC3 cells, and 26% in Hep3B cells. The assay was highly reproducible as indicated by the low variation in multiple experiments performed with each cell line. This finding suggests that the assay is robust and applicable to a large variety of cell lines.
Table 3.
Extent of lesion bypass in various cell lines
| Cell line | Gapped plasmid | Transformants ratio KanR/CmR | Bypass levels, % |
|---|---|---|---|
| H1299 | GP21 | 0.113 | |
| GP20 | 0.123 | 92% ± 6 | |
| Saos-2 | GP21 | 0.058 | |
| GP20 | 0.180 | 32% ± 2 | |
| PC3 | GP21 | 0.060 | |
| GP20 | 0.083 | 72% ± 4 | |
| Hep 3B | GP21 | 0.014 | |
| GP20 | 0.054 | 26% ± 3 |
The experiments were performed as described in the legend to Table 2, except that the indicated cell lines were used. Shown are the average results of at least four experiments.
Mostly dAMP Was Inserted Opposite the Synthetic Abasic Site in H1299 Cells.
To examine the specificity of bypass, plasmids were extracted from kanR colonies and subjected to DNA sequence analysis at the vicinity of the lesion. This process was done for gapped plasmids GP21 and GP31. These plasmids are identical except for the DNA sequence context in the vicinity of the abasic site (Fig. 1). Overall the DNA sequence of 73 mutants was determined. As can be seen in Table 4, when either GP21 or GP31 were passed through H1299 cells, a vast majority of the mutants contained base substitution mutations (36/37; 97%). Examination of the base substitutions revealed that dAMP was preferentially inserted opposite the synthetic abasic site, including 14/15 mutations in GP21 (93%) and 16/22 mutations in GP31 (73%). A similar picture was obtained with mutants arising from GP21E, a derivative of GP21 with a larger gap of approximately 350 nt (TLR with GP21E was similar to GP21; data not shown). In this case 15 of 16 mutants involved insertion of dAMP opposite the lesion (Table 4). Overall dAMP was inserted opposite the synthetic abasic site in 45/53 mutants (85%). As a control, we have determined the DNA sequence of the same gap-lesion plasmids that have not been passed through the H1299 cells. As can be seen in Table 4, under these conditions 33/36 (92%) of the mutants contained small deletions opposite the abasic site, primarily minus one deletion. This finding is similar to the published specificity of the synthetic abasic site in an E. coli recA strain (25). Moreover, it provides strong evidence that in the current assay, bypass and its associated mutagenesis indeed occurred in the H1299 cells.
Table 4.
Analysis of mutations formed during TLR through a synthetic abasic site
| Mutation type | Gapped plasmid GP21
|
Gapped plasmid GP31
|
Gapped plasmid GP21E
|
||
|---|---|---|---|---|---|
| H1299 | — | H1299 | — | H1299 | |
| Base substitution | |||||
| A | 14 | — | 16 | — | 15 |
| G | — | 1 | 2 | — | — |
| T | — | 1 | 2 | — | — |
| C | — | 1 | 2 | — | — |
| Total base substitution | 14 (93%) | 3 (19%) | 22 (100%) | — (0%) | 15 (94%) |
| Deletion | |||||
| −1 | 1 | 12 | — | 18 | — |
| −2 | — | — | — | — | 1 |
| −3 | — | 1 | — | 2 | — |
| Total deletions | 1 (7%) | 13 (81%) | — (0%) | 20 (100%) | 1 (6%) |
| Total mutants analyzed | 15 | 16 | 22 | 20 | 16 |
A DNA mixture containing the indicated gap-lesion plasmid was introduced into E. coli JM109recA, either without or with prior passage through H1299 cells as indicated. Plasmids were extracted from kanR colonies and subjected to DNA sequence analysis. Gapped plasmid GP21E is similar to GP21, except that it contains an extended gap of approximately 350 nt. Shown is the DNA sequence opposite the lesion obtained for individual clones. The most abundant type of mutation is in bold.
Recombinational Repair Does Not Contribute Significantly to Filling In of the Gap-Lesion Plasmid.
The high efficiency of bypass of the abasic site prompted us to consider whether recombinational repair is responsible for at least some of the gap-filling events. As was pointed out above, the gap-lesion can be filled in in mammalian cells only by TLR. However, once some of the plasmids molecules inside the cell were filled in by TLR, they can provide a homologous fully double-stranded plasmid copy for recombinational repair of additional gap-lesion plasmids. This may cause an overestimation of TLR efficiency by adding a component of recombinational repair. To examine this possibility we cotransfected the gap-lesion plasmid with a plasmid termed pFGP/T-amp, which mimics a repaired filled-in gap-lesion plasmid. This plasmid has no gap, and it contains an AT base pair instead of the lesion (with A on the template strand, i.e., corresponding to the lesion site). In addition, the plasmid contained the bla gene replacing part of the kanamycin gene, therefore conferring ampicillin resistance. If the gap is filled in by recombination using the homologous cotransfected plasmid, at least part of the recovered kanR plasmids are expected to have a T opposite the location corresponding to the abasic site, as in the homologous partner plasmid. DNA sequence analysis of 17 filled-in kanR GP21-derived mutants showed that 15 carried an A opposite the location corresponding to the abasic site, one had a T and one had a C. This means that at most 6% (1/17) of the mutants were obtained by recombinational repair. This is most likely an overestimation, because TLR may also lead to the incorporation of a T opposite the abasic site at such a low frequency (see Table 4). A similar experiment was performed with pFGP/C-amp as a homologous partner for GP21. In this case if recombination occurs, a C is expected opposite to the location corresponding to the abasic site, and the repaired plasmid is expected to also have the minus one deletion marker from pFGP/C-amp. DNA sequence of 22 filled-in kanR mutants revealed that 16 carried an A opposite the location corresponding to the abasic site, two had a T, three had a C, and one had a large deletion. None of the plasmids carried the nearby minus one deletion marker present in pFGP/C-amp, suggesting that the mutants with the C were obtained by TLR and not by recombination. Taken together these results indicate that the great majority of gap-lesion plasmids are repaired by TLR in this system, and the contribution of recombinational repair, if any, is low (<5%). This finding is consistent with previous studies, which showed very low levels of homologous recombination in cultured cells (37, 38).
Bypass of the Synthetic Abasic Site Does Not Require Polymerase η and Is Aphidicolin Sensitive.
A fundamental question in the TLR field is what is the identity of the DNA polymerase that carries out the bypass of a particular DNA lesion. A convenient way to study this question is by using cell lines in which particular DNA polymerases were mutated or by using specific inhibitors. In an attempt to examine whether polymerase η is involved in bypass of abasic sites in human cells, we assayed TLR in two XP-V cell lines, which are defective in polymerase η. There is convincing in vivo and in vitro evidence that polymerase η bypasses UV light-induced cyclobutyl pyrimidine dimers (reviewed in ref. 39); however, its involvement in the bypass of abasic sites in humans is not clear (40, 41). As can be seen in Table 5, bypass was high in the XP-V cell lines, reaching 88–89%, similar to bypass in the H1299 cells. This finding indicates that polymerase η is not essential for bypass of abasic sites.
Table 5.
Effect of the XP-V mutation or aphidicolin on TLR
| Cell line | Treatment | Gapped plasmid | Transformants ratio KanR/CmR | Bypass levels, % |
|---|---|---|---|---|
| H1299 | — | GP21 | 0.142 | |
| GP20 | 0.155 | 91 ± 3 | ||
| GM03618 (XP-V) | — | GP21 | 0.020 | |
| GP20 | 0.022 | 89 ± 5 | ||
| GMO3617 (XP-V) | — | GP21 | 0.024 | |
| GP20 | 0.027 | 88 ± 4 | ||
| H1299 | — | GP21 | 0.173 | |
| GP20 | 0.188 | 92 ± 4 | ||
| H1299 | Aphidicolin* in DMSO/ethanol† | GP21 | 0.034 | |
| GP20 | 0.191 | 18 ± 2 | ||
| H1299 | DMSO/ethanol solution† | GP21 | 0.186 | |
| GP20 | 0.235 | 79 ± 3 |
The experiments were performed as described in the legend to Table 2, except that the indicated cell lines and treatments were used. Shown are the average results of at least four experiments.
Aphidicolin, dissolved in DMSO/ethanol, was added to a final concentration of 25 μg/ml.
DMSO was at a final concentration of 0.05%, and ethanol was at 0.2%.
Aphidicolin is an inhibitor of DNA polymerases α, ɛ and δ, but not of the Y family DNA polymerases (7, 12, 42) and not of the yeast polymerase ζ (43); the human polymerase ζ has not been purified yet. We assayed bypass of the synthetic abasic sites in H1299 cells in the presence of aphidicolin. As can be seen in Table 5, in the control dishes in the absence of aphidicolin, bypass was 92%, as expected. This high bypass slightly decreased to 79% when the control cells were grown in the presence of DMSO/ethanol, the solvent in which aphidicolin was dissolved. When assayed in the presence of aphidicolin, bypass was only 18%, representing a 4.4-fold inhibition (Table 5). This finding indicates that at least one of the replicative polymerases, α, ɛ, or δ, is required for bypass of abasic sites in human cells.
Discussion
This study describes the development of a quantitative assay for TLR in mammalian cells. The assay includes the transient transfection of cultured cells with a gapped plasmid, which contain a synthetic abasic site in the gap region. The advantages of this assay system are: (i) Using plasmids that cannot replicate in mammalians cells allows a direct correlation between the amount of DNA rescued from the cells and the amount of gap-lesion plasmid molecules that have been repaired by TLR. (ii) TLR is assayed uncoupled from replication, providing a simpler model system. (iii) The assay can be applied to a large variety of cell lines, without being restricted to special cell lines that are permissive for episomal replication. The limitation of this system is that extrachromosomal DNA is studied, implying that some of the elements of chromosomal TLR are lost. In addition, lesion bypass during gap-filling DNA synthesis might be different from lesion bypass during replication. Nevertheless, a detailed molecular analysis using the gap-lesion plasmid system is expected to shed light on some of the fundamental properties of TLR in mammals.
Analysis of lesion bypass in several cell lines revealed that the synthetic abasic site was bypassed at a level of 26–92%. This variation in bypass among different cell lines stems, most likely, from differences in the expression or regulation of lesion bypass polymerases. Variations are quite common among different cell lines, usually because these cancer cells have undergone a variety of genetic changes (e.g., ref. 44). The lower bypass of abasic sites that we observed in some cell lines might be compensated by an increase in accurate repair of these lesions, e.g., by increased expression of repair enzymes; however, this possibility has not been examined yet. The synthetic abasic site is a model lesion for abasic sites, one of the most common spontaneous lesions (45). This lesion is highly mutagenic and constitutes a strong obstacle for DNA polymerases (46). For example, in E. coli, even when the SOS system was induced, bypass of an abasic site in vivo reached only ≈5% (25, 36). Our data imply that in mammalian cells the abasic site is bypassed at levels 5- to 18-fold higher than in E. coli, suggesting a more dominant role for TLR in mammalians cells.
Most TLR events in H1299 cells (85%) involved insertion of dAMP opposite the synthetic abasic site (Table 4). This result is consistent with a previous study performed in COS cells, with a construct carrying a similar lesion, but also carrying a simian virus 40 origin of replication (47). The similar results obtained by using vectors unable or capable of autonomous replication strengthen the physiological relevance of an assay system based on transient transfection with a nonreplicating plasmid to the study of TLR in mammalians cells. It should be mentioned that in two other studies based on shuttle vectors each carrying an abasic lesion at a specific site, there was no apparent preference for a particular nucleotide opposite the abasic site (48, 49). The reason for the discrepancy between the various studies is not clear. They may result from the difference in the systems and the substrates used.
In vitro bypass studies with purified mammalian DNA polymerases have shown that most polymerases can bypass abasic sites, at least under certain conditions (11, 24, 50–55). This underscores the need for in vivo TLR data to determine the physiological role of specific DNA polymerases in bypass of a particular lesion. Interestingly, there are conflicting results on the in vitro ability of polymerase η to bypass an abasic site (40, 41). The experiments presented here with the XP-V cell lines, which lack polymerase η, clearly show that polymerase η is not required for the bypass of abasic sites in human cell lines. This result is consistent with the finding that bypass was inhibited by aphidicolin, which inhibits DNA polymerases α, δ, and ɛ, but not η. Both polymerase α (24) and polymerase δ (52, 53) incorporate A opposite abasic sites, whereas the TLR properties of polymerase ɛ are unknown. Therefore, the sequence specificity of insertion opposite an abasic site, cannot pinpoint the polymerase(s) required for lesion bypass in vivo. Interestingly, it was suggested that in Saccharomyces cerevisiae both polymerases δ and ζ are required for bypassing an abasic site (56). In addition, it can be argued that it is polymerase δ that encounters lesions during replication. Therefore, based on its biochemical properties and on the analogy to the yeast system, polymerase δ is likely to be the DNA polymerase required for replicating abasic sites in vivo. However, an involvement of polymerase α and/or polymerase ɛ cannot be excluded at this point. In any case, this study underscores the notion that lesion bypass is not limited to the specialized DNA polymerases.
Acknowledgments
We thank Moshe Oren for his help and valuable advice. This research was supported by grants from The Israel Science Foundation (Grant 78/00) and the Minerva Foundation, Munich. Z.L. is the Incumbent of The Maxwell Ellis Professorial Chair in Biomedical Research.
Abbreviation
- TLR
translesion replication
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 2.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 4.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 5.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 6.Ohmori H, Friedberg E C, Fuchs R P P, Goodman M F, Hanaoka F, Hinkle D, Kunkel T A, Lawrence C W, Livneh Z, Nohmi T, et al. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 7.Tissier A, McDonald J P, Frank E G, Woodgate R. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yuan F, Wu X, Wang Z. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson R E, Prakash S, Prakash L. Proc Natl Acad Sci USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor J S, Geacintov N E, Wang Z. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlach V L, Feaver W J, Fischhaber P L, Friedberg E C. J Biol Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 13.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs P E, Wang X D, Li Z, McManus T P, McGregor W G, Lawrence C W, Maher V M. Proc Natl Acad Sci USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs P E, McGregor W G, Maher V M, Nisson P, Lawrence C W. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakumo Y, Roth T, Ishii H, Rasio D, Numata S, Croce C M, Fishel R. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R E, Washington M T, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodgate R. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 19.Goodman M F. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 20.Friedberg E C, Feaver W J, Gerlach V L. Proc Natl Acad Sci USA. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baynton K, Fuchs R P P. Trends Biochem Sci. 2000;25:74–79. doi: 10.1016/s0968-0004(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 22.Livneh Z. J Biol Chem. 2001;276:25639–25642. doi: 10.1074/jbc.R100019200. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z. Mutat Res. 2001;486:59–70. doi: 10.1016/s0921-8777(01)00089-1. [DOI] [PubMed] [Google Scholar]
- 24.Takeshita M, Chang C-N, Johnson F, Will S, Grollman A P. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 25.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 26.Tomer G, Reuven N B, Livneh Z. Proc Natl Acad Sci USA. 1998;95:14106–14111. doi: 10.1073/pnas.95.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomer G, Livneh Z. Biochemistry. 1999;38:5948–5958. doi: 10.1021/bi982599+. [DOI] [PubMed] [Google Scholar]
- 28.Cohen-Fix O, Livneh Z. Proc Natl Acad Sci USA. 1992;89:3300–3304. doi: 10.1073/pnas.89.8.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brower M, Carney D N, Oie H K, Gazdar A F, Minna J D. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- 30.Rodan S B, Imai Y, Thiede M A, Wesolowski G, Thompson D, Bar-Shavit Z, Shull S, Mann K, Rodan G A. Cancer Res. 1987;47:4961–4966. [PubMed] [Google Scholar]
- 31.Lindgren J, Blaszczyk M, Atkinson B, Steplewski Z, Koprowski H. Cancer Immunol Immunother. 1986;22:1–7. doi: 10.1007/BF00205709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bressac B, Galvin K M, Liang T J, Isselbacher K J, Wands J R, Ozturk M. Proc Natl Acad Sci USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehman A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H, de Weerd-Kastelein E A, Bootsma D. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleaver J E, Greene A E, Coriell L L, Mulivor R A. Cytogenet Cell Genet. 1981;31:188–192. doi: 10.1159/000131646. [DOI] [PubMed] [Google Scholar]
- 35.Graham F L, van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubnitz J, Subramani S. Mol Cell Biol. 1985;5:529–537. doi: 10.1128/mcb.5.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramani S, Rubnitz J. Mol Cell Biol. 1985;5:659–666. doi: 10.1128/mcb.5.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordonnier A M, Fuchs R P. Mutat Res. 1999;435:111–119. doi: 10.1016/s0921-8777(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 40.Masutani C, Kusumoto R, Iwai S, Hanaoka F. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haracska L, Washington M T, Prakash S, Prakash L. J Biol Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 42.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava D K, Husain I, Arteaga C L, Wilson S H. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 45.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 46.Loeb L A, Preston B D. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 47.Takeshita M, Eisenberg W. Nucleic Acids Res. 1994;22:1897–1902. doi: 10.1093/nar/22.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gentil A, Cabral-Neto J B, Mariage-Samson R, Margot A, Imbach J L, Rayner B, Sarasin A. J Mol Biol. 1992;227:981–984. doi: 10.1016/0022-2836(92)90513-j. [DOI] [PubMed] [Google Scholar]
- 49.Cabral Neto J B, Cabral R E, Margot A, Le Page F, Sarasin A, Gentil A. J Mol Biol. 1994;240:416–420. doi: 10.1006/jmbi.1994.1457. [DOI] [PubMed] [Google Scholar]
- 50.Kunkel T A, Schaaper R M, Loeb L A. Biochemistry. 1983;22:2378–2384. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- 51.Efrati E, Tocco G, Eritja R, Wilson S H, Goodman M F. J Biol Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 52.Mozzherin D J, Shibutani S, Tan C K, Downey K M, Fisher P A. Proc Natl Acad Sci USA. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daube S S, Arad G, Livneh Z. Biochemistry. 2000;39:397–405. doi: 10.1021/bi991443m. [DOI] [PubMed] [Google Scholar]
- 54.Daube S S, Tomer G, Livneh Z. Biochemistry. 2000;39:348–355. doi: 10.1021/bi9917784. [DOI] [PubMed] [Google Scholar]
- 55.Johnson R E, Washington M T, Haracska L, Prakash S, Prakash L. Nature (London) 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 56.Haracska L, Unk I, Johnson R E, Johansson E, Burgers P M, Prakash S, Prakash L. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]