Abstract
Syntaxin-5 (Sed5) is the only syntaxin needed for transport into and across the yeast Golgi, raising the question of how a single syntaxin species could mediate vesicle transport in both the anterograde and the retrograde direction within the stack. Sed5 is known to combine with two light chains (Bos1 and Sec22) to form the t-SNARE needed to receive vesicles from the endoplasmic reticulum. However, the yeast Golgi contains several other potential light chains with which Sed5 could potentially combine to form other t-SNAREs. To explore the degree of specificity in the choice of light chains by a t-SNARE, we undertook a comprehensive examination of the capacity of all 21 Sed5-based t-SNAREs that theoretically could assemble in the yeast Golgi to fuse with each of the 7 potential v-SNAREs also present in this organelle. Only one additional of these 147 combinations was fusogenic. This functional proteomic strategy thereby revealed a previously uncharacterized t-SNARE in which Sed5 is the heavy chain and Gos1 and Ykt6 are the light chains, and whose unique cognate v-SNARE is Sft1. Immunoprecipitation experiments confirmed the existence of this complex in vivo. Fusion mediated by this second Golgi SNAREpin is topologically restricted, and existing genetic and morphologic evidence implies that it is used for transport across the Golgi stack. From this study, together with the previous functional proteomic analyses which have tested 275 distinct quaternary SNARE combinations, it follows that the fusion potential and transport pathways of the yeast cell can be read out from its genome sequence according to the SNARE hypothesis with a predictive accuracy of about 99.6%.
Keywords: organelle‖syntaxin vesicle
Identifying fusogenic SNARE complexes within the Golgi stack is central to establishing the patterns of protein flow and sorting within this organelle. SNARE proteins (1) are the minimal machinery for membrane fusion (2, 3) throughout the cell and represent a multigenic family whose gene products are localized to distinct subcellular compartments (4, 5). They self-assemble into extremely stable four helix-bundles (6) composed of two cognate parts, one from each membrane. The t-SNARE marks the target membrane and contributes three helices; the v-SNARE in the vesicular membrane provides the fourth helix. The t-SNARE trimers are composed of one heavy chain (a syntaxin family member) and two distinct non-syntaxin light chains, each contributing a single helix (7). For fusion to occur, all of the subunits of the t-SNARE must reside in one bilayer, and the v-SNARE must reside in the other bilayer, termed topological restriction (8). v-SNAREs on the vesicle link with their cognate matching t-SNARE on the target membrane to generate the SNAREpins that force the two lipid bilayers into close apposition culminating in membrane fusion (2, 9).
Specificity in membrane fusion is essential to maintain the organization and identity of cytoplasmic organelles and is encoded within the membrane fusion machinery itself (4, 10, 11). The SNAREs responsible for fusion in endoplasmic reticulum (ER) to Golgi transport, vacuolar homotypic fusion, Golgi-derived vesicle to plasma membrane fusion and endosomal fusion fuse in a pattern that closely reflects the pattern of vesicle transport in the cell (11, 12).
However, there are a few cases in which the same SNARE protein may contribute to more than one SNARE complex (13, 14). How can this seeming promiscuity be reconciled with the very high specificity of membrane fusion? Syntaxin-5 (Sed5; ref. 15) is a case in point and is also pivotal for understanding the pattern of traffic within the Golgi stack. In the steady-state, Sed5 is localized to the Golgi apparatus, where it is required both for ER to Golgi and intra-Golgi vesicle transport (14, 16), and it is also the only syntaxin needed for the secretory pathway until the plasma membrane is reached (17, 18). Sed5 has been shown to form complexes with seven other SNARE proteins: Bet1, Bos1, Sec22, Gos1, Ykt6, Sft1, and Vti1 (19–22). However, although it is clear that more than one tetrameric SNARE complex must exist in this mixture, it is not known how many complexes there may be, or how much overlap may exist among them.
One tetrameric complex has been found to comprise a fusogenic SNAREpin in which the t-SNARE has Sed5 as the heavy chain, Bos1 and Sec22 as its light chains, and with Bet1 as its unique cognate v-SNARE (8). Although it has been found that several distinct quaternary complexes involving Sed5 can form by using isolated SNARE proteins in solution, it is unknown whether any of these additional complexes are fusogenic (19–22). In this study, we use a comprehensive functional proteomic approach that reveals that there are actually two distinct fusogenic SNARE complexes in the yeast Golgi, one of which is needed for entry into the Golgi stack and the other for transport within it.
Materials and Methods
Plasmid Construction, Protein Expression, and Purification.
Plasmids coding for GST-Sed5 (pFP260), GST-Gos1(pJM212), GST-Ykt6 (pJM128), GST-Bet1 (pFP284), GST-Bos1 (pFP285), GST-Sft1 (pJM129), his6-Sec22 (pFP266), His-6-Vti1 (pFP340), his6Tlg1 (pJM124), Snc1-His-6 (pJM91), Snc2-His-6 (pJM81), and protein expression have been described (7, 8, 11).
Reconstitution.
All t-SNAREs were reconstituted from individually expressed proteins. Before reconstitution, Ykt6 was lipid anchored with a geranylgeranyl lipid as described (9). For acceptor liposomes, 7.5 nmol of each SNARE constituent (15 nmol for Ykt6) were used. Liposome formation and isolation was carried out exactly as described (2). Acceptor liposomes were treated with 0.05 units/μl thrombin for 2 h to excise GST from Sed5 and Sft1. Donor liposomes were formed by using 7.5 nmol of each SNARE as described (2).
The following modifications were made when reconstituting GST-Sed5, Bos1, Sec22, Gos1, and Ykt6 in one liposome population (see Fig. 5A): (i) 7.5 nmol of GST-Sed5, Bos1, and Sec22 were added to either 7.5 (15) nmol, 3.75 (7.5) nmol, 1.9 (3.75) nmol or 0 nmol of Gos1 (Ykt6), respectively; or (ii) 7.5 nmol of GST-Sed5, Gos1, and 1.5 nmol of Ykt6 were added to 7.5 nmol, 3.75 nmol, 1.9 nmol or 0 nmol of Bos1/Sec22, respectively.
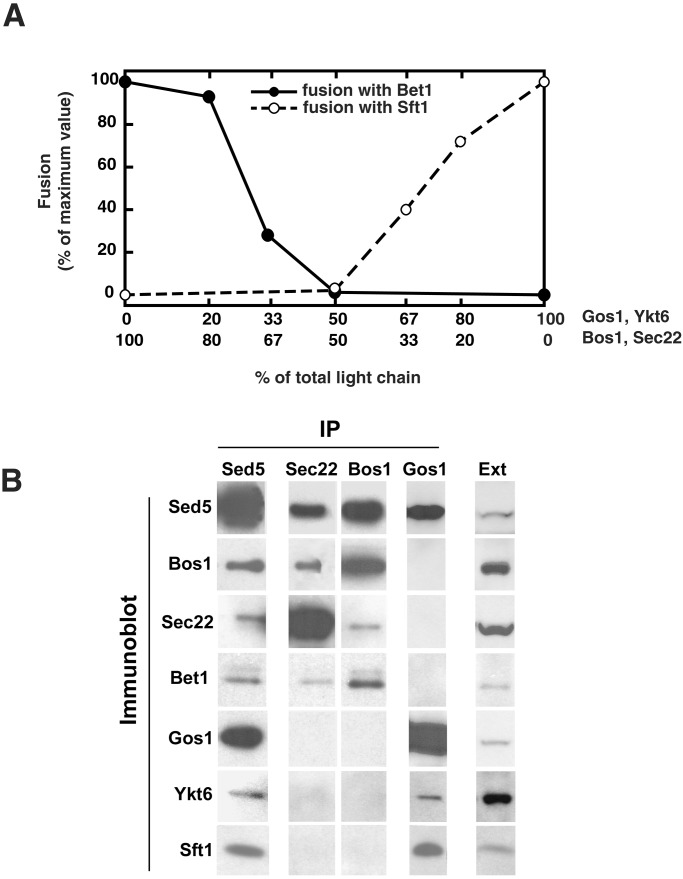
Figure 5.
(A) Shifting Sed5 between its two t-SNAREs according to the prevailing composition of light chains. Acceptor liposomes containing a constant amount of Sed5, Gos1, and Ykt6 with increasing amounts of Bos1 and Sec22 were reconstituted and mixed with Sft1 donor liposomes (○). Conversely, acceptor liposomes containing Sed5, Bos1, and Sec22 with increasing amounts of Gos1 and Ykt6 were reconstituted and mixed with Bet1 donor liposomes (●). The increase in fluorescence was monitored and converted to a measure of rounds of fusion. (B) Sed5 forms two distinct Golgi SNARE complexes in vivo. Sec18–1 spheroplasts were preincubated for 1 h at the restrictive temperature of 37°C and then lysed in detergent in the presence of 2 mM EDTA (see Materials and Methods). Cell extracts (10 mg of total protein) were incubated with antibodies to indicated SNAREs (IP) covalently coupled to protein A agarose; the resulting immunoprecipitates were washed with lysis buffer containing 0.3 M KCl. Proteins were eluted with 0.2 M glycine pH 2.7, precipitated with trichloroacetic acid, resolved by SDS/PAGE, and then subjected to an immunoblot analysis using antibodies directed against the indicated SNAREs (Immunoblot). Immunoprecipitated material (10%) and 0.1% of total cell extract (Ext) were loaded onto the gel.
Assuming an average diameter of 45 nm and ≈22,000 lipids per liposome, we estimate the copies of SNARE per donor liposome to be 360, 510, 537, 600, 580, 220, and 280 for Bet1, Bos1, Sec22, Sft1, Gos1, Ykt6, and Vti1, respectively. For acceptor liposomes, average copy numbers of candidate t-SNAREs/vesicle ranged from 70 to 110.
Fusion Assays.
We carried out standard fusion assays as described (2). NBD fluorescence was converted to rounds of fusion as described (8). In some cases, 5 μg of an Sft1 C-terminal peptide (SSVINQMTDSLGSMFTDIKNSSSRLTRSLKAGNSIW) or 0.75 nmol of the Sft1 cytosolic domain was added before fusion at 37°C.
Preparation of Yeast Cell Extracts.
Sec18–1 (23) detergent extracts were prepared as described with minor modifications (19). Typically, 4–6 liters of sec 18–1 cells (approximately 10,000 OD600) grown at 25°C in yeast extract/peptone/dextrose (YPD) medium (Bio 101) were spheroplasted at 25°C as described (19). Spheroplasts were incubated for 1 h at either 25°C or 37°C. After incubation, cells were isolated by centrifugation and immediately disrupted with glass beads in 20–30 ml of ice-cold lysis buffer (25 mM Hepes, pH 7.4/0.1 M KCl/1% Triton X-100/1% glycerol/2 mM EDTA/and protease inhibitors: 1 μg/ml leupeptin, 2 μg/ml antipain, 20 μg/ml trypsin inhibitor, 10 μg/ml benzamidine, 5 μg/ml pefabloc SC, 2 μg/ml aprotonin, 4 μg/ml chymostatin, 2 μg/ml pepstatin). Unbroken cells and debris were removed by centrifugation for 1 h at 100,000 × g. The protein concentration of supernatant was adjusted to 2 mg/ml with lysis buffer.
Antibodies and Immunoprecipitation Experiments.
Polyclonal antibodies to Sed5, Bos1, Sec22, Bet1 (19), Gos1 (17), and Ykt6 (24) have been described. Polyclonal antibodies to Sft1 have been generated by using the corresponding GST-fusion protein. Typically, 200–500 μl of antiserum was incubated with 200–500 μl of protein A agarose. After binding, the affinity resin was washed extensively with PBS and then crosslinked with disuccinimidyl suberate (Pierce) according to the manufacturer's instructions. The resin was washed first with PBS, followed by 0.2 M glycine pH 2.7 (10 bed volumes) and lysis buffer containing 1 M KCl.
In immunoprecipitation experiments, 5–10 ml of detergent extract (10–20 mg of protein) was incubated at 4°C overnight with 200 μl of affinity resin. The beads were washed first with 20 ml of lysis buffer and then with 10 ml of the same buffer containing 0.3 M KCl. The protein complexes were eluted with 1 ml of 0.2 M glycine pH 2.7 and precipitated by using 5% (vol/vol) trichloroacetic acid. Immunoprecipitated proteins were resolved by SDS/12% PAGE (Invitrogen) and then subjected to Western blot analysis using ECLPlus Western Blot Detection System (Amersham Pharmacia). Typically, 10% of the eluted material was loaded in each lane.
The stoichiometry of individual SNAREs in the complex was determined by using the corresponding recombinant proteins as quantitation standards. Both the immunoprecipitates and the increasing amounts of the recombinant proteins (typically between 0.1 ng and 100 ng) were resolved on the same gel, transferred to nitrocellulose, and then immunodetected with corresponding antibodies using ECLPlus. Densitometry of specific bands was performed with QUANTITY ONE software (Bio-Rad). Optical densities were converted into protein amounts according to the quantitation standards and then corrected for molecular mass.
Results
Two Distinct Sed5-Containing SNAREpins Can Fuse Membranes.
To understand better the SNARE interactions underlying vesicle trafficking in the Golgi, we took a functional proteomic approach to identify any and all potential fusogenic Golgi SNARE complexes encoded in the yeast genome containing Sed5. To read out the fusion potential of these SNAREs, we used a well characterized liposome fusion assay that makes use of our previous observation that v- and t-SNAREs, when reconstituted into separate fluorescent donor and nonfluorescent acceptor vesicle populations, trigger lipid bilayer fusion (2). Fusion is measured as an increase in NBD fluorescence (25) and converted to rounds of fusion, as described (26).
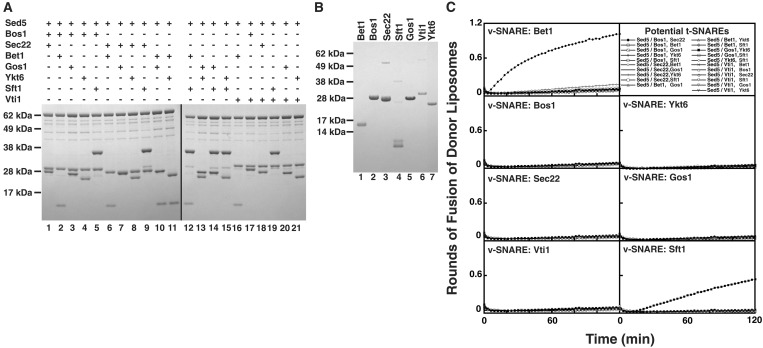
We purified and reconstituted Sed5 in liposomes in all possible combinations with two potential light chains from among the following seven SNARE proteins: Bet1, Bos1, Sec22, Gos1, Ykt6, Sft1, and Vti1, all of which are known to reside, at least in part, in Golgi membranes in yeast (4). Given seven potential Golgi light chains, there are 21 possible t-SNAREs consisting of Sed5 in combination with two distinct light chains (Fig. 1A). The same seven non-syntaxin SNAREs also were reconstituted individually in donor liposomes for testing as potential v-SNAREs (Fig. 1B).
Figure 1.
Two tetramers of Golgi-localized SNARE proteins catalyze membrane fusion. (A) Twenty-one combinations of 3 Golgi-localized SNARE proteins were reconstituted into liposomes and analyzed by SDS/PAGE (Lanes 1–21). All combinations contain the Golgi heavy chain Sed5. (B) Golgi-localized SNAREs Bet1, Bos1, Sec22, Sft1, Gos1, Vti1, and Ykt6 (Lanes 1–7, respectively) were individually reconstituted into donor liposomes; reconstituted protein was analyzed by SDS/PAGE. (C) Membrane-fusion assay identifies two fusogenic sets of donor and acceptor liposomes of 147 possible combinations. Twenty-one different acceptor liposomes (described in upper right box) were mixed with either Bet1, Bos1, Sec22, Vti1, Ykt6, Gos1, or Sft-containing donor liposomes, and the increase in fluorescence was monitored over two h. The increase in fluorescence was converted into a measure of rounds of fusion of donor liposomes.
Thus, a matrix of membrane-fusion experiments was tested whereby the 21 potential t-SNAREs were individually mixed with each of the seven potential v-SNAREs (Fig. 1C). Of 147 combinations, only two gave a positive fusion signal. One was known from our previous work (Fig. 1C and ref. 8) and consisted of the t-SNARE Sed5/Bos1,Sec22 with the v-SNARE Bet1. This result demonstrates that Bos1 and Sec22 is the only combination of light chains able to function with Sed5 for fusion with Bet1 as the v-SNARE (Fig. 1C).
The second fusogenic combination consisted of Sed5/ Gos1,Ykt6 as the t-SNARE and Sft1 as the v-SNARE (Fig. 1C). Only Gos1 and Ykt6 can act as the light chains for this second Sed5-based t-SNARE because none of the other potential light chains could substitute for either of them. The physiological importance of this previously uncharacterized Sed5-containing SNAREpin in intra-Golgi transport is supported by the following genetic and morphological data: (i) Sed5 is localized to the Golgi, and a temperature-sensitive mutation in Sed5 shows an ER to Golgi and intra-Golgi protein transport defect (15, 27, 28); (ii) Sft1 is localized to the Golgi (16, 27), and a temperature-sensitive mutation in Sft1 blocks intra-Golgi protein transport (16, 29); (iii) Gos1 is localized to the Golgi (17, 18), and a Gos1-deletion mutant in yeast shows a defect in intra-Golgi and ER to Golgi protein transport (17); and (iv) Ykt6 localization partially overlaps with Golgi markers (28), and a temperature-sensitive mutation in Ykt6 blocks intra-Golgi transport (29). However, slowly shutting off the expression of Ykt6 in yeast causes an ER-to-Golgi transport block as well (24).
Vti1 interacts genetically and biochemically with Sed5 and affects transport through the Golgi (20, 21). However, none of the potential Vti1-containing t-SNARE complexes were fusogenic with any potential Golgi v-SNARE. We tested a series of acceptor liposomes containing Sed5, Vti1, and a non-syntaxin SNARE (either Snc1, Snc2, Nyv1, Vam7, Tlg1, Sft1, Gos1, Ykt6, Bos1, Sec22, or Bet1) for fusion with each of a panel of donor liposomes containing all potential v-SNAREs. The panel of potential v-SNAREs consisted of the same set of non-syntaxin SNAREs tested as light chains except Vam7, because it lacks a transmembrane domain. In no case was fusion observed (unpublished data). Nor does Vti1 seem to act as a v-SNARE, because Vti1-containing donor liposomes did not fuse with any of the potential Golgi t-SNARE-containing acceptor liposomes (Fig. 1C). The Vti1 preparation tested is functional because it serves as a light chain required for SNARE-dependent fusion mediated by vacuolar and endosomal v- and t-SNAREs (7, 12). One possibility is that the observed interaction of Sed5 with Vti1 may not represent a SNARE complex capable of membrane fusion.
Sed5, Gos1, Ykt6, and Sft1 Are Necessary and Sufficient to Mediate Fusion in a Topologically Restricted Manner.
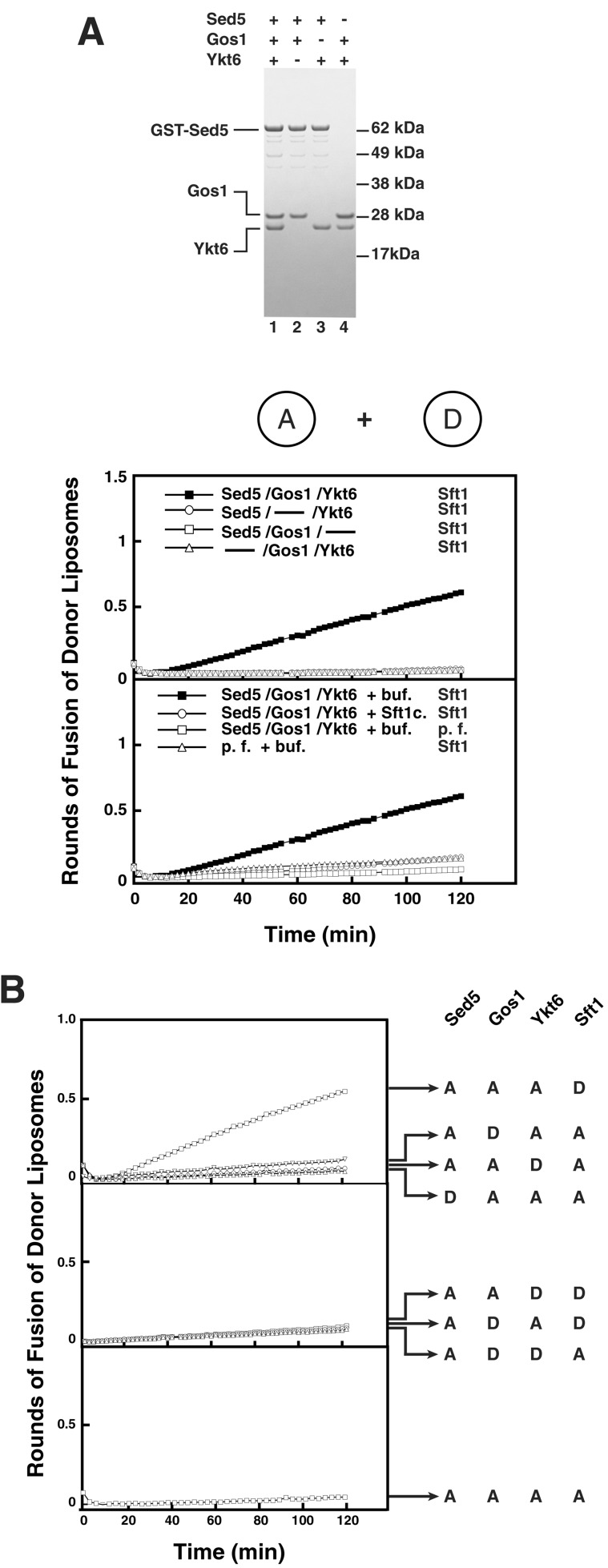
All four of these SNAREs are required for fusion (Fig. 2A, top graph). The cytosolic domain of Sft1 inhibits fusion of Sft1-donor liposomes with Sed5/Gos1,Ykt6 acceptor liposomes (Fig. 2A, bottom graph), confirming this fusion reaction requires assembly of the v-t-SNARE complex.
Figure 2.
(A) Sed5, Gos1, Ykt6, and Sft1 are necessary and sufficient to mediate lipid bilayer fusion. Sed5, Gos1, Ykt6 (Lane 1), Sed5, Gos1 (Lane 2), Sed5, Ykt6 (Lane 3), and Gos1, Ykt6 (Lane 4) were reconstituted into acceptor liposomes and analyzed by SDS/PAGE. These acceptor liposomes were mixed with Sft1 donor liposomes at 37°C, and the increase in fluorescence was monitored and converted to rounds of fusion (top graph). Fusion between acceptor and donor liposomes is v-SNARE- and t-SNARE-dependent and can be blocked by the soluble domain of Sft1 (bottom graph). Acceptor liposomes containing either Sed5/Gos1,Ykt6 or no protein (p.f.) were mixed with donor liposomes containing either Sft1 or no protein (p.f.) in the presence of either Sft1 cytosolic domain (Sft1c) or buffer (buf). (B) Topological-restriction of fusion mediated by t-Sed5/Gos1,Ykt6 and v-Sft1. A 4:0 topology (Bottom) and a 2:2 topology (Middle) of Golgi SNAREs do not catalyze membrane fusion. A distinct 3:1 topology (Top) of Golgi SNAREs mediates membrane fusion. Golgi SNAREs were reconstituted into acceptor liposomes or donor liposomes, mixed, and the increase in fluorescence was monitored over 2 h at 37°C.
In the cases of ER-to-Golgi fusion (involving Sed5, Bos1, Sec22 and Bet1) and endosomal fusion (involving Tlg2, Tlg1, Vti1 and Snc2), each SNARE is topologically restricted to function on one or the other of the two opposing lipid bilayers (12, 26). There are eight possible nonredundant distributions of four SNAREs between two bilayers: (i) one combination with four SNAREs in one and none in the other bilayers (Fig. 2B Bottom); (ii) three combinations with two SNAREs in each bilayer (Fig. 2B Middle); and (iii) four combinations where three SNAREs are in one bilayer and the fourth SNARE is in the other bilayer (Fig. 2B Top). We tested all of these possibilities. Of the eight potential topological distributions, fusion only occurred when Sed5, Gos1, and Ykt6 were in one bilayer and Sft1 was in the other (Fig. 2B Top). This topological distribution establishes that Sft1 is uniquely endowed with the capacity to function as the v-SNARE. The other three chains function as the t-SNARE in the opposite bilayer.
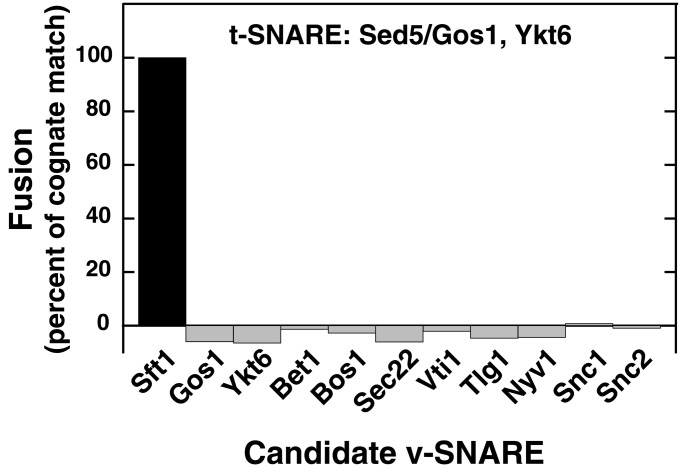
To test the specificity of fusion with the identified t-SNARE (Sed5/Gos1,Ykt6), all 11 potential v-SNAREs were individually reconstituted into donor liposomes and tested [the SNARE proteins lacking transmembrane domains (Vam7, Sec9, Spo20) were excluded]; only Sft1 could partner with Sed5/Gos1,Ykt6 as a cognate v-SNARE (Fig. 3).
Figure 3.
Specificity of fusion between the Golgi t-SNARE (Sed5/Gos1,Ykt6) and v-SNARE (Sft1) liposomes. Eleven potential v-SNAREs encoded in the yeast genome were independently reconstituted into donor liposomes and mixed with Sed5/Gos1,Ykt6 acceptor liposomes. A background of 0.072 rounds of fusion, representing fusion with protein-free liposomes, was subtracted from each experiment. The extent of fusion was subsequently normalized to represent 100% fusion for the cognate v-SNARE Sft1.
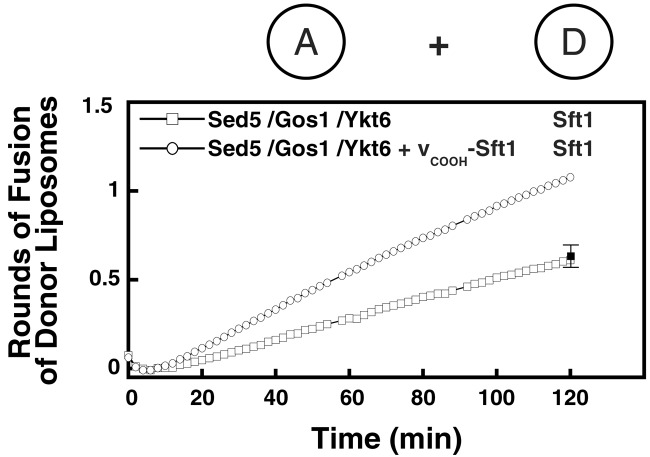
A Peptide Corresponding to the C-Terminal Portion of Sft1 Activates the Cognate t-SNARE for Fusion.
The mol ratios of lipid:protein in acceptor and donor liposomes are typically about 220 and 40, respectively (not shown). With a liposome diameter of 45 nm (2), this ratio corresponds to about 100 t-SNARE complexes per acceptor liposome and 600 v-SNAREs per donor liposome. Because the acceptor liposomes are added in 17-fold molar excess over the donor liposomes, the fluorescent lipids initially reside in donor liposomes and can theoretically undergo about six rounds of fusion before all of the v-SNAREs have been consumed by complexing with a t-SNARE (26). However, we consistently observed only 0.5–0.7 rounds of fusion with Sed5/Gos1,Ykt6,Sft1.
In other cases, autoinhibition of the t-SNARE lowers its reactivity to cognate v-SNARE (12, 26). Both the plasma membrane and the endosomal t-SNAREs can be activated either by removing the N-terminal regulatory domain of their syntaxin subunit (26) or by complexing one mol peptide [corresponding to the sequence of the C-terminal (membrane proximal) half of their cognate v-SNARE] per mol t-SNARE (12). This peptide switches the conformation of the t-SNARE by allowing the membrane-proximal portion of the t-SNARE to zip up into coiled-coil, in which the peptide provides the fourth strand (T. J. Melia et al., unpublished work).
The corresponding Sft1-derived peptide approximately doubled the extent of fusion of Sft1 donor liposomes with Sed5/Gos1,Ykt6 liposomes (Fig. 4). As a control, peptides corresponding to the equivalent C-terminal portions of the other non-syntaxin yeast SNAREs (Bet1, Bos1, Sec22, Ykt6, Gos1, Vti1, Nyv1, Tlg1, and Snc2) were unable to stimulate the rate or extent of fusion between Sed5/Gos1,Ykt6 acceptor and Sft1 donor liposomes (Fig. 4). Raising the concentration of the peptide did not result in further increases in the rate or extent of this fusion reaction (not shown). The rate of fusion is likely limited by the rate with which the bound peptide dissociates from the complex of the peptide with partly zipped-up v- and t-SNARE (T. J. Melia et al., unpublished work).
Figure 4.
A peptide corresponding to the C-terminal domain of Sft1 stimulates fusion. Soluble peptide corresponding to the C-terminal portion of the Sft1 SNARE domain, vCOOH-Sft1 (see Materials and Methods) can stimulate fusion between Sed5/Gos1,Ykt6 acceptor liposomes and Sft1 donor liposomes. As a control, vCOOH-peptides corresponding to Bet1, Bos1, Sec22, Ykt6, Gos1, Vti1, Nyv1, Tlg1, and Snc2 were tested, and the 120-min time point was averaged (black square). Sed5/Gos1,Ykt6 acceptor liposomes were mixed with Sft1 donor liposomes in the absence or presence of Sft1 peptide. Increase in fluorescence was monitored at 37°C over 2 h and converted into a measure of rounds of fusion.
The Two Golgi t-SNAREs Can Operate in the Same Membrane Without Loss of Specificity.
Sed5 thus participates in at least two distinct functional SNARE complexes: it can combine with the light chains Bos1 and Sec22 to fuse with Bet1-liposomes, or it can combine with Gos1 and Ykt6 to fuse with Sft1-liposomes. The distribution of the corresponding SNARE proteins within the Golgi of animal cells, to the extent that it is known, suggests that these two SNAREpins mediate vesicle trafficking in distinct but overlapping regions of the stack. Membrin (i.e., Bos1), rSec22b (i.e., ERS-24), and rBet1 (i.e., Bet1) are preferentially concentrated at the cis face of the animal Golgi stack (30), whereas Gos-28 (i.e., Gos1), and syntaxin-5 (i.e., Sed5) are found throughout the Golgi stack (30, 31). The orthologous t-SNARE syntaxin 5/ERS-24/membrin is needed both to receive vesicles coming from the ER and also for intra-Golgi transport (32). The relatively even distribution of Gos-28 across the stack suggests that its SNAREpin is needed for later transport between cisternae.
Because the distribution in the Golgi of the two alternative sets of light chains for syntaxin-5/Sed5 likely overlap within many cisternae, they may compete for a common pool of Sed5/syntaxin-5 heavy chain, which would allow a balance of one or the other t-SNARE to prevail, depending on the proportions of the four light chains in any given cisternae.
To see if the properties of these SNARE proteins are compatible with this hypothesis, we carried out competition experiments. A series of acceptor liposomes were prepared containing a constant amount of Sed5 with various amounts of Bos1 and Sec22 vs. Gos1 and Ykt6 (Fig. 5A). Each of these acceptor liposomes then was tested for its capacity to fuse with either Sft1 or Bet1 donor liposomes. In the first series of liposomes, the concentration of Sed5, Bos1, and Sec22 in acceptor liposomes was kept constant, whereas the concentration of the light chains Gos1/Ykt6 was progressively increased (Fig. 5A, black circles). As the concentration of the competing light chains Gos1 and Ykt6 increased in acceptor liposomes, the ability of these acceptor liposomes to fuse with Bet1 donor liposomes decreased. By contrast, the extent of fusion with Sft1 donor liposomes increased as the amount of Gos1 and Ykt6 increased (not shown). Thus Gos1 and Ykt6 compete with Bos1 and Sec22 for binding to a common population of Sed5 heavy chain, according to the balance of the two alternative pairs of light chains.
In the reciprocal experiment, Sed5, Gos1, and Ykt6 were kept at a constant amount, and Bos1 and Sec22 amounts were increased in a series of acceptor liposomes. The reciprocal behavior toward the two potential v-SNAREs was observed; as the concentration of the competing light chains Bos1 and Sec22 increased in acceptor liposomes, the ability of these acceptor liposomes to fuse with Sft1 donor liposomes decreased (Fig. 5A, white circles). As expected, the extent of fusion with Bet1 donor liposomes increased as the amount of Bos1 and Sec22 increased (not shown). Therefore, Sed5 can be effectively titrated by either set of light chains, both t-SNAREs to form within the same cisternae to engage the appropriate v-SNARE.
Sed5 Forms Two Distinct Golgi SNARE Complexes in Vivo.
Although the Golgi SNARE complex Sed5/Bos1,Sec22,Bet1 has been well documented in yeast and mammalian cells (19, 33), the discovered quaternary complex Sed5/Gos1,Ykt6,Sft1 has not been confirmed in vivo.
To determine whether this complex forms in vivo, we performed coimmunoprecipitation experiments by using cell extracts prepared from a temperature-sensitive sec18–1 (NSF) yeast strain, which accumulates SNARE complexes at the restrictive temperature of 37°C (19). Antibodies directed against different SNAREs coupled to protein A agarose were incubated with the extract and the precipitates were analyzed by immuno-decorating Western blots. As expected, antibodies to Sed5 precipitate Sed5 along with six other SNAREs—Bos1, Sec22, Bet1, Gos1, Ykt6, and Sft1 (Fig. 5B and ref. 19). To test for their presence in distinct complexes, the cell extract was subjected to immunoprecipitation with antibodies to Sec22, Bos1, and Gos1. The Sec22 and Bos1 complexes contained Sed5, Sec22, Bos1, and Bet1 but lacked Gos1, Ykt6, and Sft1 (Fig. 5B, lanes 2 and 3). As would be predicted from the fusion results, Gos1 was found in a complex with Sed5, Ykt6, and Sft1, but this complex lacked Sec22, Bos1, and Bet1 (Fig. 5B, lane 4). These results confirm the existence of two distinct nonoverlapping SNARE complexes (Sed5/Bos1,Sec22,Bet1 and Sed5/Gos1,Ykt6,Sft1) in vivo, and show that other quaternary complexes (Sed5/Sec22/Gos1/Sft1, Sed5/Sec22/Bos1/Sft1, and Sed5/Sec22/Gos1/Bet1), which can be formed from the soluble proteins (22), are not present in significant amounts in vivo.
Quantitative analysis of the Western blots using recombinant proteins as standards (see Materials and Methods for details) confirmed that the complex of Sed5/Gos1,Ykt6,Sft1 contained approximately equimolar amounts of each SNARE (Sed5, 1.00; Gos1, 0.86; Ykt6, 1.07; Sft1, 0.86; mol ratios to Sed5), as had been previously found for the Sed5/Bos1,Sec22,Bet1 complex (19). Thus, the results from immunoprecipitation experiments are in excellent agreement with the predictions from the functional analysis of fusion capacity.
Discussion
Two Distinct Golgi t-SNARE Based on the Same Heavy Chain.
Sed5, Gos1, and Ykt6 acceptor liposomes fuse with Sft1-containing donor liposomes, and only Sft1 can function as the v-SNARE in this reaction. Further, fusion is topologically restricted and occurs only when Sed5, Gos1, and Ykt6 reside in the same bilayer, and Sft1 resides in the other bilayer. This finding establishes Sed5 as the heavy chain and Gos1 and Ykt6 as the light chains of a previously uncharacterized Golgi t-SNARE and Sft1 as its unique cognate v-SNARE in the yeast cell. Based on amino acid sequence homology, the light chains Gos1 and Bos1 are closely related and are members of the “Bos1 family”, whereas Sec22 and Ykt6 are both members of the VAMP/synaptobrevin family (34). The heavy chain Sed5 thus complexes with a “Bos1-like” and with a “synaptobrevin-like” SNARE to form a functional t-SNARE. Although Bos1 and Gos1 are homologous, they cannot substitute for one another to form a functional t-SNARE (the same is true for Sec22 and Ykt6), as confirmed by the lack of fusion between Sed5/Bos1,Ykt6 with Sft1 or Sed5/Gos1,Sec22 with Bet1 liposomes. The v-SNAREs Bet1 and Sft1 show little amino acid homology (34); however, overexpression of Bet1 can rescue an otherwise lethal Sft1-gene deletion, strongly suggesting that these proteins have similar function (29).
Tsui et al. (22) have reported that several distinct quaternary complexes can be formed in solution with the heavy chain Sed5 in combination with Bos1, Sec22, Bet1, Gos1, Ykt6, Sft1, Tlg1, and Snc1. In the course of this study (Fig. 1 and unpublished data employing recombinant Tlg1 and Snc1), we have tested all of these proposed complexes and found that only two are actually fusogenic: (i) Sed5/Bos1,Sec22-containing vesicles with Bet1-containing vesicles, and (ii) Sed5/Gos1,Ykt6-containing vesicles with Sft1-containing vesicles. Evidently, not all SNAREs that can pair within a membrane can necessarily pair between membranes, and even if they do, not all such trans-SNARE complexes are necessarily fusogenic SNAREpins. Fusion imposes energetic requirements and topological restrictions that some trans-SNARE complexes may not meet.
Physiological Relevance.
Genetic, biochemical, and morphological data support distinct roles for the two t-SNAREs in Golgi trafficking and underscores their physiological relevance.
First, genetic evidence from temperature-sensitive mutations establishes that Sec22, Bos1, and Bet1 are first needed for fusion of ER-derived vesicles at the cis-Golgi (35–37); the same conclusion follows from the results of cell-free transport assays (28, 38, 39). By contrast, the phenotypes of temperature-sensitive mutants of Ykt6 (29) and Sft1 (16) cause a selective and later block in transport, and deletion of Gos1 demonstrate that these proteins are first needed for transport within the Golgi (17). Sed5 is first needed for transport from the ER to the Golgi (15, 28, 38) but also is needed for intra-Golgi transport (16).
Second, Sed5, Bos1, Sec22, and Bet1 can be coimmunoprecipitated from yeast cell extracts (19), and the same is true for their mammalian orthologues syntaxin-5, membrin, rSec22b, and rBet1 (33). Gos-28 (Gos1) and membrin/rBet1 (Bos1/Bet1) are found in mutually exclusive Sed5-containing complexes, consistent with the two distinct SNAREpins we have found (33). Our immunoprecipitation experiments now identified Ykt6 and Sft1 as the two previously unidentified components of the Sed5/Gos1 complex.
Third, morphological studies reveal that the mammalian orthologues have different distributions across the Golgi stack. Although Sed5 is present in every Golgi cisternae (30, 31), Bos1, Sec22, and Bet1 are highest at the cis face and lowest, or absent, at the trans face (30). Thus, the t-SNARE syntaxin-5/rSec22b, membrin is mainly a cis-Golgi t-SNARE. Gos-28, like syntaxin-5, is present in every Golgi cisternae (31, 40), suggesting that the identified t-SNARE Sed5/Ykt6,Gos1 also may be found later in the stack. Data on the distributions of hYkt6 (Ykt6) and GS-15 (Sft1) have not been reported.
Our data showing that Sed5 participates in either of two SNAREpins, each possessing a distinct set of light chains and a v-SNARE, is therefore supported by the in vivo and in vitro observations mentioned above. Because Sed5 can partition into one or the other t-SNARE according to which set of light chains is present, the cis-trans distribution of Bos1, Gos1, Sec22, and Ykt6 should determine the relative concentrations of the two t-SNAREs through the stack.
Specificity of Fusion Encoded in SNARE Proteins.
Together with earlier work (11, 12), five syntaxins representing the Golgi, early endosomes, vacuoles, and plasma membrane have now been tested for fusion in a total of 275 quaternary combinations, with the 11 potential v-SNAREs or light chains encoded in the yeast genome. Of these, only nine combinations (i.e., about 3%) are fusogenic, and all but one (i.e., 0.4%; Sec22 as v-SNARE with the plasma membrane t-SNARE) correspond to known transport pathways in the cell.
Put differently, a physical chemist armed only with the DNA sequence of yeast could use the SNARE fusion assay to read out the fusion potential and transport pathways of the cell according to the SNARE Hypothesis (1) with an accuracy exceeding 99.6%.
Acknowledgments
This research was supported by a National Institutes of Health grant (to J.E.R.) and postdoctoral fellowships from the Medical Research Council of Canada (to F.P.), the European Molecular Biology Organization (to K.P.), and the National Institutes of Health (to J.M.).
Abbreviations
- GST
glutathione S-transferase
- ER
endoplasmic reticulum
References
- 1.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Söllner T H, Rothman J E. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 3.Nickel W, Weber T, McNew J A, Parlati F, Söllner T H, Rothman J E. Proc Natl Acad Sci USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelham H R. Trends Cell Biol. 2001;11:99–101. doi: 10.1016/s0962-8924(01)01929-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y A, Scheller R H. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 6.Sutton R B, Fasshauer D, Jahn R, Brunger A T. Nature (London) 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda R, McNew J A, Weber T, Parlati F, Engel T, Nickel W, Rothman J E, Söllner T. Nature (London) 2000;407:198–202. doi: 10.1038/35025084. [DOI] [PubMed] [Google Scholar]
- 8.Parlati F, McNew J A, Fukuda R, Miller R, Söllner T H, Rothman J E. Nature (London) 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- 9.McNew J A, Weber T, Parlati F, Johnston R J, Melia T J, Söllner T H, Rothman J E. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scales S J, Chen Y A, Yoo B Y, Patel S M, Doung Y C, Scheller R H. Neuron. 2000;26:457–464. doi: 10.1016/s0896-6273(00)81177-0. [DOI] [PubMed] [Google Scholar]
- 11.McNew J A, Fukuda R, Parlati F, Johnston R J, Paz K, Paumet F, Söllner T H, Rothman J E. Nature (London) 2000b;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 12.Paumet F, Brugger B, Parlati F, McNew J A, Söllner T, Rothman J E. J Cell Biol. 2001;155:961–968. doi: 10.1083/jcb.200104092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelham H R. Exp Cell Res. 1999;247:1–8. doi: 10.1006/excr.1998.4356. [DOI] [PubMed] [Google Scholar]
- 14.Nichols B J, Pelham H R. Biochim Biophys Acta. 1998;1404:9–31. doi: 10.1016/s0167-4889(98)00044-5. [DOI] [PubMed] [Google Scholar]
- 15.Hardwick K G, Pelham H R. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banfield D K, Lewis M J, Pelham H R. Nature (London) 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- 17.McNew J A, Coe J G, Sogaard M, Zemelman B V, Wimmer C, Hong W, Söllner T H. FEBS Lett. 1998;435:89–95. doi: 10.1016/s0014-5793(98)01044-8. [DOI] [PubMed] [Google Scholar]
- 18.Holthuis J C, Nichols B J, Dhruvakumar S, Pelham H R. EMBO. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Søgaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Söllner T. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 20.von Mollard G F, Nothwehr S F, Stevens T H. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupashin V V, Pokrovskaya I D, McNew J A, Waters M G. Mol Biol Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui M M, Tai W C, Banfield D K. Mol Biol Cell. 2001;12:521–538. doi: 10.1091/mbc.12.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser C A, Schekman R. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 24.McNew J A, Sogaard M, Lampen N M, Machida S, Ye R R, Lacomis L, Tempst P, Rothman J E, Söllner T H. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- 25.Struck D K, Hoekstra D, Pagano R E. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 26.Parlati F, Weber T, McNew J A, Westermann B, Söllner T H, Rothman J E. Proc Natl Acad Sci USA. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wooding S, Pelham H R. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao X, Barlowe C. J Cell Biol. 2000;149:55–66. doi: 10.1083/jcb.149.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsui M M, Banfield D K. J Cell Sci. 2000;113:145–152. doi: 10.1242/jcs.113.1.145. [DOI] [PubMed] [Google Scholar]
- 30.Hay J C, Klumperman J, Oorschot V, Steegmaier M, Kuo C S, Scheller R H. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orci L, Ravazzola M, Volchuk A, Engel T, Gmachl M, Amherdt M, Perrelet A, Söllner T H, Rothman J E. Proc Natl Acad Sci USA. 2000;97:10400–10405. doi: 10.1073/pnas.190292497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Wong S H, Tang B L, Xu Y, Hong W. Mol Biol Cell. 1999;10:435–453. doi: 10.1091/mbc.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay J C, Chao D S, Kuo C S, Scheller R H. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- 34.Weimbs T, Mostov K, Low S H, Hofmann K. Trends Cell Biol. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- 35.Newman A P, Shim J, Ferro-Novick S. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim J, Newman A P, Ferro-Novick S. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman A P, Ferro-Novick S. J Cell Biol. 1987;105:1587–1594. doi: 10.1083/jcb.105.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spang A, Schekman R. J Cell Biol. 1998;143:589–599. doi: 10.1083/jcb.143.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao X, Ballew N, Barlowe C. EMBO. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao D S, Hay J C, Winnick S, Prekeris R, Klumperman J, Scheller R H. J Cell Biol. 1999;144:869–881. doi: 10.1083/jcb.144.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]