Abstract
It is unclear whether the core promoter is involved in developmental regulation. To address this question, we mutated the TATA box of the human γ-globin gene, produced transgenic mice, and examined the effect of the mutation during the course of mouse development. In our test system, the γ-globin gene is expressed at similar levels in the embryonic and adult erythroid cells. The TATA box mutation dramatically reduced expression of the γ-globin gene in the adult but not in embryonic erythroid cells. In addition, the disruption of the γ TATA box significantly reduced the recruitment of TATA box-binding protein (TBP) in the adult cells, but not in embryonic cells, suggesting that the recruitment of TBP to the γ gene promoter is developmentally specific. Similarly, the recruitment of transcription factor II B and RNA polymerase II to the γ promoter was affected in the adult but not in embryonic cells. The distinct effects of the TATA mutation in the embryonic and adult developmental stages suggest that the basal transcription apparatus can be recruited to a core promoter in a developmental stage-dependent manner. The TATA mutation resulted in a shift of transcription initiation site 6 bp or longer upstream to the cap site both in the embryonic and adult erythrocytes. We conclude that the TATA box determines the initiation site but not the efficiency of transcription of the γ-globin gene.
The general transcriptional apparatus has been viewed as a nonregulated basal component because the RNA polymerase II (Pol II) machinery is largely thought to be invariant in its composition or expression. The core promoter is thought to be neutral in developmental regulation. Recent evidence suggests that the basal transcriptional machinery itself might display tissue-specific or gene-selective properties (refs. 1–7; for review, see refs. 8–11). However, it is unclear whether the core promoter is involved in developmental regulation. Indirect evidence for developmental specificity has been produced by studies showing that enhancers stimulate promoters by a TATA box-independent mechanism in undifferentiated embryonic cell types, but they change to a TATA box-dependent mode as cell differentiation proceeds (12, 13). We decided to test directly the possibility of developmental specificity of the core promoter by using embryonic and adult cells from transgenic mice carrying a TATA box-mutant γ-globin gene promoter.
The human β-globin cluster provides a model system for studying developmental regulation of gene expression. The high-level erythroid-specific and developmental stage-specific expression of the genes of the β−globin locus is thought to be under the control of the locus control region (LCR) and the proximal promoter elements of individual genes. The cis regulatory elements (such as CACCC box, GATA-1 sites, CCAAT box, NF-E2/Ap1 sites, and YY1 sites) present in both the LCR and the proximal promoter regions, and their associated trans factors (such as EKLF family proteins, GATA-1 family proteins, NF-E2, NF-Y/CP1, and YY1) have been extensively investigated (reviewed in ref. 14). Only a few studies have been done to assess the role of core promoter elements in the regulation of β-like-globin genes. The core promoter usually consists of three kinds of cis elements that may not be simultaneously present in a promoter: the TATA box, the initiator element (Inr), and the downstream promoter element. All of the promoters of β−globin locus genes contain a TATA box. A functional initiator element and an extra downstream promoter element (DPE) are also present in the human β-globin promoter (15, 16). The TATA box of chicken β-globin gene plays a role in mediating promoter–enhancer communication through the binding of GATA-1 (17). The association of transcription factor II D (TFIID) with the TATA is not a limiting factor in the rate of initiation of the human β-globin promoter transcription (18). The recruitment of EKLF to 5′HS2 and 5′HS3 of LCR depends on the TATA box of the human β-globin promoter (19).
The experimental approach was to mutate the γ gene TATA box and test whether the mutation affected the recruitment of the basal transcriptional machinery and γ gene expression in embryonic and adult erythroid cells of transgenic mice carrying the γ TATA box mutation. To obtain such information, transgenic mice showing significant levels of γ gene expression are required, and such mice are produced when a micro-LCR cassette is linked with a γ gene containing a promoter truncated to −382γ (20). In such μLCR(−382) Aγ mice, the γ gene is expressed at high level in all of the developmental stages. By introducing the γ TATA box mutation in the μLCR(−382)Aγ construct, we expected to readily detect and quantitate any negative effects of the mutation on Aγ gene expression. By using this system, we found that (i) in vivo, the TATA box is necessary for γ gene expression during definitive erythropoiesis but is redundant during embryonic erythropoiesis; (ii) TATA box-binding protein (TBP) is recruited to the γ gene promoter and is required for γ gene expression in vitro and in vivo; and (iii) the recruitment of TBP and the general transcriptional machinery depends on the intact TATA box in definitive erythroid cells but not in embryonic erythroid cells. Taken together, our results suggest that the mechanism for recruiting the general transcriptional machinery to the γ gene promoter in embryonic erythroid cells is different from that in definitive erythroid cells.
Materials and Methods
Constructs, Transgenic Mice, RNA and DNA Analysis.
Construct μLCR(−382)Aγ, which contained an μLCR-linked human Aγ gene from −382 (StuI) to +1950 (HindIII), has been described (20). Plasmid μLCR(−382)Aγ(mut TATA) was constructed by substituting the TATA box sequence “AATAAA” with the sequence “GCTAGC” (a NheI site) through PCR-based mutagenesis. Production of transgenic mice, RNase protection assay, and copy number measurement were described (21). Briefly, the construct μLCR(−382)Aγ(mut TATA) was microinjected into mouse eggs, and transgenic founders were identified by slot blot hybridization. Copy number of the transgenic gene in the offspring was determined by Southern hybridization with human genomic DNA as standard and a mouse α-globin probe as loading control. Total RNA was prepared from the tissues containing the primitive erythrocytes (d10, d12 blood and yolk sac) and the tissues containing the definitive erythrocytes (d12, d16 fetal liver, and d16 adult blood). The γ-globin mRNA expression was detected by RNase protection assay and quantified by a PhosphorImager. To minimize experimental error, samples from individual animals were quantified independently and multiple measurements were performed both in RNase protection and Southern hybridization assays.
Preparation of Nuclear Extracts.
HeLa nuclear extracts were purchased from Promega. Nuclear extracts from K562 and murine erythroleukemia (MEL) cells were prepared as described either by Mantovani (22) or by Dignam et al. (23).
Gel Retardation Assay.
Labeled oligonucleotides (1 × 104 cpm) encompassing the wild-type or mutated γ TATA boxes were used as probes and incubated with purified TBP (Promega) for 10 min at room temperature in the binding buffer containing 10 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, 0.5 mM DTT, 10% (vol/vol) glycerol, 1 μg poly(dI-dC), and 0.05% Nonidet P-40. For supershifting assay, antibodies against TBP (Santa Cruz Biotechnology) were added in the amount of 100 ng and 500 ng, respectively, and incubated for another 10 min. Samples were electrophoresed in 4.5% polyacrylamide gel in 0.5× Tris-borate/EDTA (TBE) buffer (45 mM Tris-borate/1 mM EDTA, pH 8.0) containing 4 mM Mg2+ at 4°C. The sequences of the double-stranded oligonucleotides used as probes are: γ TATA box, 5′GGATGAAGAATAAAAGGAAGCACCCT3′; γ mut TATA box, 5′GGATGAAGAGCTAGCGGAAGCACCCT3′.
In Vitro Pol II Transcription Assay.
In vitro transcription assays were performed as described by Mantovani (22). Plasmid used as transcription template was prepared by using NucleoSpin Extraction kit (CLONTECH). In TBP-depletion assays, the HeLa nuclear extracts were treated at 47°C for 10 min before in vitro transcription assay. Purified TFIID (TBP; 1 pfu per reaction, Promega) was used. Superscript II RNase H− Reverse Transcriptase (GIBCO/BRL) was used for primer extension.
Chromatin Immunoprecipitation (ChIP) Assay.
ChIPs were performed as described (24, 25) with minor modifications. One spleen, or brain, or six yolk sacs were used per condition. The tissues from the transgenic animals (identified by γ-globin specific antibody) were passed through a 21-gauge needle in 5–10 ml of RPMI medium 1640, and then crosslinked immediately. Anti-TBP [TFIID(TBP) (SI-1), sc-273], anti transcription factor II B [TFIIB(SI-1), sc-274], anti-Pol II [Pol II(N-20), sc-899], antibodies, and normal rabbit IgG were obtained from Santa Cruz Biotechnology. Protein A and Protein G-Sepharose 4B Conjugate was obtained from Zymed.
Quantitative PCR.
Real-time quantitative PCR was performed on the purified chromatin samples by using the Roche LightCycler system (Roche Molecular Biochemicals). PCRs were performed by using a SYBR-Green Master kit (Roche Molecular Biochemicals). The primer pairs used were designed according to the specific sequence of target DNA based on human and mouse β-globin sequences (GenBank accession numbers U01317 and X14061) by using PRIMER3 software for primer design (available online at http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi).
The following primer pairs were designed and used in this work: human Aγ TATA box, 5′GGCTGGCTAGGGATGAAGA3′ (upper strand) and 5′GGCGTCTGGACTAGGAGCTT3′ (lower strand). Mouse βmaj gene promoter region: 5′GTCATCACCGAAGCCTGATT3′ (upper strand), and 5′TGTCTGTTTCTGGGGTTGTG3′ (lower strand). Mouse ɛy gene promoter region: 5′GTTGAAGGAGGAGCCAAAAA3′ (upper strand) and 5′TGCTAGAAGTGGTGGCCTTT3′ (lower strand).
Results
An Intact TATA Box and TBP Recruitment Are Required for in Vitro γ-Globin Gene Transcription.
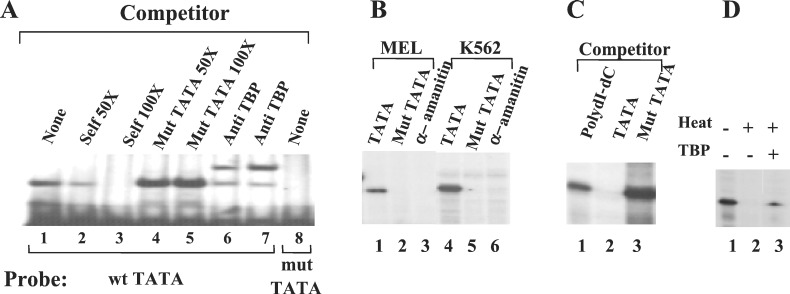
To investigate the role of the core promoter of the γ-globin gene in the recruitment of the basal transcription machinery, we mutated the canonical TATA box by substituting the sequence “AATAAA” with the sequence “GCTAGC” in the context of the construct μLCRAγ(−382) (20). The γTATA box is shown by gel mobility shift assays to bind TBP (Fig. 1A, lane 1). The retarded band can be competed away with an excess of nonradiolabeled wild-type probe (lanes 2 and 3) but not by the mutant TATA box probe (lanes 4 and 5). The complex of the TATA box and TBP can be supershifted by the antibody against TBP (lanes 6 and 7). When the mutated TATA box was used as probe, no retarded band was observed, suggesting that the mutated TATA box is incapable of being bound by TBP (lane 8).
Figure 1.
The in vitro γ-globin gene expression depends upon an intact TATA box and recruitment of TBP. (A) The γTATA binds TBP, and the mutation of the γTATA abrogates the TBP binding. Gel shift assays were carried out as described (20). The mutated TATA box fails to bind TBP (lane 8) and to compete away the TBP binding on the wild-type TATA box (lane 4 and 5). The sequences of the double-stranded oligonucleotides used as probes were: γ TATA box, 5′GGATGAAGAATAAAAGGAAGCACCCT3′; γ mut TATA box, 5′GGATGAAGAGCTAGCGGAAGCACCCT3′. (B) The TATA box mutation abolishes transcription of the γ-globin gene. In vitro RNA transcription assays were performed by using nuclear extracts from MEL or K562 cells. The DNA templates used in the assays were μLCR(−382)Aγ (lanes 1, 3, 4, and 6) and μLCR(−382)Aγ(mut TATA) (lanes 2 and 5). α−Amanitin was added in the assays shown in lanes 3 and 6. (C) In vitro γ-globin gene transcription is inhibited by addition of the TATA box oligonucleotide (lane 2) into the assay system but not by poly(dI-dC) or the oligonucleotide with mutated TATA box. (D) TBP heat inactivation. The plasmid HS2γLuc and HeLa nuclear extract were used for in vitro transcriptions (lane 1). Lane 2: HeLa nuclear extracts were treated at 47°C for 10 min before in vitro transcription assay. Lane 3: purified TFIID (TBP) was added into the heat-treated HeLa nuclear extract.
To investigate whether the TATA mutation affects γ gene transcription, we performed in vitro transcription assay by using nuclear extracts from HeLa, MEL, and K562 cells. Similar results were obtained with the three kinds of nuclear extracts. Fig. 1B shows representative results from MEL and K562 nuclear extracts. Both extracts were capable of transcribing the wild-type Aγ gene (Fig. 1B, lanes 1 and 4). The transcription was Pol II-specific because it was suppressed by the addition of α-amanitin (Fig. 1B, lanes 3 and 6). The Aγ gene transcription was abolished by the TATA box mutation (Fig. 1B, lanes 2 and 5). This transcription was independent of the presence of the LCR because similar results were obtained by using an Aγ construct without the μLCR (data not shown). The TATA box dependency of γ gene transcription was supported further by the results of TATA box competition assay (Fig. 1C). In an in vitro transcription assay that used HeLa cell nuclear extract and the wild-type Aγ template, addition of TATA box oligonucleotide inhibited Aγ gene transcription (Fig. 1C, lane 2). By contrast, addition of TATA box-mutated oligonucleotide failed to suppress Aγ gene transcription (Fig. 1C, lane 3), similar to the addition of a nonspecific competitor poly(dI-dC) (Fig. 1C, lane 1). To test the requirement of TBP for γ gene transcription, a TBP-depletion assay was carried out (Fig. 1D). TBP was inactivated by treating the HeLa nuclear extract at 47°C for 10 min before in vitro transcription assay. This treatment abolished the γ promoter-directed transcription (Fig. 1D, lane 2). When purified TBP was added to the heat-treated nuclear extract, the transcription was partially restored (lane 3).
These results indicate that an intact γ TATA box and TBP recruitment are essential for in vitro transcription of the γ-globin gene. Taken together, these results demonstrate that, as expected, the γ-globin gene contains a typical TATA box- and TBP-dependent Pol II promoter.
The TATA Box Is Necessary for γ Gene Expression During Definitive Erythropoiesis, but It Is Dispensable in Embryonic Erythropoiesis.
To assess the effects of the TATA box mutation on γ gene expression in erythroid cells undergoing normal development, we produced transgenic mice with the construct μLCR(−382)Aγ(mut TATA). For developmental studies, timed pregnancies were interrupted at 10, 12, and 16 days, and yolk sac, blood, and fetal liver samples were collected for measurement of human γ and murine α and ζ mRNA by RNase protection assay. Quantitative data of γ gene expression at different developmental stages are summarized in Table 1. The day-10 blood and yolk sac represent an early stage of embryonic erythropoiesis. At day 12, the yolk sac still contains large numbers of nucleated embryonic red cells, and the fetal blood is composed predominantly of nucleated erythrocytes of yolk sac origin. The day-12 fetal liver is the site of adult erythropoiesis, and it is composed of adult erythroblasts and a small proportion (less than 10%) of hematogenously contaminating embryonic erythroblasts. At day 16, the fetal blood contains mostly adult-type red cells and the liver adult erythroblasts. The adult blood contains erythroid cells originating from bone marrow. As shown in Table 1, the introduction of the TATA box mutation did not affect Aγ gene expression in embryonic cells; the level of γ mRNA in day-10 and day-12 blood and yolk sac samples were essentially the same as those of the control μLCR(−382)Aγ transgenic mice carrying a γ promoter with the wild-type TATA box. In contrast, in the adult blood, Aγ gene expression in the TATA box-mutant mice was about 5-fold lower compared with the control mice. From these experiments, we conclude that an intact TATA box is not required for the normal level of γ gene expression in the embryonic cells, whereas it is essential for full-level γ expression in the adult cells.
Table 1.
Expression of the human γ gene in the transgenic mice carrying the μLCR (−382)Aγ(mut TATA) construct
| Line | Copy number | Human γ % of murine α-like mRNA per
copy
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Embryonic
erythropoiesis
|
Definitive erythropoiesis
|
|||||||||
| Day 10
|
Day 12
|
Day
12
|
Day 16
|
Adult
|
||||||
| Blood | Yolk sac | Blood | Yolk sac | Fetal liver | Blood | Fetal liver | Blood | |||
| μLCR(−382)Aγ(mut TATA) | ||||||||||
| A | 6 | 4.2 ± 0.3 | 7.6 ± 1.0 | 13.5 ± 4.8 | 19.3 ± 1.2 | 12.0 ± 1.1 | 12.0 ± 3.4 | 7.7 ± 1.1 | 4.3 ± 1.2 | |
| B | 24 | 3.5 ± 0.2 | 6.5 ± 1.1 | 8.8 ± 1.2 | 16.0 ± 1.9 | 7.1 ± 1.9 | 4.4 ± 0.9 | 4.1 ± 0.4 | 2.0 ± 0.5 | |
| C | 16 | 6.5 ± 0.8 | 9.4 ± 1.7 | 17.6 ± 1.8 | 27.5 ± 6.4 | 14.8 ± 1.1 | 9.1 ± 2.9 | 7.5 ± 1.1 | 2.6 ± 0.8 | |
| D | 8 | 4.3 ± 0.7 | 7.8 ± 1.1 | 8.8 ± 2.6 | 19.0 ± 3.1 | 12.1 ± 2.3 | 9.2 ± 1.0 | 9.9 ± 1.9 | 4.9 ± 0.9 | |
| Mean | 4.6 ± 1.3 | 7.8 ± 1.2 | 12.2 ± 4.2 | 20.5 ± 4.9 | 11.5 ± 3.2 | 8.7 ± 3.2 | 7.3 ± 2.4 | 3.5 ± 1.4 | ||
| μLCR(−382)Aγ* | ||||||||||
| Mean | 5.7 ± 6.3 | 5.2 ± 3.3 | 10.4 ± 9.5 | 21.0 ± 7.4 | 11.8 ± 2.8 | 13.4 ± 4.7 | 15.7 ± 4.8 | 16.5 ± 4.5 | ||
The Recruitment of TBP to the γ Gene Promoter Is Developmentally Specific.
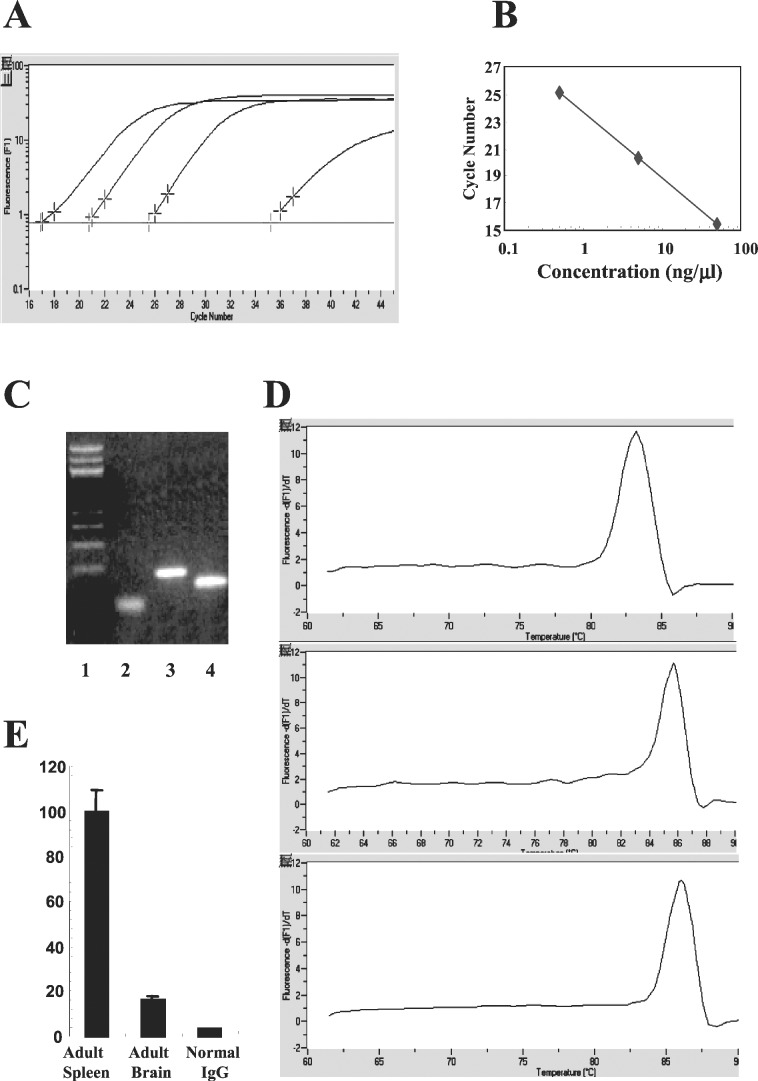
The binding of TBP to TATA box initiates the assembly of a transcription initiation complex. However, the dispensability of TATA box for γ gene expression in embryonic erythroid cells suggests that either TBP is not required for the recruitment of the general transcription machinery or that TBP is required, but its recruitment is independent of the TATA box. To discriminate between the two possibilities, we measured TBP recruitment in embryonic blood and yolk sac of day-11 transgenic fetuses carrying the TATA box-mutated γ gene by real-time PCR quantitative ChIP assay. The DNA brought down by the TBP antibody was quantitated by real-time PCR with a set of primers spanning the TATA box regions of the human Aγ promoter. A representative panel of data is shown in Fig. 2A. Linear correlation between the amount of template and PCR product is well maintained between 0.5 to 50 ng of DNA (Fig. 2B). Fig. 2C shows the result of electrophoresis of the PCR product, demonstrating the correct size; Fig. 2D shows the melting curve, indicating the purity of the product. To minimize experimental imprecision, TBP recruitment in the murine ɛy-globin promoter was quantitated simultaneously in the same TBP-immunoprecipitated samples and served as internal control. The effect of the TATA box mutation on TBP recruitment was calculated by dividing the ratio of TBP recruitment on the mutated γ promoter over that of the ɛy promoter in the TATA-mutated mice by the ratio in the control mice. All data were corrected by the copy numbers of the transgenes. The specificity of TBP antibody is demonstrated in Fig. 2E. Normal IgG barely brought down the γ TATA box. Only 15–20% of γ TATA box were immunoprecipitated by the TBP antibody in the mouse brain, the tissue that does not express the γ-globin gene. These results confirmed the specificity and quantitative nature of the real-time PCR-based ChIP assay.
Figure 2.
Real-time PCR-based ChIP assay. (A) Representative data generated by LightCycler (Roche Molecular Biochemicals) showing the amplification curve of three different amounts of DNA along with a control (extreme Right). Human genomic DNA was amplified by using a pair of primers covering the TATA box of the γ-globin promoter. The x axis indicates the cycle number and the y axis indicates the intensity of fluorescence (F1) in logarithmic scale (arbitrary units). The plot of DNA concentration vs. the cycle threshold number is shown in B, indicating the linear correlation between DNA template (0.5 to 50 ng) and products. (C) Sizing of the products with agarose electrophoresis. Lane 1, size ladder; lane 2, γ TATA box region (100 bp); lane 3, murine ɛy promoter region (200 bp); lane 4, murine βmaj promoter region (170 bp). (D) Melting curves of the PCR products of γ TATA box region (lane 1), ɛy promoter (lane 2), and βmaj promoter (lane 3). x axis, temperature (°C); y axis, dF1/dT. The specificity of TBP antibody was tested by comparing the recovery of TATA box immunoprecipitated by TBP antibody to that by nonspecific antibody (E). Mouse brain served as control tissue nonexpressing the γ-globin gene (E).
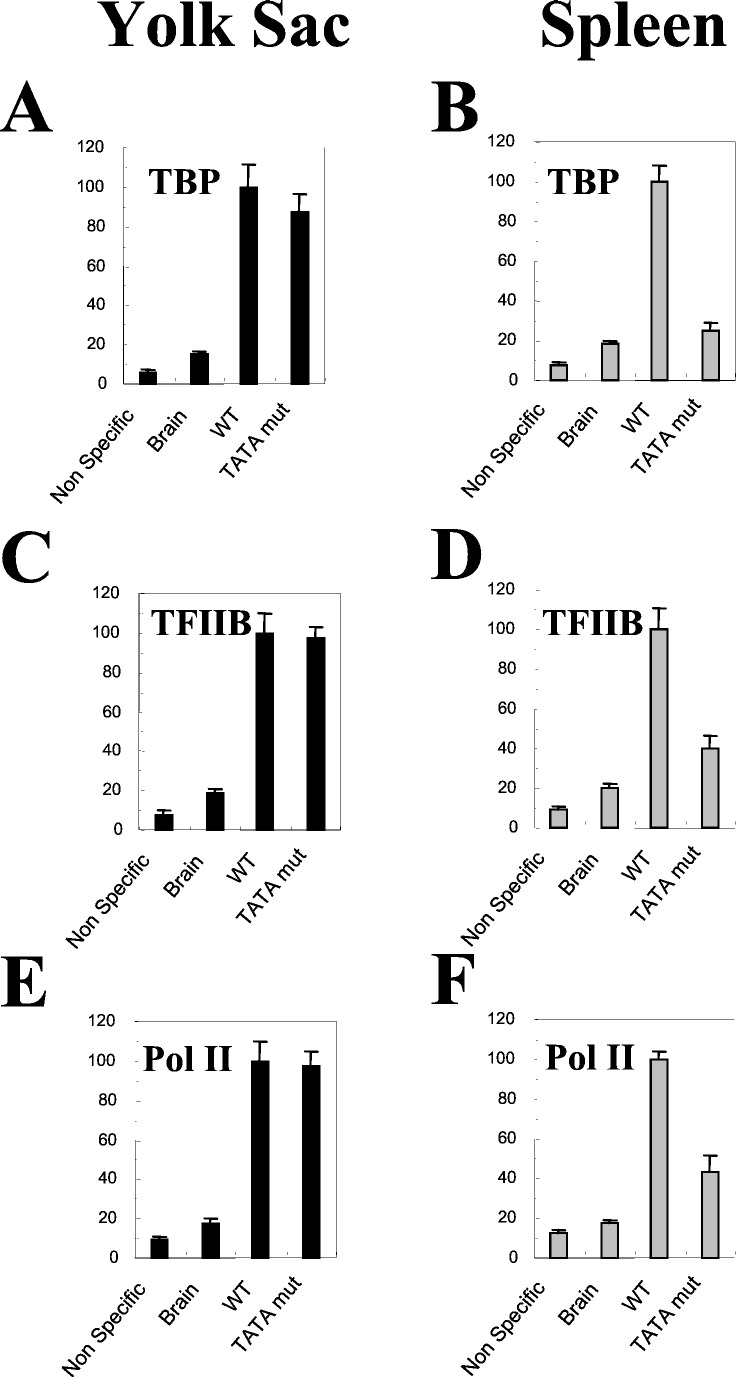
By using this method, we quantitated the effect of the TATA box mutation on recruitment of TBP during the course of mouse development. The yolk sac of transgenic mice carrying either the wild-type γ-globin gene or the TATA box mutated γ-globin gene was subjected to real-time PCR quantitative ChIP assay. In the yolk sac of transgenic mice carrying the TATA box-mutated γ-globin gene, TBP recruitment on the mutated γ TATA box was at 90% of the wild-type control (Fig. 3A). This observation suggested that, in embryonic erythrocytes, the recruitment of TBP is independent of the intactness of the γ TATA box.
Figure 3.
Recruitment of TBP, TFIIB, and Pol II in the transgenic mice carrying the TATA box-mutated γ-globin gene. The day-11 yolk sac and adult spleen were harvested from transgenic mice carrying the wild-type γ construct [μLCR(−382)Aγ] or the TATA box mutated construct [μLCR(−382)Aγ(mut TATA)] and were subjected to real-time PCR quantitative ChIP assay with antibodies against TBP (A), TFIIB (B), and Pol II (C), respectively. Triplex real-time PCR was performed, and the recruitment of each factor to the endogenous ɛy promoter in yolk sac or βmajor promoter in spleen was examined as internal control, respectively. The relative recruitment level of a factor to the TATA box-mutated γ promoter was calculated by the following formula: (γ-ChIP/γ-copy number)/control-ChIP, where “γ-ChIP” and “control-ChIP” mean the recoveries of the specific anti-body IP of the γ TATA box and ɛy (or βmaj) promoter, respectively. The recruitment in the wild-type control (black bar) was converted to 100%, and the recruitment in the TATA box mutated γ promoter (gray bar) was expressed as a percentage of the control.
To examine whether the TATA mutation, which significantly reduced γ gene transcription in definitive erythroid cells, affects TBP recruitment on the γ gene in definitive erythroid cells, we performed the real-time PCR quantitative ChIP assays with disaggregated cells from adult spleens isolated from phenylhydrazine-treated transgenic mice. The majority of splenic cells of phenylhydrazine-treated animals are adult erythroblasts. In this experiment, we used the murine βmaj-globin gene as an internal control. The disruption of the γ TATA box reduced the recruitment of TBP to 25% in the adult spleen, compared with the control (Fig. 3B). These results suggest that, in contrast to embryonic cells, the γ TATA box is required for the TBP recruitment in adult cells.
Disruption of TATA Box Affects TFIIB and Pol II Recruitment in Adult but Not in Embryonic Erythroblasts.
As TATA box-dependency of TBP recruitment is subjected to developmental regulation, we were interested to know whether recruitment of other components of the basal transcription machinery is associated with the TATA box. We measured the recruitment of TFIIB and Pol II in the TATA box-mutant transgenic mice. The disruption of the γ TATA box reduced the recruitment of TFIIB and Pol II to 40% and 44% of the control in adult erythroblasts, respectively (Fig. 3 D and F). These results suggest that the γ TATA box is important for the recruitment of the basal transcriptional machinery to the γ promoter in adult erythroid cells. In contrast, disruption of the γ TATA box had no effects on the recruitment of TFIIB and Pol II in the embryonic erythrocytes (Fig. 3 C and E).
The TATA Box Determines the Transcription Initiation Site in both Primitive and Definitive Erythrocytes.
Because the TATA box is thought to dictate the position of the transcription initiation site in TATA-containing promoters, we examined whether or not the TATA box mutation has differential effects on determination of the initiation site in primitive and in definitive erythroid cells. The results from RNase protection assays in the yolk sac and adult blood of transgenic mice carrying the TATA box-mutant construct showed that in both types of cells only a minority (≈20–24%) of the Aγ gene transcripts had the normal initiation site; 40–48% of the transcripts started at positions −6 to −10, and 28–39% started at position −25 or at a region further upstream (Fig. 4). Similar results were obtained by primer extension assay (data not shown). This observation is consistent with previous findings in cultured cells transfected with the TATA box-mutated γ-globin constructs (26).
Figure 4.
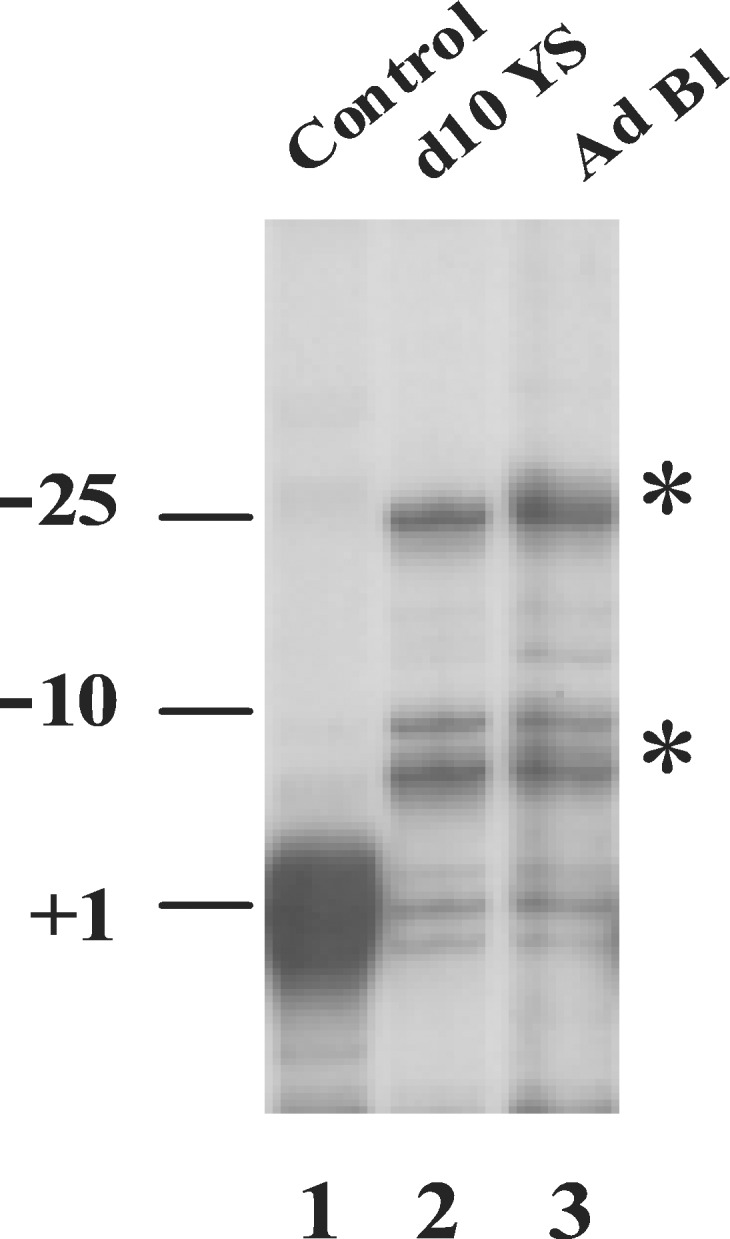
Transcription initiation site of the TATA box-mutated γ-globin gene. Total RNA from yolk sac or adult blood was hybridized with a 32P-labeled RNA probe spanning exon 1 to −382 promoter of the γ-globin gene. After RNase digestion, the protected fragments were separated on a 6% denatured polyacrylamide gel. Lane 1 is the wild-type control, and the protected band represents the +1 position. The initiation site of the mRNA transcribed from the TATA box-mutated γ-globin gene is shown in lane 2 (d10 yolk sac) and lane 3 (adult blood). The abnormal initiation sites are marked by asterisks.
Discussion
The studies presented here demonstrate that transcription efficiency is correlated with the TBP recruitment, but the mechanism whereby TBP is recruited to the γ gene promoter is distinct at the embryonic and adult developmental stages. A large body of data from in vitro and in vivo experiments is consistent with the model that assembly of the initiation complex starts with the binding of TBP to TATA box. This model also applies to the adult stage of development of the transgenic animals, because TBP occupancy in the γ promoter strongly correlates with levels of in vivo transcription. However, the model may be oversimplified, because disruption of the γ TATA box does not have any apparent effects on TBP recruitment and on transcription level of the γ-globin gene in embryonic erythrocytes. There are three possible interpretations for the TATA box-independence of γ gene expression at the embryonic stage: (i) other cis elements in the γ promoter are able to compensate for the absence of the TATA box; or (ii) gene-specific factors and/or cofactors affect recruitment of the basal transcription apparatus in a development-specific manner; or (iii) the basal transcription apparatus is distinct in the cells at the early developmental stage from that in the adults.
It has been reported that 1% of random DNA could functionally substitute for a TATA box in the his3 promoter of yeast (27). It also has been suggested that the GATA-1 site at −38 and Sp1 sites around −50 might serve as the TATA box replacements in the γ promoter (26). To test this possibility, we deleted the sequence from −50 to −35 of the γ promoter in the context μLCR(−382)Aγ (mut TATA), but the transcription level and the transcription initiation sites of this double-mutated γ promoter were the same as those of the single TATA box mutant in stably transfected K562 cells (data not shown). Because the β gene promoter contains an initiator element (Inr) in addition to the TATA box (16), we mutated the sequences around the normal initiation site (from ACACA to TTAGG) in the context of μLCR(−382)Aγ (mut TATA). This combinatorially mutated γ promoter also displayed similar transcription activity as the single TATA box mutant in stably transfected K562 cells (data not shown). Thus, the sequences in the vicinity of the γ TATA box are unable to function as a TATA box replacement. If other cis elements are responsible for TBP recruitment in the TATA box-deficient γ promoter, it is also necessary to postulate that these elements would differentially recruit TBP in the embryonic and adult environments.
TBP could be recruited to a promoter through various modes. The existence of TATA-less promoters suggests that the recruitment of TBP can be TATA box independent in some promoters. Indeed, in the case of human β-globin gene, two other core promoter elements, Inr and DPE, have been demonstrated to be functionally necessary for the recruitment of TFIID (15, 16). In addition, formation of the transcription initiation complex may start with assembly of factors other than TBP. Usheva and Shenk (28) have shown that transcriptional factor YY1 can bind to the core promoter and replace TBP to direct basal transcription in vitro. Holmes and Tjian (1) reported that Drosophila TRF-1 could replace TBP and direct transcription of the tudor gene in a tissue-specific fashion. Two studies in Caenorhabditis elegans showed that a TBP-related factor, CeTLF, was required to express a subset of RNA Pol II genes and was also necessary to establish bulk transcription during early embryogenesis (4, 5). These findings suggest that the choice of usage of TBP could be one of the mechanisms that regulate gene expression. The γ-globin gene carries a typical Pol II promoter encompassing the CACCC, CCAAT, and TATA boxes. The distinct performance of the same TATA box-deficient γ promoter in embryonic and adult erythrocytes suggests that the responsible elements for this difference are of trans rather than of cis nature. This finding raises the possibility that the transfactors and/or cofactors bound the γ-globin gene promoter and the LCR change during the course of development. Such changes may result in an altered mechanism of recruitment of the basal transcription apparatus, which has different dependence than the TATA box.
The third possibility is that differences in the composition of the basal transcription apparatus or modifications of its component account for the distinct behaviors of the TATA box at the embryonic and adult developmental stages. An experiment that can directly support this interpretation is not feasible because of the restricted amount of yolk sac. However, several observations may favor this interpretation. Disruptions of the γCACCC or the γCCAAT box have only minor effects on γ gene expression at the embryonic stage, whereas they are necessary for γ gene expression in the adult stage (unpublished work). These observations suggest that the basal transcription machinery of the embryonic erythroid cells may be distinct from that of cells of later developmental stages.
Acknowledgments
We thank Drs. Patrick Navas, Michael McArthur, Chao-Zhong Song, and Wendy Gombert for their criticism of this study. This work was supported by grants from the National Institutes of Health.
Abbreviations
- Pol II
RNA polymerase II
- LCR
locus control region
- TBP
TATA box-binding protein
- TFIID
transcription factor II D
- TFIIB
transcription factor II B
- MEL
murine erythroleukemia
- ChIP
chromatin immunoprecipitation
References
- 1.Holmes M C, Tjian R. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- 2.Kuras L, Kosa P, Mencia M, Struhl K. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 3.Li X Y, Bhaumik S R, Green M R. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 4.Dantonel J C, Quintin S, Lakatos L, Labouesse M, Tora L. Mol Cell. 2000;6:715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 5.Kaltenbach L, Horner M A, Rothman J H, Mango S E. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 6.Willy P J, Kobayashi R, Kadonaga J T. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 7.Zhang D, Penttila T L, Morris P L, Teichmann M, Roeder R G. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 8.Dantonel J C, Wurtz J M, Poch O, Moras D, Tora L. Trends Biochem Sci. 1999;24:335–339. doi: 10.1016/s0968-0004(99)01436-x. [DOI] [PubMed] [Google Scholar]
- 9.Berk A J. Cell. 2000;103:5–8. doi: 10.1016/s0092-8674(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 10.Pugh B F. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- 11.Smale S T. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 12.Majumder S, DePamphilis M L. Mol Cell Biol. 1994;14:4258–4268. doi: 10.1128/mcb.14.6.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder S, DePamphilis M L. BioEssays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- 14.Stamatoyannopoulos G, Grosveld F. In: Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Majerus P W, Perlmutter R M, Varmus H, editors. Philadelphia: Saunders; 2000. pp. 135–182. [Google Scholar]
- 15.Lewis B A, Kim T K, Orkin S H. Proc Natl Acad Sci USA. 2000;97:7172–7177. doi: 10.1073/pnas.120181197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis B A, Orkin S H. J Biol Chem. 1995;270:28139–28144. doi: 10.1074/jbc.270.47.28139. [DOI] [PubMed] [Google Scholar]
- 17.Fong T C, Emerson B M. Genes Dev. 1992;6:521–532. doi: 10.1101/gad.6.4.521. [DOI] [PubMed] [Google Scholar]
- 18.Antoniou M, de Boer E, Spanopoulou E, Imam A, Grosveld F. Nucleic Acids Res. 1995;23:3473–3480. doi: 10.1093/nar/23.17.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J S, Lee C H, Chung J H. Proc Natl Acad Sci USA. 1999;96:10051–10055. doi: 10.1073/pnas.96.18.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatoyannopoulos G, Josephson B, Zhang J W, Li Q. Mol Cell Biol. 1993;13:7636–7644. doi: 10.1128/mcb.13.12.7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Stamatoyannopoulos J A. Mol Cell Biol. 1994;14:6087–6096. doi: 10.1128/mcb.14.9.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantovani R. Methods Mol Biol. 1994;31:289–298. doi: 10.1385/0-89603-258-2:289. [DOI] [PubMed] [Google Scholar]
- 23.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlando V, Strutt H, Paro R. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 25.Duan Z, Stamatoyannopoulos G, Li Q. Mol Cell Biol. 2001;21:3083–3095. doi: 10.1128/MCB.21.9.3083-3095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sargent T G, DuBois C C, Buller A M, Lloyd J A. J Biol Chem. 1999;274:11229–11236. doi: 10.1074/jbc.274.16.11229. [DOI] [PubMed] [Google Scholar]
- 27.Singer V L, Wobbe C R, Struhl K. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 28.Usheva A, Shenk T. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]