Abstract
An improved understanding of the evolution of gene function at the molecular level may provide significant insights into the origin of biological novelty and adaptation. With the approach of ancestral protein reconstruction, we here address the question of how a dramatically enhanced ribonucleolytic activity and the related antiviral activity evolved in a recently duplicated ribonuclease (eosinophil-derived neurotoxin) gene of higher primates. We show that the mother gene of the duplicated genes had already possessed a weak antiviral activity before duplication. After duplication, substitutions at two interacting sites (Arg-64→Ser and Thr-132→Arg) resulted in a 13-fold enhancement of the ribonucleolytic activity of eosinophil-derived neurotoxin. These substitutions are also necessary for the potent antiviral activity, with contributions from additional amino acid changes at interacting sites. Our observation that a change in eosinophil-derived neurotoxin function occurs only when both interacting sites are altered indicates the importance of complementary substitutions in protein evolution. Thus, neutral substitutions are not simply “noises” in protein evolution, as many have thought. They may play constructive roles by setting the intramolecular microenvironment for further complementary advantageous substitutions, which can lead to improved or altered function. Overall, our study illustrates the power of the “paleomolecular biochemistry” approach in delineating the complex interplays of amino acid substitutions in evolution and in identifying the molecular basis of biological innovation.
It has long been recognized that gene duplication plays an important role in evolution by providing opportunities for the emergence of novel gene functions (1–9). The molecular evolutionary mechanisms underlying the functional divergence of duplicated genes, however, are not well understood, in part because it is difficult to reconstruct the sequences and functional characteristics of ancestral genes and proteins. Recent advances in computational inference (10–16) and experimental reconstruction (17, 18) of ancestral proteins provide significant hope for resolving some of these fundamental questions (19). With this approach, we here reveal the importance of complementary substitutions in the functional divergence of a pair of recently duplicated eosinophil-associated RNase genes of primates.
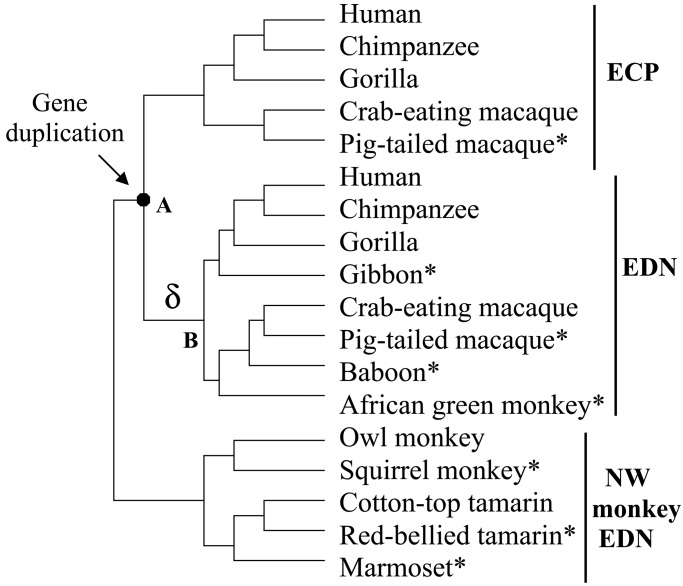
These two RNase genes are eosinophil-derived neurotoxin (EDN, or RNase 2) and eosinophil cationic protein (ECP, or RNase 3). They belong to the RNase A gene superfamily (20). It has been established that this gene pair emerged from a duplication event about 31 million years ago in the evolutionary lineage of hominoids and Old World (OW) monkeys (7, 21, 22). Only one copy of the EDN/ECP gene exists in the genomes of New World (NW) monkeys and prosimians examined so far (22; H.F.R., unpublished data), and this gene was named EDN by convention (Fig. 1). In humans, EDN and ECP proteins are found in the large specific granules of eosinophilic leukocytes (reviewed in ref. 23). In vitro studies showed that human EDN reduces the infectivity of certain RNA viruses including respiratory syncytial virus (RSV) and HIV (24, 25), through an RNase-dependent process (24). Antiviral activity is also found in OW monkey EDN (26). Human ECP, however, only shows a weak antiviral activity even at a relatively high concentration (27), but it has a cell-membrane-disruptive function that is likely responsible for its toxicity to bacteria and parasites (28–30). The EDNs of NW monkeys lack both antibacterial and antiviral activities (26, 31). As RNases, EDN and ECP can digest RNA, with EDNs of hominoids and OW monkeys being substantially more catalytically efficient than ECP and NW monkey EDN (27, 31–33), which suggests significant enhancement of RNase activity in the EDN lineage after gene duplication (Fig. 1). Because the observed antiviral activity depends on RNase activity, it is possible that the enhancement of RNase activity in EDN may contribute to its antiviral activity. Here we focus on the amino acid substitutions that are responsible for the enhanced RNase activity in EDN and their roles in the related antiviral activity.
Figure 1.
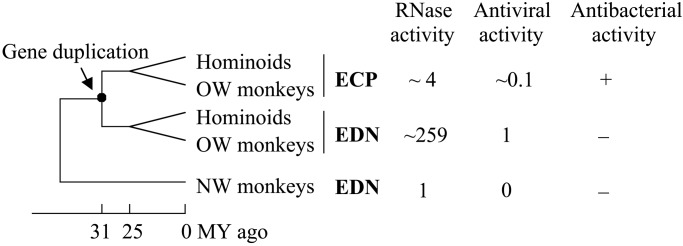
Gene duplication and functional changes in the evolution of EDN and ECP genes of higher primates. The RNase activities shown are from the present study, except that of ECP, which was transformed from the originally reported value to reflect the activity expected under the present experimental conditions. Antiviral activities relative to that of human EDN are shown. For antibacterial activity, + and − indicate activity detected or undetected. MY, million year (7, 22–24, 26, 27, 31, 33).
Materials and Methods
Sequencing of Additional EDN/ECP Genes.
The EDN gene was amplified by PCR from the genomic DNAs of the gibbon (Hylobates leucogenys), pig-tailed macaque (Macaca nemestrina), African green monkey (Cercopithecus aethiops), baboon (Papio hamadryas), red-bellied tamarin (Saguinus labiatus), marmoset (Callithrix jacchus), and squirrel monkey (Saimiri sciureus), with the primers derived from the published human EDN sequences as follows: 5′-ATGGTTCCAAAACTGTTCACTTCC-3′ and 5′-GAGGAGTGCTGATACAGGAGC-3′. The N terminus primer encodes the first 8 amino acids of the signal peptide, whereas the C terminus primer is positioned downstream of the coding region. PCRs were performed under conditions recommended by the manufacturer (Life Technologies, Rockville, MD). The PCR products were cloned into pCR II TA cloning vector (Invitrogen) and sequenced from both directions by the dideoxy chain termination method with the Perkin–Elmer 377 automatic sequencer. The ECP gene of the pig-tailed macaque was similarly obtained. The EDN and ECP sequences of the human (Homo sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), crab-eating macaque (Macaca fascicularis), and the EDN sequences of the cotton-top tamarin (Saguinus oedipus) and owl monkey (Aotus trivirgatus) were obtained from refs. 22 and 33.
Inference of Ancestral Sequences.
Ancestral protein sequences of all interior nodes in the phylogenetic tree of Fig. 2 were statistically inferred from the present-day sequences by using the distance-based Bayesian method (13) and parsimony method (34). The species tree as shown in Fig. 2 and the JTT-f model of amino acid substitution (35) were used in the Bayesian inference.
Figure 2.
The phylogenetic tree used for the inference of ancestral sequences. Sequences determined in this study are marked with *.
Mutagenesis.
The QuikChange site-directed mutagenesis kit of Stratagene was used to generate designed mutations in the EDN gene constructs according to the manufacturer's instructions. The mutations were confirmed by DNA sequencing.
Isolation of Recombinant Protein.
The expression constructs of human EDN and owl monkey EDN were as described (33). Both constructs were in pFLAG CTS bacterial expression vector (Kodak) with a bacterial signal peptide sequence at the N terminus and an octapeptide DYKDDDDK (FLAG) sequence at the C terminus. Previous studies showed that the FLAG octapeptide does not interfere with the folding or the catalytic activity of recombinant ribonucleases (31). Recombinant proteins were isolated from 6 liters of bacterial cultures after a 4-h induction with isopropyl-1-thio-β-galactoside (10 μM). After harvest and cell lysis by freeze-thaw and sonication, recombinant proteins were concentrated and isolated by M2 anti-FLAG monoclonal antibody affinity chromatography (Sigma). The concentration of the recombinant protein was determined by quantitative Western blot analysis with a FLAG-conjugated bacterial alkaline phosphatase protein at known concentration (Sigma).
Ribonuclease Assay.
The RNase activity of the recombinant proteins against a standard yeast tRNA substrate was measured in 40 mM sodium phosphate buffer (pH 7.4) at 25°C. Purified RNase (0.1–1.0 pmol) was added into 0.8 ml of the aforementioned buffer with 1.42 nmol of tRNA. The reaction was stopped by 0.5 ml of 20 mM lanthanum nitrate with 3% perchloric acid, and insoluble tRNA was removed by centrifugation. The amount of solubilized tRNA was determined by UV absorbance at 260 nm. The catalytic activity of the RNase was determined as the nanomol of RNA digested per second per nanomol of RNase used (31).
Antiviral Assay.
The activity of recombinant RNases in reducing the infectivity of RSV on Hep-2 human epithelial cells was examined by the quantitative shell vial amplification technique according to the published procedure (36). Recombinant RNases were added to Hep-2 monolayers growing on coverslips (50,000 cells per coverslip) followed by ≈2000 plaque-forming units (infectious units) of RSV-B (American Type Culture Collection). After a 60-min spin amplification, the vials were incubated at 37°C overnight, after which the coverslips were washed, acetone fixed, and stained with FITC-anti-RSV with methylene blue counterstain (Chemicon). Infected cells were identified by fluorescence microscopy.
Results
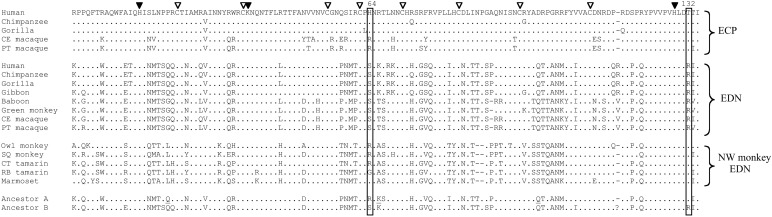
To identify the amino acid changes that are responsible for the enhancement of the RNase activity in EDN evolution, it is necessary to have EDN sequences from many primate species. We sequenced the coding region of the EDN gene from one additional hominoid (gibbon), three OW monkeys (pig-tailed macaque, baboon, and African green monkey), and three NW monkeys (red-bellied tamarin, marmoset, squirrel monkey). In addition, we obtained the ECP gene from the pig-tailed macaque. An expected phylogenetic tree was obtained when these gene sequences and previously obtained EDN and ECP sequences (22, 33) were analyzed together (data not shown). By using a known tree of these species (ref. 37; Fig. 2), we inferred the protein sequence for the most recent common ancestor of EDN and ECP (Fig. 2, node A) and that for the most recent common ancestor of hominoid and OW monkey EDNs (Fig. 2, node B) with the Bayesian method. The present-day and inferred ancestral sequences are presented in Fig. 3. We did not use the EDN or ECP sequences from the orangutan Pongo pygmaeus (22) in this analysis because our unpublished study suggested extra gene duplication and possible gene conversions in this species, which may confound the present analysis. The overall posterior probability for the inference is 96.4% for node A and 98.3% for node B. Nevertheless, three sites of ancestral protein A existed at which the posterior probabilities are lower than 50% (Fig. 3; and see Discussion). Furthermore, the Bayesian inference uses the JTT-f model of amino acid substitution, which may not be appropriate for all sites (13, 38). We thus also used the parsimony ancestral inference, and found that the result is in general agreement with the Bayesian inference. One of the equally parsimonious ancestral states often coincides with the Bayesian inference. Our further analysis is mainly based on the Bayesian inference, except that the ancestral states at positions involving gaps were inferred by parsimony (Fig. 3).
Figure 3.
The present-day and ancestral sequences of EDN and ECP. Only the mature peptides (without signal sequences) are shown. The two important sites discussed in the text are boxed. The three major catalytic residues and eight structural cysteines are marked with solid and open arrows, respectively. The ancestral amino acids with <50% posterior probabilities are underlined. The parsimony alternative ancestral states are P, or G, or L at site 21; R, or T, or A at site 65; and S or D at site 99. Experiments show that these three sites are unimportant to the RNase activity. “?” in ancestor B indicates equally parsimonious inferences of Arg or Val. CE macaque, crab-eating macaque; PT macaque, pig-tailed macaque; SQ monkey, squirrel monkey; CT tamarin, cotton-top tamarin; RB tamarin, red-bellied tamarin.
Comparison between the two ancestral sequences (A and B) suggests that 9-aa substitutions occurred in EDN after duplication but before the separation of hominoids and OW monkeys (branch δ, Fig. 2). Because EDNs from both humans and crab-eating macaque have enhanced RNase and antiviral activity (refs. 26 and 33; Fig. 1), it is likely that protein B already had these properties and that functional changes in EDN are attributable to some of the nine substitutions in branch δ (Fig. 2).
Our previous study indicated that the final three amino acids of EDN (Fig. 3) are necessary but not sufficient for the enhanced RNase activity (33). The inferred ancestral sequences (Fig. 3) suggest that only one amino acid change in this 3-aa region occurred in branch δ, and it was Thr-132→Arg. Both the original and resultant amino acids for this change have a >98% posterior probability, and parsimony gives the same inference, suggesting that it is a reliable inference. In fact, EDNs of all hominoids and OW monkeys have Arg-132, whereas EDNs of NW monkeys and all ECPs have Thr-132 (Fig. 3). Earlier sequence analysis by Beintema (39) also predicted the importance of this position, because Arg-132 may replace a basic residue at position 63, which is in proximity to a catalytic residue of the enzyme. We first tested the importance of residue 132 on human EDN by changing its Arg-132 back to Thr. The RNase activity of this mutant protein (GZ-C) was reduced by 12-fold (Table 1). We further examined the importance of this change in the EDN of owl monkey, a NW monkey, and our result shows that the Thr-132→Arg change does not improve the RNase activity, but decreases it further by 40% (GZ-3; Table 1). These findings strongly suggest that Arg-132 is necessary for the enhanced RNase activity of human EDN, and that Arg-132 contributes by interacting with at least another substitution.
Table 1.
RNase activities of native and mutant EDN proteins
| EDN proteins | Site 64 | Site 132 | RNase activity, s−1 | Relative activity |
|---|---|---|---|---|
| Owl monkey | Arg | Thr | 0.0063 ± 0.0002 | 1 |
| GZ-3 | Arg | Arg | 0.0035 ± 0.0003 | 0.6 |
| GZ-5 | Ser | Thr | 0.0097 ± 0.0025 | 1.5 |
| GZ-4 | Ser | Arg | 0.0920 ± 0.0040 | 15 |
| Human | Ser | Arg | 1.6300 ± 0.0300 | 259 |
| GZ-C | Ser | Thr | 0.1330 ± 0.0110 | 21 |
| GZ-F | Arg | Arg | 0.0218 ± 0.0012 | 3.5 |
| GZ-G | Arg | Thr | 0.1310 ± 0.0040 | 22 |
| Ancestor A | Arg | Thr | 0.0242 ± 0.0012 | 3.8 |
| AS-29 | Arg | Arg | 0.0191 ± 0.0006 | 3.0 |
| AS-28 | Ser | Thr | 0.0130 ± 0.0002 | 2.1 |
| AS-30 | Ser | Arg | 0.3160 ± 0.0060 | 50 |
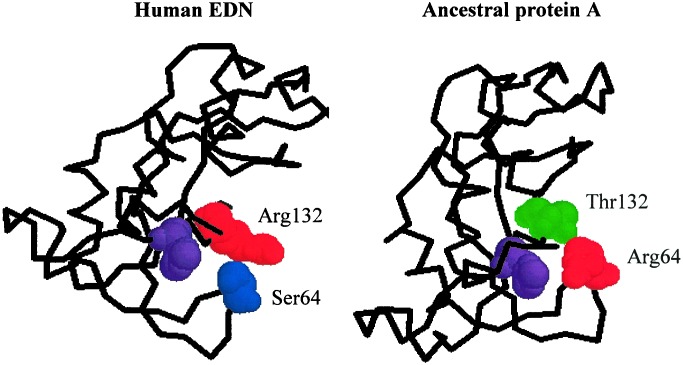
We examined the other 8-aa substitutions that occurred in branch δ in an attempt to identify residues that are likely to be interacting with Arg-132. The crystal structure of human EDN has been solved (40), and we mapped all nine aforementioned residues onto this structure. This analysis identified residue 64 as in close vicinity of residue 132 (Fig. 4). The distance between Ser-64 and Arg-132 is about 0.6 nm, which is shorter than the length of the side chain of an Arg residue. The crystal structure also shows that Ser-64 and Arg-132 are close to His-129, one of the three catalytic sites of the enzyme (Fig. 4), which provides structural evidence for possible roles of residues 64 and 132 in influencing the catalytic efficiency of the enzyme. The ancestral inference suggests Arg (posterior probability = 94.3%) for node A and Ser (99.6%) for node B at position 64 and an Arg-64→Ser substitution in branch δ. The same inference is given by parsimony. We modeled the structure of the ancestral protein A by using the human EDN structure (Fig. 4), and confirmed that Arg-64 and Thr-132 are in proximity to each other in this ancestral protein as well. Residue 64 has an invariant Ser in the EDNs of hominoids and OW monkeys, but it varies (His, Arg, and Gly) in ECPs and NW monkey EDNs. This pattern of variation is consistent with the hypothesis that Ser-64 becomes important in EDN only after the gene duplication. We subsequently investigated the role of this residue by experimentation.
Figure 4.
Crystal structures of human EDN and ancestral protein A, showing the physical interaction between residues at positions 64 and 132. The protein backbones are shown with the spacefill presentation of the residues at the two important sites as well as that of the catalytic His-129 (in purple). The structure of the ancestral protein A was modeled according to the human EDN structure (PDB Id: 1HI2) by SWISS-MODEL (53). Both structures are depicted with the RASMOL program (54).
When Ser-64 of human EDN is mutated back to Arg, the RNase activity decreases 74-fold (see GZ-F in Table 1), confirming that Ser-64 is important for the enhanced RNase activity in human EDN. On the contrary, when Arg-64 of owl monkey EDN is mutated to Ser (GZ-5; Table 1), the RNase activity does not change significantly. We then examined the activity of double mutants at positions 64 and 132. As expected, when Arg-64 and Thr-132 are changed to Ser and Arg, respectively, in owl monkey EDN, the RNase activity increases 15-fold (see GZ-4 in Table 1); the reciprocal changes in human EDN reduce the RNase activity 12-fold (GZ-G; Table 1).
Our results from the RNase activity assay (Table 1) and protein structural analysis (Fig. 4) strongly suggest the importance of the interacting residues Ser-64 and Arg-132 for the high RNase activity of human EDN. To demonstrate the role of these two substitutions directly in evolution, we reconstructed the inferred ancestral protein of node A (Fig. 3) by introducing 19-aa changes into human EDN. If our ancestral inference is reasonably good, we would expect to find that the ancestral protein is a functional RNase. Our result showed that this expectation is indeed met, and that the RNase activity of the ancestral protein is ≈4 times that of owl monkey EDN and equals that of human ECP. We then introduced Arg-64→Ser (AS-28) and Thr-132→Arg (AS-29) mutations to the ancestral protein, respectively. As expected from previous results, neither AS-28 nor AS-29 are ribonucleolytically more active than the ancestral protein (Table 1). However, when both substitutions are introduced (AS-30), the RNase activity increases 13-fold (Table 1). These results are in good agreement with those obtained from the present-day proteins (owl monkey and human EDNs), and demonstrate the role of the two complementary substitutions in the evolutionary enhancement of RNase activity in EDN.
As mentioned earlier, hominoid and OW monkey EDNs have antiviral activity against RSV in vitro by a process that depends directly on its RNase activity. We thus hypothesized that the two substitutions at positions 64 and 132 may also contribute to the origin (or improvement) of the antiviral activity in EDN after gene duplication (Fig. 1). We found that when both Arg-64 and Thr-132 are introduced into the human EDN (GZ-G), the antiviral activity decreases significantly (P < 0.01, Table 2). Owl monkey EDN is not known to be antiviral (26). When we introduced Ser-64 and Arg-132 into owl monkey EDN (GZ-4), we could not detect significant antiviral activity (Table 2), despite the fact that its RNase activity increased 15-fold (Table 1). The existence of a weak antiviral activity in ECP (27) suggests that the common ancestor of EDN and ECP may also possess a weak antiviral activity. Our result from the ancestral protein A confirms this prediction (Table 2). We then examined if the antiviral activity would increase when Ser-64 and Arg-132 are introduced into the ancestral protein. The resultant protein (AS-30) is found to have antiviral activity similar to that of the ancestral protein A (P > 0.2) despite enhanced RNase activity. This finding, combined with that of GZ-G, suggests that Ser-64 and/or Arg-132 are necessary, but not sufficient for a potent antiviral activity, and that additional amino acids contribute to the enhancement of the antiviral activity in EDN by interacting with one or both of these two residues.
Table 2.
Antiviral activity of EDN proteins
| RNases, ng/μl | Infectious units/ml ± SEM | % of control |
|---|---|---|
| Control buffer | 2,409 ± 83 | 100 |
| Owl monkey EDN (37) | 2,851 ± 304 | 118 |
| GZ-4 (17) | 2,409 ± 414 | 100 |
| Human EDN (25) | 1,122 ± 73* | 47 |
| GZ-G (33) | 1,621 ± 90* | 67 |
| Ancestor A (66) | 1,888 ± 26* | 78 |
| AS-30 (66) | 1,814 ± 157* | 75 |
, P < 0.001, when compared with the control.
Discussion
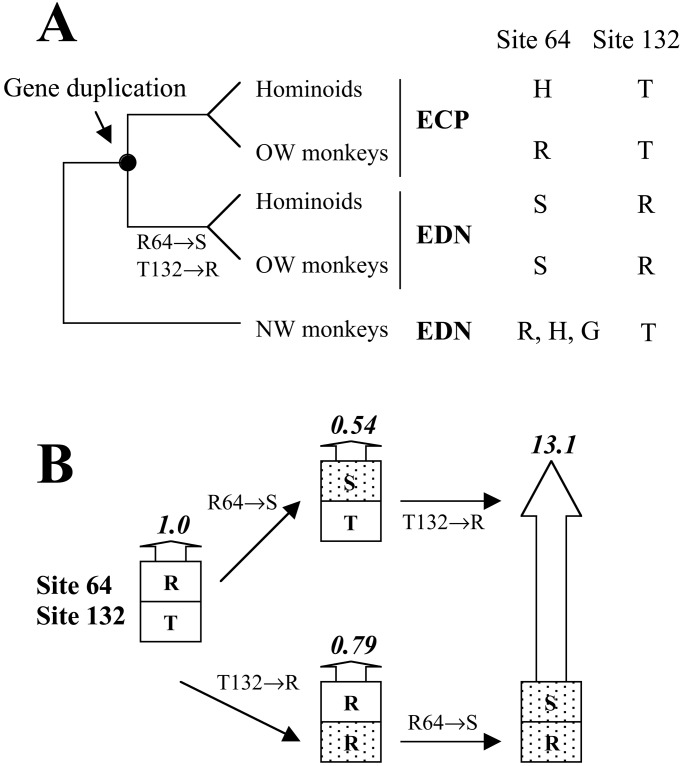
Combining protein structural and functional analysis with site-directed mutagenesis and reconstruction of ancestral protein, we identified two amino acid substitutions (Arg-64→Ser and Thr-132→Arg) that contributed to the evolutionary enhancement of the RNase activity of EDN after gene duplication. Our data strongly suggest that a functional as well as a physical interaction exists between these residues in the enzymatically active form of EDN. We further show that the mother gene of the duplicated EDN and ECP genes had a weak antiviral activity, and the aforementioned two substitutions, by interacting with yet-to-be-identified amino acid substitutions (hereby denoted as X), contributed to the increase of the antiviral activity in EDN evolution. The temporal order of these substitutions, however, is unknown because they all occurred in branch δ (Fig. 2), and no extant species can break this branch. As such, two scenarios are possible with regard to the order of the substitutions at positions 64 and 132 (Fig. 5). In the first scenario, Arg-64→Ser occurred first, which decreased the RNase activity by 46%, and then Thr-132→Arg occurred, which by interacting with Ser-64, raised the RNase activity 24-fold. In the second scenario, Thr-132→Arg occurred first and reduced the RNase activity by 21%. Then, Arg-64→Ser occurred and increased the RNase activity 17-fold. Although we do not know how much effect a 46% or 21% reduction in RNase activity of EDN could have had on the fitness of the organisms that bore the mutation, the fact that these mutations were fixed in population suggests that they were probably neutral or at most slightly deleterious. Indeed, the existence of a duplicate of EDN (i.e., ECP) at this stage may also reduce the fitness effect of these individual mutations. The second mutation, however, by interacting with the first one, increased the RNase activity 17- to 24-fold. It would also improve the antiviral activity of EDN if the aforementioned substitutions X had already occurred at this time. It seems reasonable to assume that the fitness effect of EDN depends primarily on its antiviral potency. Then, the second of the two substitutions becomes complementary advantageous, on the basis of earlier neutral or nearly neutral substitutions. On the other hand, if substitutions X occurred after those at positions 64 and 132, X will become complementary advantageous in raising the antiviral activity. In either case, it is clear that no single substitution alone served to enhance the RNase activity or antiviral activity, and the fitness effect and fate of a mutation can depend on the nature of previous substitutions. With this notion in mind, one realizes the importance of what have been called neutral substitutions during the evolution of protein function. This contribution may be particularly prominent in the evolution of new functions after gene duplication, because purifying selection is likely to be relaxed to some extent because of genetic redundancy, and previously prohibited substitutions become acceptable (9, 41–44). These neutral substitutions may change the protein sequence in such a way that further mutations are complementary advantageous. In this respect, neutral substitutions are not simply “noises”, as many have thought (e.g., ref. 45, p. 178); they can play a constructive role in the evolution of protein function. It is interesting to note that Kimura (ref. 43, p. 325) envisioned the importance of neutral substitutions in adaptive protein evolution by coining the term of “Dykuizen–Hartl” effect (46), which refers to the situation in which an otherwise neutral allele becomes advantageous under an altered environment. Our observation supports Kimura's idea and extends his meaning of “environment” to include all amino acid residues that interact in some fashion with the one under consideration. Because most proteins have complex crystal structures with many physically interacting residues, it is likely that, in the broadest sense, the “Dykuizen–Hartl” effect due to complementary advantageous substitutions is a more common theme in protein evolution than considered previously.
Figure 5.
Complementary substitutions in the evolutionary enhancement of RNase activity in EDN. (A) The present-day and ancestral amino acids at the two critical sites. (B) Evolutionary scenarios of the complementary substitutions. The height of and the number on each arrow show the relative RNase activity of the protein.
On the basis of the function and biology of EDN, we here reasoned that possession of EDN with potent antiviral activity is advantageous to the organism because it provides better protection against viral infections. Our previous analysis of the EDN and ECP sequences showed that the rate of nonsynonymous nucleotide substitution is about 1.6 times that of synonymous substitution in branch δ (7). Although we were not able to reject the neutral evolution hypothesis statistically because of the relatively small number of substitutions involved, the analysis did suggest the likelihood of positive selection occurring in branch δ, which is consistent with the results obtained in this study. The nucleotide substitution pattern in EDN is more complex than what existing mathematical models have assumed because of nonindependence among amino acid residues. How this complexity affects the test of positive selection is still unclear. At any rate, nucleotide substitutions involved in functional changes of duplicated genes are likely under more than one evolutionary force, as clearly demonstrated in RNASE1B, a duplicate of the pancreatic RNase gene of a leaf-eating monkey (9).
As mentioned, three amino acid residues have posterior probabilities lower than 50% in the inferred sequence of ancestor A (see Fig. 3). We examined possible influences of these three sites on the RNase activity on the background of either human EDN or owl monkey EDN, and our results showed that these sites have no significant effect (data not shown). As such, the function of the inferred ancestral protein A is likely unaffected by the uncertainty of the ancestral states at the three sites. Our RNase assays show that the activity of human EDN is about 68 times that of ancestral protein A (Table 1). Therefore, in addition to substitutions at positions 64 and 132, which explain a 13-fold increase in activity, other substitutions have also contributed to another ≈5-fold increase. Ancestral protein A has a RNase activity similar to that of ECP, but four times that of owl monkey EDN, suggesting that the RNase activity has not changed in ECP evolution since gene duplication. It also suggests either that the RNase activity has been reduced in the NW monkey lineage since its separation from OW monkeys and hominoids, or that the RNase activity had increased in the OW monkey and hominoid lineage before the gene duplication. Similar hypotheses can also be made with regard to the evolution of the antiviral activity. It might be possible to test these hypotheses in the future by reconstructing the EDN of the common ancestor of higher primates (hominoids, OW monkeys, and NW monkeys). This reconstruction may also help answer the puzzling question on the function of EDN in NW monkeys, because it does not seem to have a strong activity in any functional aspect seen in the ECPs or EDNs of hominoids and OW monkeys. It will also be interesting to examine the function of EDN in prosimian primates and non-primate mammals. Rodent eosinophil-associated RNase genes, which, collectively, are the orthologue of the primate EDN and ECP genes, have been cloned (47, 48). These genes are products of recent gene duplication in rodents (48, 49), and they show varied RNase activity (49, 50).
A final interesting result of this work is the observation that EDN antiviral activity does not correlate precisely with the specific degree of RNase activity, at least within the range of activities represented by the extant and ancestral forms studied here. In some sense, this result is not unexpected, because we have already shown that several highly enzymatically active members of the RNase A superfamily, including RNase 6 and pancreatic RNase, have no antiviral activity against RSV (27). Our finding that potent RNase activity is necessary, but not sufficient for antiviral activity further emphasizes the complex interplays of amino acid substitutions in functional changes of EDN. Host–pathogen interactions are usually dynamic, and RNA viruses such as RSV evolve rapidly (51). We assumed and subsequently demonstrated that the 31-million-year-old ancestral protein A could reduce the infectivity of present-day RSV for its present-day target cells, although this infectivity may represent the relic of activity directed against a more ancient form of this or a related virus. However, this result is also consistent with the hypothesis that the antiviral mechanism of EDN does not depend on direct interaction of the ribonuclease with the extracellular form of the virion, a hypothesis that has gained support from DNA polymorphic data of human EDN (52) and the observation that EDN also has activity in vitro against at least one other unrelated RNA virus (25). Although delineation of the antiviral mechanism is currently under study, it seems likely that EDN interacts with both target cell and virus surface components such that viral genomic RNA (and/or the cellular transcription apparatus) is brought into close proximity to EDN at some stage of the primary infection (23). As such, the two steps of the process—interaction with cell surface and/or viral targets, followed by ribonucleolytic deactivation of RNA—are clearly complementary events. Indeed, it is intriguing to consider the possibility that the motifs mediating the primary cell–virus–ribonuclease interactions may also evolve in a fashion complementary to that leading to increased RNase activity, and together serve to enhance the overall antiviral efficiency of EDN. Future work aimed at identifying critical amino acid substitutions in EDN will help uncover the molecular mechanism of its antiviral activity and may provide further insights into the evolutionary forces behind it.
Acknowledgments
We thank Dr. Kimberly Dyer for excellent technical assistance. We also thank the member editor and two anonymous referees for constructive comments on an earlier version of the manuscript. This work was supported in part by a start-up fund and a Rackham grant of University of Michigan (to J.Z.).
Abbreviations
- EDN
eosinophil-derived neurotoxin
- ECP
eosinophil cationic protein
- RSV
respiratory syncytial virus
- OW
Old World
- NW
New World
Footnotes
References
- 1.Bridges C B. Teaching Biol. 1936. November, 17–23. [Google Scholar]
- 2.Stephens S G. Advan Genet. 1951;4:247–265. doi: 10.1016/s0065-2660(08)60237-0. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Sex Chromosomes and Sex-Linked Genes. New York: Springer-Verlag; 1967. [Google Scholar]
- 4.Nei M. Nature (London) 1969;221:5175–5177. doi: 10.1038/221040a0. [DOI] [PubMed] [Google Scholar]
- 5.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 6.Li W H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 7.Zhang J, Rosenberg H F, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes A L. Adaptive Evolution of Genes and Genomes. New York: Oxford Univ. Press; 1999. [Google Scholar]
- 9. Zhang, J., Zhang, Y.-p. & Rosenberg, H. F. (2002) Nat. Genet., in press. [DOI] [PubMed]
- 10.Schluter D. Nature (London) 1995;377:108–109. doi: 10.1038/377108a0. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Kumar S, Nei M. Genetics. 1995;141:1641–1651. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koshi J M, Goldstein R A. J Mol Evol. 1996;42:313–320. doi: 10.1007/BF02198858. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Nei M. J Mol Evol. 1997;44, Suppl. 1:S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- 14.Pagel M. Syst Biol. 1999;48:612–622. [Google Scholar]
- 15.Galtier N, Tourasse N, Gouy M. Science. 1999;283:220–221. doi: 10.1126/science.283.5399.220. [DOI] [PubMed] [Google Scholar]
- 16.Pupko T, Pe'er I, Shamir R, Graur D. Mol Biol Evol. 2000;17:890–896. doi: 10.1093/oxfordjournals.molbev.a026369. [DOI] [PubMed] [Google Scholar]
- 17.Jermann T M, Opitz J G, Stackhouse J, Benner S A. Nature (London) 1995;374:57–59. doi: 10.1038/374057a0. [DOI] [PubMed] [Google Scholar]
- 18.Chandrasekharan U M, Sanker S, Glynias M J, Karnik S S, Husain A. Science. 1996;271:502–505. doi: 10.1126/science.271.5248.502. [DOI] [PubMed] [Google Scholar]
- 19.Golding G B, Dean A M. Mol Biol Evol. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- 20.D'Alessio G, Riordan J F. Ribonucleases: Structures and Functions. New York: Academic; 1997. [Google Scholar]
- 21.Hamann K J, Ten R M, Loegering D A, Jenkins R B, Heise M T, Schad C R, Pease L R, Gleich G J, Barker R L. Genomics. 1990;7:535–546. doi: 10.1016/0888-7543(90)90197-3. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg H F, Dyer K D, Tiffany H L, Gonzalez M. Nat Genet. 1995;10:219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg H F, Domachowske J B. J Leukocyte Biol. 2001;70:691–698. [PubMed] [Google Scholar]
- 24.Domachowske J B, Dyer K D, Bonville C A, Rosenberg H F. J Infect Dis. 1998;177:1458–1464. doi: 10.1086/515322. [DOI] [PubMed] [Google Scholar]
- 25.Lee-Huang S, Huang P L, Sun Y, Huang P L, Kung H F, Blithe D L, Chen H C. Proc Natl Acad Sci USA. 1999;96:2678–2681. doi: 10.1073/pnas.96.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domachowske J B, Bonville C A, Dyer K D, Rosenberg H F. Nucleic Acids Res. 1998;26:5327–5332. doi: 10.1093/nar/26.23.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domachowske J B, Dyer K D, Adams A G, Leto T L, Rosenberg H F. Nucleic Acids Res. 1998;26:3358–3363. doi: 10.1093/nar/26.14.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young J D E, Peterson C G B, Venge P, Cohn G J. Nature (London) 1986;321:613–616. doi: 10.1038/321613a0. [DOI] [PubMed] [Google Scholar]
- 29.Waters L S, Taverne J, Tai P C, Spry C J, Targett G A, Playfair J H. Infect Immun. 1987;55:877–881. doi: 10.1128/iai.55.4.877-881.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehrer R I, Szklarek D, Barton A, Ganz T, Hamann K J, Gleich G J. J Immunol. 1989;142:4428–4434. [PubMed] [Google Scholar]
- 31.Rosenberg H F, Dyer K D. J Biol Chem. 1995;270:21539–21544. doi: 10.1074/jbc.270.37.21539. [DOI] [PubMed] [Google Scholar]
- 32.Slifman N R, Loegering D A, McKean D J, Gleich G J. J Immunol. 1986;137:2913–2917. [PubMed] [Google Scholar]
- 33.Rosenberg H F, Dyer K D. Nucleic Acids Res. 1997;25:3532–3536. doi: 10.1093/nar/25.17.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitch W M. Syst Zool. 1971;20:406–416. [Google Scholar]
- 35.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 36.Domachowske J B, Bonville C A. BioTechniques. 1998;25:644–647. doi: 10.2144/98254dt01. [DOI] [PubMed] [Google Scholar]
- 37.Goodman M, Porter C A, Czelusniak J, Page S L, Schneider H, Shoshani J, Gunnell G, Groves C P. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 39.Beintema J J. FEBS Lett. 1989;254:1–4. doi: 10.1016/0014-5793(89)80996-2. [DOI] [PubMed] [Google Scholar]
- 40.Mosimann S C, Newton D L, Youle R J, James M N. J Mol Biol. 1996;260:540–552. doi: 10.1006/jmbi.1996.0420. [DOI] [PubMed] [Google Scholar]
- 41.Ohno S. Nature (London) 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- 42.Li W-H, Gojobori T. Mol Biol Evol. 1983;1:94–108. doi: 10.1093/oxfordjournals.molbev.a040306. [DOI] [PubMed] [Google Scholar]
- 43.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 44.Lynch M, Conery J S. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 45.Hartl D L, Clark A G. Principles of Population Genetics. 3rd Ed. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 46.Dykhuizen D, Hartl D L. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson K A, Olson E V, Madden B J, Gleich G J, Lee N A, Lee J J. Proc Natl Acad Sci USA. 1996;93:12370–12375. doi: 10.1073/pnas.93.22.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Dyer K D, Rosenberg H F. Proc Natl Acad Sci USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singhania N A, Dyer K D, Zhang J, Deming M S, Bonville C A, Domachowske J B, Rosenberg H F. J Mol Evol. 1999;49:721–728. doi: 10.1007/pl00006594. [DOI] [PubMed] [Google Scholar]
- 50.McDevitt A L, Deming M S, Rosenberg H F, Dyer K D. Gene. 2001;267:23–30. doi: 10.1016/s0378-1119(01)00392-4. [DOI] [PubMed] [Google Scholar]
- 51.Woelk C H, Holmes E C. J Mol Evol. 2001;52:182–192. doi: 10.1007/s002390010147. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Rosenberg H F. Genetics. 2000;156:1949–1958. doi: 10.1093/genetics/156.4.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 54.Sayle R A, Milner-White E J. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]