Abstract
Endosperm of cereal grains is one of the most important renewable resources for food, feed, and industrial raw material. It consists of four triploid cell types, i.e., aleurone, starchy endosperm, transfer cells, and cells of the embryo surrounding region. In maize, the aleurone layer is one cell layer thick and covers most of the perimeter of the endosperm. Specification of maize aleurone cell fate is proposed to occur through activation of the tumor necrosis factor receptor-like receptor kinase CRINKLY4. A second maize gene essential for aleurone cell development is defective kernel 1 (dek1). Here we show that DEK1 shares high homology with animal calpains. The predicted 2,159-aa DEK1 protein has 21 transmembrane regions, an extracellular loop, and a cysteine proteinase domain that shares high homology with domain II of m-calpain from animals. We propose that DEK1 functions to maintain and restrict the aleurone cell fate imposed by CR4 through activation of its cysteine proteinase by contact with the outer endosperm surface. DEK1 seems to be the only member of the calpain superfamily in plants, Arabidopsis DEK1 sharing 70% overall identity with maize DEK1. The expression of dek1 in most plant tissues in maize and Arabidopsis, as well as its presence in a variety of higher plants, including angiosperms and gymnosperms, suggests that DEK1 plays a conserved role in plant signal transduction.
Endosperm of cereal grains is a valuable resource for food, feed, and industrial raw material. Endosperm is one product of the double-fertilization event in flowering plants and contains four cell types, i.e., aleurone, starchy endosperm, transfer cells, and cells of the embryo-surrounding region (1–3). In fully developed maize grains, the aleurone is strictly limited to one cell layer covering the perimeter of the inner mass of starchy endosperm cells. Endosperm initially develops as a coencyte in the central cell, consisting of a narrow layer of cytoplasm with dividing nuclei covering a large central vacuole (1). Cellularization of the endosperm is initiated by the formation of cell walls forming alveoli around each nucleus with the open end facing inward toward the central vacuole. Specification of maize aleurone cell fate occurs after the first round of periclinal divisions in the endosperm alveoli, the outer or peripheral cells forming aleurone initials, and the inner cells or alveoli representing starchy endosperm initials.
We previously proposed that aleurone cell fate specification occurs by activation of the tumor necrosis factor receptor-like receptor kinase CRINKLY4 (CR4) (4) by a hitherto unidentified positional cue (5). Morphologically, the aleurone cell fate is manifested by the presence of preprophase bands, a microtubular array lacking in starchy endosperm cells (6). During early stages of grain development, aleurone cells divide both anticlinally, expanding the surface area of the endosperm, and periclinally. The inner daughter cells of these periclinal divisions redifferentiate to become starchy endosperm cells, restricting the aleurone layer to exactly one layer of cells (7, 8). A second maize locus affecting aleurone cell fate specification is defective kernel 1 (dek1) (9). In an elegant study with Ds-induced chromosome breakage in a dek1/+ genotype, Becraft and Asuncion-Crabb (10) recently demonstrated that somatic loss of dek1 function at late developmental stages leads to loss of aleurone cell identity and redifferentiation to a starchy endosperm cell fate. Conversely, by using the unstable Mutator (Mu) dek1-PIA allele, somatic gain of the cell autonomous dek1 function leads to conversion of starchy endosperm cells into aleurone cells (10). These studies demonstrate that the fate of endosperm cells, even at late developmental stages, is flexible, and that aleurone cell fate must be actively maintained throughout grain development. The exact role of the dek1 gene in this process is unknown. In addition to aleurone cell development, the Cr4 gene is implicated in leaf development, homozygous mutant plants having crinkly leaves. Similarly, the phenotype of homozygous dek1 embryos show that dek1 function is not restricted to aleurone cell differentiation, dek1 embryos sometimes contain a root primordium, but lack shoot structures (11).
The purpose of this study was to identify the dek1 gene. To achieve this goal, we screened a population of maize lines containing a high-copy number of Mu elements, identified and cloned a Mu element cosegregating with the dek1 mutant phenotype, and used the sequence flanking the Mu insertion to clone maize dek1.
Materials and Methods
Mutant Screening and Microscopy.
Visual inspection of the 41,000 ears in Pioneer Hi-Bred International's Trait Utility System in Corn collection (12) revealed 12,600 ears that contained a mixture of plump (wild type) and nonplump grains. Mature nonplump grains from all segregating ears, representing potential endosperm mutant grains, were fixed overnight at room temperature in 2% glutaraldehyde/0.3 M sodium phosphate buffer, pH 6.5. Hand sections were stained with Toluidine blue and mounted on glass slides with 15% Mowiol/35% glycerol/0.07 M Tris (pH 7.5). Preparation for resin embedding of dek1-mum1 kernels followed published protocols (13).
Allelism Test, Cosegregation Analysis, and Cloning of dek1.
The identity of the dek1-mum1 mutant gene as dek1 was established in a cross between dek1-mum1/+ plants and plants carrying the dek1–1394 reference allele (kindly provided to O.-A.O. by P. Becraft, Iowa State University, Ames). Cosegregation analysis was performed in a population of plants segregating for the dek1-mum1 allele. Plants carrying ears with only wild-type grains were classified as wild type, plants with 25% dek1-mum1 kernels as dek1-mum1/+. Homozygous dek1-mum1 embryos do not germinate. Seeds from the dek1-DR1129 stock (130E) was obtained from the Maize Genetics Cooperator Stock Center, Urbana, IL.
DNA for the cosegregation analysis was isolated from freeze-dried leaf material according to the cetyltrimethylammonium bromide protocol (14), digested with restriction enzymes (Roche Molecular Biochemicals) and separated on 0.8% agarose gels. DNA fragments were transferred to Hybond-N+ membranes in 10× SSC. Genomic DNA gel blots were probed with 32P-labeled Mu-element-specific probes (15) kindly provided by V. Chandler, Univ. of Arizona, Tucson. A subgenomic library of the polymorphic DNA fragment from dek1-mum1/+ plants (see Fig. 2 A and B) was prepared in Asp-718-digested ZAP-Express (Stratagene) vector. A single clone containing the cosegregating 3-kb fragment was identified by using the Mu1 probe. The DNA flanking the Mu1 insertion was identified by sequencing the insert. The Mu insertion in dek1-DR1129 was identified by PCR by using dek1 primer AACCTGCATCAGGGATTGCCATCGTAAG in combination with Mu-TIR primer AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC.
Figure 2.
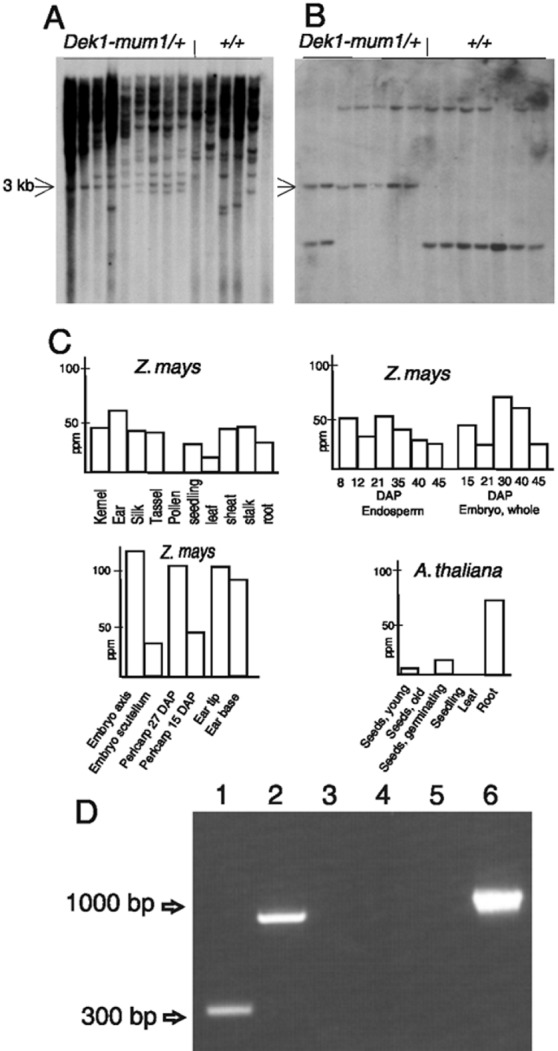
Cosegregation and expression analysis of dek1. (A) DNA blot from heterozygous dek1-mum1/+ and homozygous +/+ plants digested with KpnI and probed with a Mu1-specific probe. The blot identifies a 3-kb band cosegregating with the dek1-mum1 phenotype (arrow). (B) DNA blot from the same segregating population as in A probed with dek1 cDNA. (C) Massively parallel signature-sequencing experiments measuring dek1 transcript abundance in ppm in maize and Arabidopsis tissues. (D) Reverse transcription–PCR analysis of the dek1 transcript in dek1-mum1 homozygous endosperms. Combinations with a Mu-TIR primer and a dek1-specific primer from the 5′ side (lane 1) and the 3′ side (lane 2) of the Mu1 insertion site give bands of expected sizes, showing the presence of Mu1 in the dek1-mum1 transcript. A combination of the two dek1-specific primers from both sides of the insertion site (lane 3) fails to give a product, demonstrating the lack of wild-type dek1 transcript without Mu1 in dek1-mum1 homozygous mutant endosperm. Lanes 4–6 contain control reactions with poly(A)+ RNA from B73 endosperm with use of the same primer combinations as in lanes 1–3, respectively.
dek1 Transcript Isolation and Sequencing.
Total RNA was isolated from 9 days after pollination (DAP) maize kernels with Purescript (Gentra Systems) and from 7 DAP Arabidopsis thaliana Columbia seed as described (16). PCR cloning of overlapping regions of cDNA into pCR 2.1 TOPO (Invitrogen) was according to standard molecular protocols. cDNA sequences were compared against corresponding genomic sequences to determine exon-intron arrangement. A comparison of the A. thaliana dek1 gene in GenBank (accession nos. AC027034, F7A10.23) and the experimentally determined dek1 cDNA sequence of A. thaliana revealed several incorrect intron-exon splice predictions in the annotations of the predicted coding sequence. The corrected sequence of the A. thaliana cDNA has GenBank accession no. AY061803.
dek1 Isolation and Sequencing.
dek1 maize cDNA fragments were used as probes to screen a maize Mo17 genomic bacterial artificial chromosome (BAC) library. The HindIII and EcoRI subfragments homologous to calcium-requiring cysteine proteinases (calpain) cDNA were identified in selected BAC clones by gel blot hybridization, cloned into a plasmid vector pBSKS (Stratagene), and sequenced with the help of the EZ∷TN transposon insertion system (Epicentre Technologies, Madison, WI).
dek1 Expression Analysis.
Poly(A)+ RNA from dissected tissue samples were submitted to Lynx Therapeutics (Foster City, CA) for massively parallel signature-sequencing analysis (17). The signature sequences for maize and A. thaliana dek1 orthologues, GATCCATGGTTCTTTGG (GenBank AY061806, position 6758) and ATCTTGAGACACGCAT (GenBank AY061803, position 6905), respectively, were calculated on a ppm transcript basis. The presence of dek1 transcript in homozygous dek1-mum1 endosperm was assayed by using poly(A)+ RNA extracted from dissected 15 DAP endosperms from dek1-mum1 and B73 wild-type kernels. Poly(A)+ RNA was isolated with oligo(dT) magnetic beads (Dynal, Great Neck, NY) and first-strand cDNA was made from 100 ng poly(A)+ RNA with Superscript System (GIBCO/BRL) according to the manufacturer's instructions. The dek1-specific primers from the 5′ and 3′ side of the Mu1 insertion site in the dek1-mum1 allele have the sequence 5′-CCACCCATGGACTTAGATGCCTTTGG-3′ and 5′-AGATCCAGAAGGACTTCCAGCACCAAG-3′, respectively. The Mu-TIR primer used was as described above. PCR was performed with annealing temperature at 62°C for 40 cycles, and 20 μl of the amplification products was visualized on a 1.5% Agarose gel.
Results
Isolation of a Mu-Tagged dek1 Allele from a Large-Scale Microscopy Screen.
As a step toward a broader understanding of the mechanisms involved in aleurone cell development, we conducted a microscopy-based screen of a subset of lines in Pioneer Hi-Bred International's Trait Utility System in Corn maize population (13). In this screen, we identified two lines lacking aleurone cells, both of which represented novel alleles of dek1 as demonstrated by crosses to heterozygous dek1 plants carrying the dominant gene for purple aleurone cells. In this genetic background, the lack of aleurone cells is revealed by the white color of the mutant grains (Fig. 1A). In a subsequent analysis, we showed that only one of the two alleles was tagged by the Robertson's Mutator (18). We therefore used this mutant, which we call dek1-mum1, to characterize the dek1 mutant phenotype and to clone the dek1 gene. Similar to dek1 alleles described (9, 10), mature homozygous dek1-mum1 mutant grains lack aleurone cells, as apparent from hand sections stained with Sudan red, a dye that detects the characteristic oil bodies of aleurone cells (Fig. 1 B and C). However, closer examination of sections of mature grains revealed that the periphery of mutant endosperms contains cell wall material similar to aleurone cell walls (data not shown).
Figure 1.
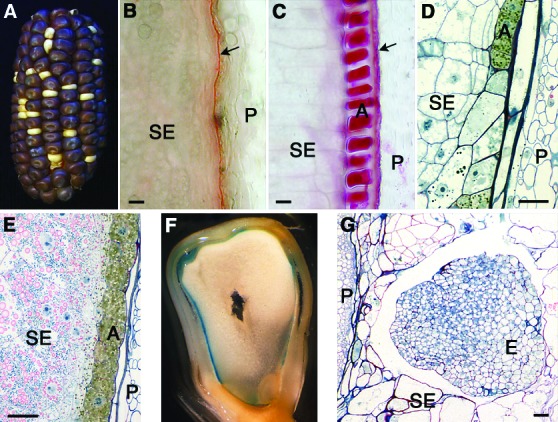
The phenotype of dek1-mum1 homozygous maize kernels. (A) F1-ear from the cross dek1-mum1/+ × dek1–1394/+. The ear segregates 3:1 for wild-type (dark) and dek1 (white) grains, demonstrating that the two mutations are both in the dek1 gene. (B) Hand section of dek1-mum1 kernel stained with the lipid stain Sudan red 7B (31). Starchy endosperm cells (SE) are located in the periphery of the endosperm close to the remnants of the nucellus (arrow). The maternal pericarp (P) is seen to the right. (C) Transverse hand section of wild-type kernel with peripheral aleurone cells (A) adjacent to the remnants of the maternal nucellus (arrow). (D) Transverse section of 15 DAP homozyogous dek1-mum1 endosperm embedded in Spurr resin (13) showing partial presence of aleurone cells (A) with dark cytoplasm in the periphery of the endosperm. (E) Wild-type endosperm (15 DAP) with normal aleurone layer. (F) dek1-mum1 grain showing blue stain from the Gus reporter driven by the aleurone-specific barley Ltp2-promoter. (G) Homozygous dek1-mum1 embryo (E) at 15 DAP. The shape of the embryo is globular, lacking the embryo axis found in wild-type embryos.
To investigate the possibility that dek1-mum1 grains initiate aleurone cell formation at an early stage and then subsequently fail to maintain this cell fate as the grain matures, we sectioned developing grains and looked for aleurone cells. A typical situation is shown in Fig. 1D, where many of the peripheral endosperm cells have dark staining cytoplasm typical of wild-type aleurone cells (Fig. 1E). To verify independently that these cells are true aleurone cells, we crossed heterozygous dek1-mum1/+ plants with maize plants carrying the barley aleurone-specific Ltp2 promoter fused to the Gus-reporter gene (19). Homozygous dek1-mum1 grains harvested 15 DAP frequently contained peripheral cells with aleurone cell morphology and stained blue from Gus expression (Fig. 1F). From these experiments we conclude that dek1-mum1 grains initially specify aleurone cell fate, but lack the ability to maintain it as the grains mature.
Similar to other dek1 alleles (9), dek1-mum1 embryos arrest at the juvenile globoid stage and are devoid of shoot structures (Fig. 1G). dek1-mum1 endosperm develops normal transfer cells (data not shown), so two of the four endosperm cell types are unaffected by the dek1 mutation.
Cloning of the Maize dek1 Gene.
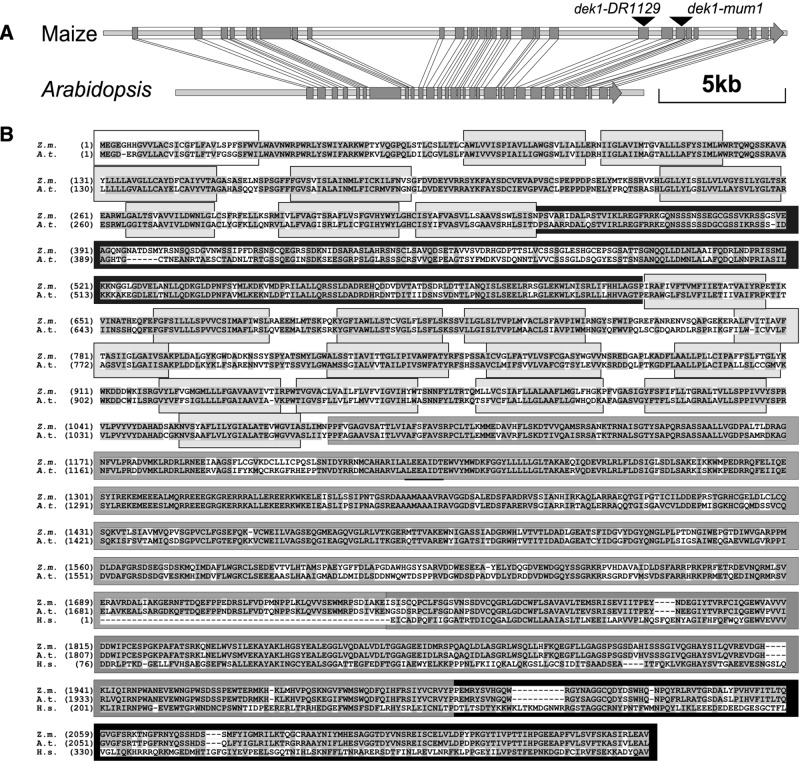
To identify the dek1 gene, cosegregation analysis (15) was performed with Mu probes on DNA gel blots from plants derived from a cross between a heterozygous dek1-mum1/+ plant and the maize inbred line B73. Hybridizations with a Mu1-specific probe revealed a polymorphic fragment exclusively in the heterozygous plants (Fig. 2A). The cloned flanking sequence was used as a probe in a population of 300 segregating individuals (Fig. 2B). No break of linkage was observed between the polymorphic fragment and the dek1-mum1 phenotype, indicating that the Mu element was inserted either in the dek1 gene itself or in a tightly linked locus. Sequence analysis revealed that the DNA used as a probe is positioned in exon 24 of a gene spanning 24,000 bp (GenBank accession no. AY061804) (Fig. 3A). The corresponding cDNA was cloned and found to contain a 6,477-bp coding sequence (GenBank accession no. AY061805) (Fig. 3A). Sequencing of this gene was assisted by comparison with the A. thaliana orthologue, which shares a very high degree of sequence conservation with the maize gene, including an identical structure of 31 exons (GenBank accession no. AC027034, F7A10.23) (Fig. 3A). To verify independently the identity of the cloned gene as dek1 we analyzed the dek1-DR1129 allele. With PCR, we detected a Mu8 insertion in exon 22 of dek1-DR1129 (Fig. 3A). The dek1-DR1129 mutant was used because reverse genetics screens of the Trait Utility System in Corn population (12) failed to detect additional Mu-tagged dek1 alleles. From these analyses, we conclude that the cloned gene represents maize dek1.
Figure 3.
dek1 gene and protein structure. (A) Intron–exon structure of maize and Arabidopsis dek1 genes. Exons are shown in gray. The dek1-mum1 allele has a Mu1 inserted between base pairs 18954 and 18955, dek1-DR1129 a Mu8-insertion between base pairs 17827 and 17828. (B) Alignment of predicted maize and A. thaliana DEK1 proteins. Alignment with human calpain (H.s) domain II and III is shown (Lower). Similar and identical residues are shaded. Shades in grayscale identify the sequence of the DEK1 domains shown in Fig. 4.
Maize and Arabidopsis dek1 Transcripts Are Present in Most Plant Tissues.
The dek1 transcript is present at a low steady-state level in both maize and Arabidopsis and is not reliably detected with conventional RNA blotting methods. Analysis of dek1 expression was therefore accomplished by massively parallel signature sequencing (17) on microbead arrays from a total of 37 maize and 11 Arabidopsis tissues (Fig. 2C). These experiments showed the dek1 gene is expressed in most maize tissues at a level ranging from 30 to 55 ppm. In endosperm, the transcript is elevated during early developmental stages and declines near maturity. In embryos, transcripts peak at middevelopment. Dissection of embryos at middevelopment into their main components revealed 4-fold higher expression in the embryonic axis, supporting the proposed role for dek1 in shoot formation (9). Elevated transcript levels are also present in young pericarp and immature ear tip and base. In A. thaliana, low levels of the dek1 transcript occur in young and germinating seeds (10 ppm), but the transcript was undetected in older seeds and seedlings. The highest level of dek1 transcript in A. thaliana is in seedling roots (67 ppm). Together, these results suggest dek1 functions in diverse developing tissues.
By using reverse transcription–PCR, the wild-type dek1 transcript is detectable in diverse maize tissues in a spatial and temporal pattern that is in full agreement with the results of the massively parallel signature-sequencing analysis described above (data not presented). When the same method is applied to poly(A)+ RNA extracted from dissected homozygous dek1-mum1 endosperms, a Mu1 containing dek1 transcript is detected by using gene-specific primers from both sides of the Mu-insertion site in combination with a Mu-TIR primer (Fig. 3 D and A). No wild-type dek1 transcript could be detected by using only the gene-specific primers. On the basis of these results we conclude that the transcriptional machinery is able to read through the Mu1 element in the dek1-mum1 allele, and that this extended transcript is stable enough to allow detection by reverse transcription–PCR. Based on the occurrence of several stop codons in Mu1 in all frames, the dek1-mum1 homozygous endosperm and embryos are expected to lack a functional DEK1 protein.
Maize DEK1 Is a Member of the Calpain Gene Superfamily.
The predicted maize DEK1 protein (2,159 aa residues; Fig. 3B) was identified as a member of the calpain gene superfamily by the presence of a conserved cysteine proteinase domain (II), shared by all calpains, and by domain III, found in most calpains (20) (Figs. 3B and 4A). In human m-calpain, domain III resembles the C2 domain of Ca2+-regulated proteins (20). Conventional calpains, typified by human m-calpain, are cytosolic enzymes activated by a rise in intracellular Ca2+, as well as by signaling that includes the protein kinase C-, tyrosine kinase-, or adhesion molecule-derived cascade (21). Upon activation, the enzyme associates with proteins or phospholipids in the plasma membrane and undergoes autolysis (21). The substrates of calpains are typically part of signal transduction cascades, such as protein kinase Cs (22, 23). Through these mechanisms, conventional calpains fulfill essential functions in multiple developmental pathways in animals (24, 25).
Figure 4.
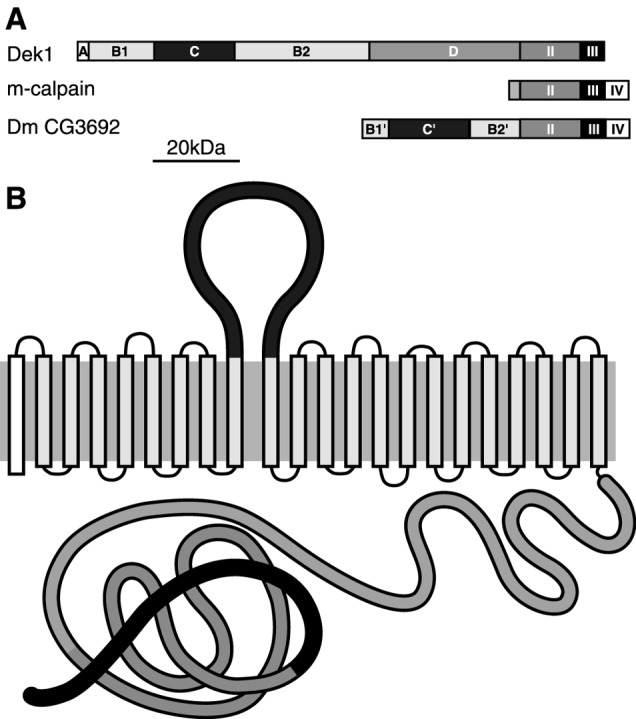
Domain structure of DEK1. (A) Domain structure of maize DEK1, m-calpain, and Drosophila CG3692 calpain. (B) Predicted maize DEK1 structure showing the loop region on the outside of the cell and the cysteine proteinase domain on the inside. The model is based on the TMHMM2.0 program (32).
Despite its similarities, maize DEK1 deviates considerably from conventional calpains (Fig. 4A). DEK1 lacks domain IV, which contains five calpain EF hands (26), and has both a novel and extensive N terminus. This portion of DEK1 contains five distinguishable domains (Fig. 3B). Domain A has predicted endoplasmic reticulum and membrane targeting signals. Domains B1 and B2 have eight and 13, respectively, predicted transmembrane stretches. These regions are interrupted by a predicted 300-aa loop region, domain C. Domain D is a hydrophilic, charged region found between domains B2 and II (Figs. 4A and 3B). With respect to the membrane orientation of these domains, it is important to note that all structural prediction algorithms place the loop region of domain C on a side opposite the domains distal to B2 (Fig. 4B).
dek1 Seems to be the Only Member of the Calpain Gene Superfamily in Plants.
The Arabidopsis DEK1 orthologue has a high level of amino acid identity (70%) with the maize protein, the highest sequence conservation occurring in domains II (88%) and III (83%) (Fig. 3B). High identity is also present in the membrane-spanning domains, B1 (72%) and B2 (64%). The lowest identity is in domain C (57%). The predicted Drosophila melanogaster calpain CG3692 (27) combines features of both conventional calpains and DEK1, possessing domains II, III, and IV, and an N-terminal extension similar to DEK1 (Fig. 4A), which suggests that membrane-embedded calpains are not exclusive to plants. In contrast to the diversity of calpains found in animal systems (20), however, we observe a stark contrast in plant systems, where membrane-embedded calpains seem to be the only subtype found. This conclusion is based on several observations, foremost among them our inability to detect any evidence of alternate calpain types in searches of all available plant databases, including the fully sequenced Arabidopsis genome and the nearly completed sequence of the rice genome. Moreover, by using probes from the most conserved part of the dek1 cDNA, no cross-hybridization on genomic DNA blots has been detected in maize at low-stringency conditions. Instead, we only find evidence in support of highly conserved, dek1-like orthologues. Examples range from a predictably close relative in rice (92% identical; GenBank accession nos. AP004161 and AY062272) to an intriguing clone from loblolly pine, a gymnosperm (79% identity; GenBank accession no. AW043258). This level of identity in domain II seems diagnostic of DEK1 and distinguishes it from other cysteine proteinases in plants. In addition, DEK1 proteins have an unusually large number of highly conserved transmembrane segments (Fig. 3B). The fully sequenced Arabidopsis genome is predicted to encode more than 4,500 proteins with transmembrane segments. Of these, DEK1 has the highest number of transmembrane domains for proteins with an assigned function, a fact that underscores its unusual nature.
Discussion
The wild-type maize aleurone layer is strictly one cell layer thick and is sometimes referred to as the epithelial layer of the endosperm (Fig. 1C). As shown by Becraft et al. (4), the CR4 protein receptor-like kinase is implicated in aleurone cell fate specification, homozygous cr4 endosperm lacking aleurone cells in patches of variable sizes on the endosperm surface. This gene functions cell-autonomously (28). dek1, also working cell autonomously in aleurone development, is required throughout endosperm development for continued presence of aleurone cells (10). As shown here for homozygous dek1-mum1, mutant endosperm initiate aleurone cell fate specification at a high frequency, as shown by the presence of peripheral cells containing the darkly stained cytoplasm diagnostic of aleurone cells. The fact that at least some of these cells possess true aleurone cell identity is shown here by activation of the barley aleurone specific Ltp2-promoter (Fig. 1F). In mature endosperm, aleurone cells are almost completely lacking (Fig. 1 A–C). One possible explanation for the presence of aleurone cells in young endosperm could be reversion of the dek1-mum1 allele wild-type dek1. We believe this explanation is unlikely for two reasons. First, because of the high incident of aleurone cells in these endosperms. Second, a high frequency of Mu reversions in the endosperm at an early developmental stage would be highly atypical for Mu, usually displaying a strong preference for excisions at late developmental stages (29). An alternative explanation for the presence of aleurone cells at early stages could be partial function of the dek1-mum1 allele. We hold this possibility to be unlikely both based on the lack of wild-type transcript in mutant endosperm (Fig. 2D) and the presence of several stop codons in all reading frames the dek1-mum1 transcript. Thus, even if translated, the DEK1 protein would lack the cytoplasmic domain. Instead, we favor an interpretation of these data that the initiation of aleurone cell fate does not depend on DEK1.
As proposed earlier, the CR4 (4) is a likely candidate for the initiation of aleurone cell fate specification (5). On the basis of the presence of the proposed external domain of CR4, it was surprising to discover that DEK1 also seems to be a membrane protein with a putative external domain. However, in light of the finding in other systems that precise cellular restriction of cell fate determination usually involves multiple genes (30), we now propose a model for aleurone cell development in which DEK1 plays a role in maintaining the aleurone cell fate specified at an early developmental stage in the endosperm. On the basis of the homology to calpains, we infer that DEK1 fulfills this function by acting as a cysteine proteinase on a substrate in aleurone cell signaling that is yet to be identified. In addition to the role of maintaining aleurone cell fate, we propose that DEK1 serves to restrict aleurone cell fate to the surface layer of the maize endosperm. We speculate that this function is mediated through interaction with molecules on the endosperm surface by the membrane portion and extracellular domain of DEK1, leading to activation of its cysteine proteinase activity.
Comparison of maize and Arabidopsis DEK1 reveals a high degree of conservation, 70% overall identity, even though the two plants represent the two main classes of flowering plants, monocots and dicots, respectively. Even more striking is the finding that the high degree of similarity extends beyond flowering plants and also includes gymnosperms such as loblolly pine. Indeed, extensive searches in available databases strongly suggest that DEK1 is the only calpain prototype found in plants, all plants currently investigated containing a single copy of DEK1. Clearly, the wide occurrence of DEK1, its highly conserved structure, its presence in many plant tissues (Fig. 2D), and its effect on maize embryogenesis in the form of a failure to generate a proper embryo axis, strongly point to an important role for DEK1 in plant signal transduction.
In conclusion, DEK1 is a member of the calpain gene superfamily, containing a cysteine proteinase domain conserved among all members of this gene family. In addition, DEK1 contains a high number of transmembrane segments suggestive of a complex three-dimensional structure capable of conveying contact with the extracellular environment. We propose that DEK1 maintain and restrict aleurone cell fate to the outer endosperm cell layer by activation of its cysteine proteinase through interacting with the extracellular environment.
Acknowledgments
We thank Jane Spauldine, Berit Morken, Peter Sekkelsten, and Karin O. Stenberg Olsen for the mutant screening, Karla Kurth for molecular analysis, and Lizabeth C. John for field support. The Biotechnology Program of the Bioprocessing and Production Division of The Research Council of Norway is acknowledged for support throughout this project.
Abbreviations
- Mu-mutator
dek1-defective kernel 1, mum1-mutator-induced mutant 1
- DAP
days after pollination
- CR4
tumor necrosis factor receptor-like receptor kinase CRINKLY4
- calpain
calcium-requiring cysteine proteinases
Footnotes
References
- 1.Olsen O-A. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- 2.Lopes M A, Larkins B A. Plant Cell. 1993;5:1383–1399. doi: 10.1105/tpc.5.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger F. Curr Opin Plant Biol. 1999;2:28–32. doi: 10.1016/s1369-5266(99)80006-5. [DOI] [PubMed] [Google Scholar]
- 4.Becraft P W, Stinard P S, McCarty D. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 5.Olsen O-A, Lemmon B E, Brown R C. Trends Plant Sci. 1998;3:168–169. [Google Scholar]
- 6.Brown R C, Lemon B E, Olsen O-A. Plant Cell. 1994;6:1241–1252. doi: 10.1105/tpc.6.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becraft P W, Brown R C, Lemmon B E, Olsen O-A, Opsahl-Ferstad H-G. Developmental Biology of Endosperm Development. Dordrecht, The Netherlands: Kluwer; 2000. [Google Scholar]
- 8.Morrison I N, Kuo J, O'Brian T P. Planta. 1975;123:105–116. doi: 10.1007/BF00383859. [DOI] [PubMed] [Google Scholar]
- 9.Neuffer M G, Sheridan W, Bendbow E. Maize Genet Cooperation News Lett. 1978;52:84–88. [Google Scholar]
- 10.Becraft P W, Asuncion-Crabb Y. Development (Cambridge, UK) 2000;127:4039–4048. doi: 10.1242/dev.127.18.4039. [DOI] [PubMed] [Google Scholar]
- 11.Neuffer M G, Coe E H, Wessler S R. Mutants of Maize. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. p. 71. [Google Scholar]
- 12.Bensen R J, Johal G S, Crane V C, Tossberg J T, Schnable P S, Meeley R B, Briggs S P. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamberlin Mark A, Horner Harry T, Palmer Reid G. Int J Plant Sci. 1994;155:421–436. [Google Scholar]
- 14.Saghai Maroof M A, Biyashev R M, Yang G P, Zhang Q, Allard R W. Proc Natl Acad Sci USA. 1994;91:5466–5470. doi: 10.1073/pnas.91.12.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomet P S. In: The Maize Handbook. Freeling M, Walbot V, editors. New York: Springer; 1994. pp. 243–249. [Google Scholar]
- 16.Schultz D J, Craig R, Cox-Foster D L, Mumma R O, Medford J I. Plant Mol Biol Rep. 1994;12:310–316. [Google Scholar]
- 17.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd D H, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 18.Robertson D R. Mutat Res. 1978;51:21–28. [Google Scholar]
- 19.Kalla R, Shimamoto K, Potter R, Nielsen P S, Linnestad K, Olsen O-A. Plant J. 1994;4:849–860. doi: 10.1046/j.1365-313x.1994.6060849.x. [DOI] [PubMed] [Google Scholar]
- 20.Sorimachi H, Suzuki K. J Biochem. 2001;129:653–664. doi: 10.1093/oxfordjournals.jbchem.a002903. [DOI] [PubMed] [Google Scholar]
- 21.Sato K, Kawashima S. Biol Chem. 2001;382:743–751. doi: 10.1515/BC.2001.090. [DOI] [PubMed] [Google Scholar]
- 22.Kishimoto A, Mikawa K, Hasimoto K, Yasuda I, Tanaka S, Tominaga M, Kuroda T, Nishizuka Y. J Biol Chem. 1989;264:4088–4092. [PubMed] [Google Scholar]
- 23.Tremblay R, Chakravarthy B, Hewitt K, Tauskela J, Morley P, Atkinson T, Durkin J P. J Neurosci. 2000;20:7183–7192. doi: 10.1523/JNEUROSCI.20-19-07183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthur J S C, Elce J S, Hegadorn C, Williams K, Greer P A. Mol Cell Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 26.Strobl S, Fernandez-Catalan C, Braun M, Huber R, Masumoto H, Nakagawa K, Irie A, Sorimachi H, Bourenkow G, Bartunik H, et al. Proc Natl Acad Sci USA. 2000;97:588–592. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 28.Becraft P W, Kang S H, Suh S G. Plant Physiol. 2001;127:486–496. [PMC free article] [PubMed] [Google Scholar]
- 29.Levy A A, Britt A B, Luehrsen K R, Chandler V L, Warren C, Walbot V. Dev Genet. 1989;10:520–531. doi: 10.1002/dvg.1020100611. [DOI] [PubMed] [Google Scholar]
- 30.Tabata T. Nat Rev Genet. 2001;2:620–630. doi: 10.1038/35084577. [DOI] [PubMed] [Google Scholar]
- 31.Brundrett M C, Kendrick B, Peterson C A. Biotechnol Histochem. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
- 32.Krogh A, Larsson B, von Heijne G, Sonhammer E L L. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]