Abstract
Using single-cell sequence analysis, we discovered that a high proportion of cells in tissues as diverse as buccal epithelium and heart muscle contain high proportions of clonal mutant mtDNA expanded from single initial mutant mtDNA molecules. We demonstrate that intracellular clonal expansion of somatic point mutations is a common event in normal human tissues. This finding implies efficient homogenization of mitochondrial genomes within individual cells. Significant qualitative differences observed between the spectra of clonally expanded mutations in proliferating epithelial cells and postmitotic cardiomyocytes suggest, however, that either the processes generating these mutations or mechanisms driving them to homoplasmy are likely to be fundamentally different between the two tissues. Furthermore, the ability of somatic mtDNA mutations to expand (required for their phenotypic expression), as well as their apparently high incidence, reinforces the possibility that these mutations may be involved actively in various physiological processes such as aging and degenerative disease. The abundance of clonally expanded point mutations in individual cells of normal tissues also suggests that the recently discovered accumulation of mtDNA mutations in tumors may be explained by processes that are similar or identical to those operating in the normal tissue.
Keywords: somatic mutation‖clonal expansion‖single cell‖aging‖cancer
Somatic mitochondrial mutations have been studied intensively because of their proposed involvement in the aging process (1–3). One of the early recognized problems of the mitochondrial theory of aging is the fact that because each cell contains a large number of mtDNA molecules, a mutant mtDNA can start influencing the physiology of the cell only once it has accumulated to a significant fraction of all mtDNA in the cell (4). Indeed, the fraction of mtDNA mutations necessary for their “phenotypic expression” is on the order of 90%, as determined in in vitro studies (5, 6). The only logical possibility for a somatic mutation to accumulate in the cell is through “clonal expansion,” i.e., accumulation of the progeny of the single initial mutated DNA molecule.
It has been recognized for a while that large somatic mtDNA deletions are capable of clonally expanding in individual cells (7–11), reaching levels of physiological significance. In contrast to deletions, somatic point mutations have not been shown to be capable of expanding and/or reaching homoplasmy (i.e., approaching 100%) in individual somatic cells in vivo, although some have been shown to do so in cell culture (5, 12, 13) and in the germ line (14, 15). Indirect evidence supporting the possibility that somatic point mutations are likely to expand clonally also comes from studies of the distribution of the two haplotypes among the colon crypts of an engineered heteroplasmic mouse (16). The lack of direct in vivo data is particularly disturbing given the recent evidence of accumulation with increasing age of a large number of mtDNA point mutations at the tissue level (17, 18). The expected incidence of point mutations apparently significantly exceeds that of deletions and approaches one per mtDNA copy, thus making clonal accumulation of point mutations in mtDNA an attractive potential aging mechanism. Interestingly, a high rate of somatic point mutations in a transgenic mouse expressing an error-prone mtDNA polymerase gamma in the heart resulted in a severe, early onset cardiomyopathy (19).
Recently, somatic mtDNA point mutations came to the focus of attention when it was discovered that a high proportion of human tumors contained one or more of such mutations (20–25). In addition to a very important practical implication for diagnostic detection of tumor cells, this discovery has posed a conceptual paradox. The striking fact that the whole tumor carries mtDNA molecules of a single genotype that is different from the genotype of the host organism prompted the hypothesis that certain mtDNA mutations may facilitate tumor development and thus may be selected for in tumors (21, 24). These concepts are undermined by the fact that some of the somatic mutations found in tumors apparently are identical to certain neutral polymorphisms and thus are unlikely to have emerged through selection. An alternative explanation would be that homoplasmic cells are present already in normal tissue. When one such cell becomes transformed, the resulting tumor automatically becomes homoplasmic. Data on the incidence of intracellular homoplasmy in normal tissues is necessary to evaluate these explanations.
To assess whether somatic mtDNA point mutations expand in normal human tissues, we used a cell-by-cell sequence-analysis approach originally developed for studying mtDNA deletions (11). Our studies of human buccal epithelial cells and cardiomyocytes indicate that in an aged human body, there is approximately one expanded mtDNA point mutation per cell on average. These results are discussed with respect to the mitochondrial theory of aging and the possible mechanisms leading to mtDNA homoplasmy in normal tissues and tumors.
Materials and Methods
Tissue Samples.
Cardiomyocytes were obtained from autopsy heart samples snap-frozen in liquid nitrogen within 24 h after death and stored at −80°C. The six samples involved were from unrelated individuals 0.3, 2, 31, 51, 104, and 109 years old. Buccal cells were obtained via standard buccal swabs from seven unrelated individuals 2, 5, 5, 7, 76, 85, and 93 years old. At least 12 individual cells were analyzed per sample/swab.
Isolation of Single Cells and PCR Amplification of mtDNA.
Tissue was dissociated into cell suspension, cells were collected individually, and a small portion of each cell's DNA was subjected to long-distance PCR such that almost the entire mitochondrial genome was amplified as a single PCR fragment ≈16 kb long (base pairs 161–16,510; ref. 11). Long PCR was performed in all cardiomyocytes and in approximately one half of the buccal cells analyzed. DNA from every cell was subjected to at least two independent replicate shorter PCRs (1.2-kb fragment, base pairs 15,972–609, amplified with the corresponding 20-mer primers). This fragment provided a readable sequence approximately between base pairs 16,010 and 590. The 1.2-kb fragment covered the region of base pairs 16,485–195 not included in the 16-kb PCR fragment and was used to detect mutations in cells where long PCR either failed or was not attempted and to quantify mutations. The PCR-amplified DNA as well as the remaining original unicellular DNA were stored permanently for future analyses. Special precautions were taken to avoid contamination of cellular DNA; pre- and post-PCR procedures were carried out in separate rooms located on different floors with one-way flow of materials and reagents between the rooms. PCRs without input DNA were performed as controls on a regular basis and were consistently negative.
We did not include RNase treatment on a routine basis, because no regular mtRNA is expected to be amplified by the 15,972–609 primers. The analysis including RNase treatment was done for selected cells with prominent mutations (lanes 21, 22, 23, 29, 30, and 31 in Table 1). RNase treatment did not affect the presence of mutation in either of the cells tested.
Table 1.
Clonally expanded mutations detected in single cells
| Lane no. | Position, bp | Nucleotide change | Cell type | Age, years | Mutant fraction*, % | SD†, % |
|---|---|---|---|---|---|---|
| 1 | 73 | G→A | Buccal | 76 | 39 | 5 |
| 2 | 189 | A→G | Myocyte | 109 | 71 | 8 |
| 3 | 189 | A→G | Myocyte | 109 | 58 | 5 |
| 4 | 214 | A→G | Buccal | 85 | 60 | 14 |
| 5 | 251 | G→C | Buccal | 93 | 98 | 2 |
| 6 | 252 | T→C | Buccal | 85 | 69 | 23 |
| 7 | 303–310 | ins C | Buccal | 5 | 30 | 11 |
| 8 | 303–310 | ins C | Buccal | 76 | 61 | 3 |
| 9 | 303–310 | ins C | Buccal | 76 | 28 | 3 |
| 10 | 303–310 | ins 1,2C | Buccal | 76 | 83 | 4 |
| 11 | 303–310 | ins 1,2C | Buccal | 76 | 43 | 4 |
| 12 | 303–309 | ins 2,3,4C | Buccal | 85 | 100 | 0 |
| 13 | 303–309 | ins C | Buccal | 85 | 43 | 15 |
| 14 | 303–310 | del C | Buccal | 93 | 95 | 7 |
| 15 | 303–310 | ins C | Buccal | 93 | 29 | 1 |
| 16 | 303–310 | del C | Buccal | 93 | 88 | 1 |
| 17 | 303–310 | ins 1,2C | Buccal | 93 | 53 | 9 |
| 18 | 303–310 | ins 1,2,3C | Buccal | 93 | 94 | 1 |
| 19 | 303–310 | del C | Myocyte | 104 | 28 | 3 |
| 20 | 16,028 | T→C | Myocyte | 51 | 65 | 7 |
| 21 | 16,029 | T→C | Myocyte | 109 | 53 | 4 |
| 22 | 16,029 | T→C | Myocyte | 109 | 57 | 4 |
| 23 | 16,033 | G→A | Myocyte | 109 | 50 | 14 |
| 24 | 16,034 | G→A | Myocyte | 109 | 25 | 1 |
| 25 | 16,035 | G→A | Myocyte | 104 | 58 | 4 |
| 26 | 16,035 | G→A | Myocyte | 109 | 100 | 0 |
| 27 | 16,036 | G→A | Myocyte | 104 | 73 | 10 |
| 28 | 16,036 | G→A | Myocyte | 109 | 91 | 8 |
| 29 | 16,049 | G→A | Myocyte | 109 | 80 | 8 |
| 30 | 16,052–3 | del C | Myocyte | 104 | 78 | 11 |
| 31 | 16,054 | A→G | Myocyte | 51 | 65 | 1 |
Note that generally there was one mutation per cell except for the four cells in which there were two mutations (mutations in lanes 1 and 10, 5 and 15, 2 and 21, and 3 and 29).
The averages are of at least two independent measurements from two independent PCR amplifications from the same cell.
Formal standard deviations are given to illustrate the extent of reproducibility of the data.
The cells that yielded 16-kb PCR products were tested for the presence of large mtDNA deletions, as detected by agarose gel electrophoresis (11). Large deletions can be detected by this approach down to 1% and lower. In this study, the cells with detectable clonally expanded deletions were excluded from further analysis to avoid the possibility that expanded point mutations were linked to and coexpanded with such deletions.
Target Sequence.
Our goal was to test the hypothesis that mtDNA point mutations frequently attain homoplasmy in somatic cells of the human body. The search therefore was focused on mtDNA sequences with the highest a priori likelihood to find mutations, i.e., the 1,121-bp-long control region (base pairs 16,024–576). This region was chosen as the target sequence on the basis of the reported high incidence of somatic mutations within this area in normal fibroblasts (17), muscle (18), and various tumors (22–25). In addition, our initial screening of ≈25% of the mitochondrial genome in cardiomyocytes using two-dimensional gene screening (26) identified the 16,025–16,055-bp area as a putative hotspot for expanded mutations in the heart (data not shown). Sequencing of PCR fragments was performed at MWG Biotech, Charlotte, NC. In our experience, direct sequencing provided for a detection limit of ≈20% or higher.
Identification and Quantification of Clonally Expanded mtDNA Mutations.
To identify mutations in individual cells, we first determined the “bulk” genotype of each sample involved in this study. DNA was isolated by SDS/proteinase K digestion from bulk amounts of tissue (i.e., at least 10,000 cells), and the control region was amplified and sequenced. The control region as amplified from DNA of every cell under study (at least 12 cells per sample were analyzed) was sequenced completely and compared with the bulk sequence of the corresponding tissue sample. Any differences between the bulk sequence and the sequence obtained from an individual cell were considered to be somatic mutations.
As expected, multiple polymorphisms were identified in bulk sequences that distinguished each sample from other samples and/or from the Anderson sequence (27; data not shown). In addition to homoplasmic polymorphisms, two cases of apparently inherited heteroplasmy were detected. Specifically, the 2-year-old heart contained 55% of the common T16,356C transition (MITOMAP, www.gen.emory.edu/mitomap.html). The buccal swab from the 7-year-old girl presented with a 35% C-tract polymorphism. DNA from the mother was available in this case, and it showed 45% of the same polymorphism, supporting the idea that the mutation was inherited. In both heteroplasmy cases, every cell analyzed was polymorphic.
Heteroplasmy is not restricted to cells from young individuals; screening of a wider group of individuals for their bulk genotype revealed many cases of inherited heteroplasmy in the old samples, with a frequency similar to that observed in the young. In contrast to the heteroplasmic samples, the bulk “old” buccal samples showed low, difficult-to-quantify levels of insertion and/or deletion mutants at the 303–309/310 C-tract in addition to the predominant genotype. This feature is consistent with the anticipated high mutational rate at the C-tract, not observed in bulk heart DNA (n = 14), and thus was not considered a sign of inherited heteroplasmy.
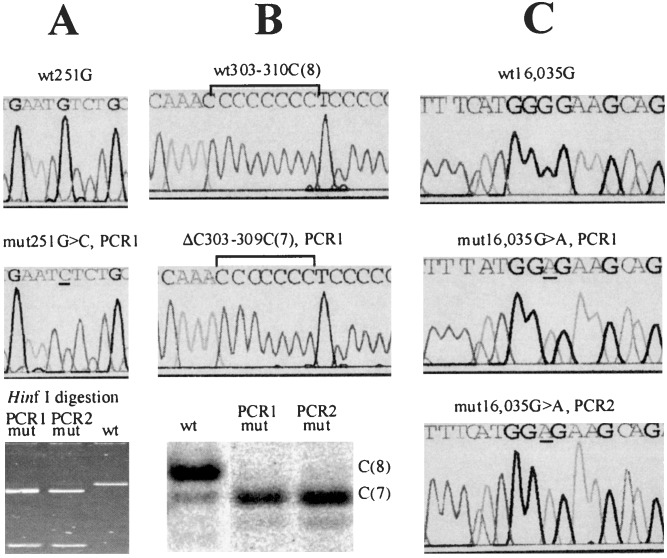
When a mutation was detected in a cell, an independent PCR (base pairs 15,972–609) was performed from the DNA of the same cell once or twice such that the total number of such PCRs from any cell with suspected mutation was at least two. If the mutation was detected in all PCR amplifications, it was considered to be confirmed. This replicate PCR from the same cell rules out any PCR artifacts of a stochastic nature and confirms reproducibility of the quantification procedure as discussed previously (11) and below. It is very important therefore that our approach allows replicate PCR amplifications from the same cell. The fractions of the mutations were quantified as described in the Fig. 1 legend and Table 1.
Figure 1.
Detection, quantification, and confirmation of point mutations in single cells. Somatic nucleotide changes are detected first by direct sequencing of PCR product from single cells. Mutations (Middle) are determined as changes in the sequence compared with the sequences from the majority of cells of the same individual (Top). After an initial identification of a mutation, a duplicate PCR was performed from the DNA of each presumably mutant cell (PCR 2). The mutations then were confirmed and quantified by one of the following methods, depending on the type of the mutation. Note that in each case, the two duplicate experiments yielded very similar results. mut, mutant; wt, wild type. (A) If the mutation resulted in a nucleotide change within a restriction site (in this example, a G → C change creates a HinfI restriction site, G′ANTC), direct restriction analysis was used to confirm and quantify the mutation by gel densitometry (Bottom). (B) A deletion/insertion within a C-tract was confirmed by a variant of the T-PCR approach (15). Briefly, a PCR fragment was restriction-digested to provide a labeled PCR/restriction fragment ≈50-nt long, which included the C-tract. The DNA was run on a sequencing gel. A deletion or insertion in the C-tract manifested itself as a band shift. The weaker subbands do not represent mutations but rather aberrant PCR products that ultimately limit the assay sensitivity. The image was quantified by using a PhosphorImager (Molecular Dynamics). (C) If a mutation did not alter a restriction site or the C-tract length, direct sequencing of the PCR fragments was used.
Results and Discussion
Clonally Expanded mtDNA Mutations Are Abundant in the Cells of Aged but Not Young Human Tissues.
Analysis of 36 cells from buccal swabs from three individuals 76, 85, and 93 years old (12 cells each) revealed a total of 15 expanded mutations in 13 cells. In most cases, there was one (rarely two) mutation(s) per cell, and different cells contained different mutations. Evidently, each of these mutations had expanded clonally from a single initial mutational event. The complete list of mutations and their cellular fractions is presented in Table 1. Note that the insertion of one C in the C-tract sometimes is accompanied by the insertion of two or three nucleotides. It is possible that insertion of the first nucleotide creates instability in the sequence leading to consecutive secondary mutations.
Similarly, analysis of 36 cardiomyocytes from three individuals 51, 104, and 109 years old (12 cells each) yielded 15 clonally expanded mutations in 13 cells. This result is even more impressive than that for buccal cells. A cardiomyocyte contains over 10 times as many mtDNA copies as a buccal cell. Hence, expanding to a significant level in such a cell should have been even more dramatic an achievement for a single initial mutant copy than it would be in an epithelial cell.
Analysis of a similar number of young cells/individuals (cardiomyocytes and buccal cells) revealed only one buccal cell with ≈30% insertion of a C in the C-tract of a 5-year-old boy. The difference in mutant fraction between young and old samples is highly statistically significant (P < 0.005 according to Fisher's exact test, assuming that all old samples and one young sample are “mutant-bearing,” whereas the rest of young samples are “mutant-free”). This result implies that clonally expanded mutations accumulate with age. The data are not sufficient, however, to draw conclusions about the kinetics of this process. Recently, Attardi and coworkers concluded that C-tract alterations do not accumulate with age (17). We see a number of reasons for the difference between the two studies. Attardi and coworkers studied fibroblasts rather than epithelial cells, and their young samples were on average 5 times older than those in this study. Also, in our experience the mutation rate at the C-tract critically depends on the length of the tract; thus old persons with a C7 will present consistently with low mutant fractions, thus potentially masking the age dependence in the fibroblast data, in which C7 and C8 variants were pooled together. Furthermore, an occasional young individual with a C-tract heteroplasmy could also mask the age dependence. Reassuringly, longitudinal studies by Attardi's group revealed considerable and consistent increases of the mutant fractions with age within the same person (17), which is in accord with our conclusion.
The above data imply that clonal expansion of single mtDNA molecules is a widespread process in somatic cells. Most probably expansions affect all or almost all cells in a tissue, and we do not detect expanded mutations in all cells because only a very small fraction of the mitochondrial genome (≈5%) was scanned. Had we analyzed the whole mitochondrial genome, we most probably would have found more (different) expanded mutations. In fact, our preliminary data indicate that this is indeed the case; expanded mutations are observed in the coding regions of mtDNA as well as in the control region. In any case, our results clearly indicate that a very large proportion of somatic cells in human tissues normally contain significantly expanded mtDNA mutations. Furthermore, preliminary single-cell/fiber analysis of heart and muscle of individuals with neutral heteroplasmies demonstrated extreme segregation of the two genotypes between different cells/fibers (Y.K., N.D.B., and K.K., unpublished data). These data lend additional support to our conclusion that intracellular clonal expansion of mitochondrial genomes is a common event in the adult human organism.
Tests for Potential Artifacts.
A potential problem of quantification of PCR-amplified DNA is allelic preference, i.e., preferential amplification of one of the templates, either mutant, or wild type. Although it is unlikely that a single base-pair change would affect the efficiency of amplification of a 16- or 1.2-kb-long PCR fragment, we have checked the most prominent mutations (lanes 4, 5, 6, 8, 21, 22, 23, 29, 30, and 31 in Table 1) for allelic preference. Specifically, the corresponding PCR fragments were mixed to produce an easy-to-quantify 1:1 mutant/wild type mixture, diluted 1 million times, and then reamplified to the initial copy number. Mutant fractions were determined in the resulting PCR products, and no significant deviation from the original mutant fraction was observed in any of the cases that were subjected to the test.
It has been shown that mtDNA sequences with somatic mutations in some cases may be confused with nuclear pseudogenes of mtDNA sequences (28, 29), although this required strong selection against mtDNA (28, 30). To make sure that the mutations we discovered did not originate in pseudogenes, we searched the public version of the human genome (BLAST, www.ncbi.nlm.nih.gov/BLAST/) for homologies to the mtDNA sequence of base pairs 15,900–700. Approximately 100 pseudogene-like sequences homologous to the various parts of the control region are present in the human nuclear DNA. None of pseudogene sequences, however, spanned the whole control region and thus could not be the source of our PCR products, which do span the whole region. Moreover, the nuclear sequence with the highest homology to mtDNA was less than 90% homologous, which is consistent with the established relatively ancient origin of pseudogenes (31–33). In contrast, mutant sequences found in this study contain mostly one (in a few cases two) base change(s) in more than 1 kb, i.e., >99.8% homology.
We cannot completely exclude the possibility that some of the mutations found in individual cells were generated in a germ-line cell rather than in a somatic cell. The fact that almost no mutations were detected by our approach in tissues (both buccal swabs and the heart) of young individuals, however, implies that almost all mutations were generated (or at least expanded) somatically. This conclusion is supported also by the observation that mutations are highly tissue-specific; germ-line mutations, in contrast, would be expected to affect all tissues. Furthermore, even if some of the mutations reported here were in fact low percentage germ-line mutations, they still must have undergone clonal expansion in individual cells to produce the frequency distribution observed, and thus they support the main conclusions of this paper just as well as if they were somatic mutations.
Rapid Dynamics of mtDNA in Somatic Cells: Intracellular “Homogenization” of Mitochondrial Genomes.
mtDNA is present in somatic cells in the numbers of hundreds (e.g., in typical epithelial cells including buccal cells studied here) to hundred-thousands (e.g., cardiomyocytes). The in vivo dynamics of these “populations” of mitochondrial genomes in somatic cells have been studied in only a few special cases. Somatic mtDNA mutations can be used as markers to trace such dynamics. For example, the observation that almost all cells contain clonally expanded somatic mutations implies that most mtDNA copies in a typical somatic cell should have been derived from a single mtDNA copy (which happened to carry that particular mutation). Based on the results of modeling experiments (34), it is expected that in the case of epithelial cells, a common ancestor for all current mtDNA copies in a typical epithelial cell should have existed only ≈70 cell generations earlier. In other words, the typical number of generations necessary for a nascent mutant mtDNA copy that is destined to become expanded to achieve such a state is ≈70 generations of the corresponding stem-cell lineage.
In the case of cardiomyocytes, the time necessary for clonal expansion is less clear. In most cases, expansion is likely to take place after the formation of the heart is essentially complete; otherwise one would expect to see clones of cardiomyocytes bearing the same expanded mutation, which was not what we observed, although more data are needed for a well justified conclusion. Most likely, expansion in cardiomyocytes takes place after they become postmitotic and is driven by mtDNA synthesis coupled to DNA repair and/or turnover (ref. 35; see details below). In conclusion, in a high proportion (if not all) of somatic cells, mitochondrial genomes are subject to homogenization apparently by replacement with the progeny of a single mtDNA genome.
Mutant Spectra of Cardiomyocytes and Buccal Cells Are Significantly Different.
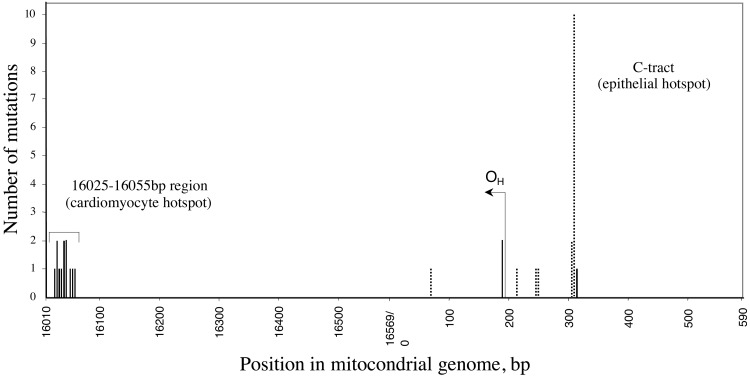
The spectra of expanded mutations are shown in Fig. 2. Note that the set of mutations found in epithelial cells differs dramatically from that present in cardiomyocytes. The most significant difference involves a strong epithelium-specific mutational hotspot located in the homopolymeric C (7–8)-tract located between base pairs 303 and 309 or 310. Although as many as 11 mutations were detected at this hotspot among 36 epithelial cells, only one cell containing 28% mutation was detected among the 36 heart cells (Table 1). Another important feature is that almost all cardiomyocyte mutations fall within a compact area 30 bp long between base pairs 16,025–16,055 of the control region, whereas none of the mutations detected in the epithelial cells appears to reside even close to this area.
Figure 2.
Spectra of clonally expanded point mutations in cardiomyocytes and buccal epithelial cells. The number of cells bearing a particular expanded mutation is plotted against the position of the mutation within the control region of the mitochondrial genome. Solid bars, mutations detected in cardiomyocytes; dotted bars, mutations detected in buccal cells; OH, origin of replication of the heavy strand.
Statistical analysis of the data demonstrates that the two spectra are highly significantly different. To test whether our data are sufficient to support such a hypothesis, the observed mutations were divided into two groups according to location (between either base pairs 0 and 590 or 16,010 and 16,569). The statistical significance of the observed differences between cell types with respect to the frequency of mutations within each segment was assessed first by using the two-sided Fisher's exact test (36). The spectra proved to be highly significantly different in either of the segments (segment 0–590, P < 0.0001; segment 16,000–16,569, P < 0.0002). Next, assuming independence of mutation processes in these regions, a standard metaanalysis (37) was applied to obtain an overall P value (P < 10−6), which confirms with very high confidence that the spectra in different cell types indeed are different.
Our data are in accord with the recent demonstration by Attardi and coworkers that spectra of mtDNA mutations differ significantly between skeletal muscle and fibroblasts (17, 18), although the comparison should be made with caution, because these studies involved homogenates rater than single cells. None of the prominent hotspots reported in fibroblasts and muscle cells have counterparts in either buccal cells or cardiomyocytes, which further supports tissue specificity of the spectra. In the heart, however, we do observe the A189G mutation that Attardi and coworkers report as a major hotspot in the muscle (18). We propose that the differences between the mutational spectra of cardiomyocytes and epithelial cells reflect a difference in the mechanisms of clonal expansion in the two cell types.
Epithelial Mutational Hotspot: Clonal Expansion via Random Segregation?
Length alterations in the C-tract apparently are silent mutations, because they are identical to the corresponding common polymorphisms and thus are likely to be “passive riders” in the process of clonal expansion. They are likely to originate from polymerase errors, because homopolymer tracts are known to be error hotspots for DNA polymerases in vitro (38). The mutational rate at the C-tract should correlate with the intensity of mtDNA replication, and the fact that these mutations are virtually absent in cardiomyocytes thus implies that the mtDNA in myocytes may replicate at a lower rate than in epithelial cells, consistent with their postmitotic status. An alternative explanation for the absence of the C-tract hotspot in myocytes, i.e., that C-tract mutants are abundant in myocytes but do not expand and thus evade our analysis, is invalid. Indeed, we know that expanded mutations do exist in cardiomyocytes at a frequency similar to that in buccal cells, thus C-tract mutations, if they were available at a comparable fraction, should have been expanded passively just as they were in buccal cells. Interestingly, the remaining four non-C-tract mutations detected in buccal cells are also identical to common neutral polymorphisms (MITOMAP and BLAST).
It has been suggested by a number of investigators that in proliferating cells, the mechanism of clonal expansion of mtDNA mutations may be random genetic drift via unbiased mtDNA replication and sorting during cell division (34, 39–41). A random segregation mechanism is consistent with our current finding that the major mutational hotspot in these cells consists of neutral length alterations in the C-tract. It should be noted, however, that a clearly nonrandom selection was observed for/against ostensibly neutral haplotypes in the mouse (16). A similar mechanism could have worked in the case of buccal cells instead of or in addition to random segregation.
Cardiomyocyte Hotspot: Clonal Expansion by Selection?
We see two potential explanations for the observed differences in the 16,025–16,055 hotspot, which was seen almost exclusively in cardiomyocytes but was absent completely in buccal cells. First, mtDNA bearing these mutations may possess some selective advantage, with selection mechanisms being active in myocytes only. Interestingly, an elegant mechanism has been proposed, which provides for preferential survival of subfunctional mitochondria in postmitotic but not in proliferating cells (42). This mechanism postulates that dysfunctional mitochondria accumulate less oxidative damage than the working ones and thus avoid degradation in the lysosomes, which otherwise is a normal course of mitochondrial turnover in nondividing cells. The involvement of such a mechanism would imply that the 16,025–16,055 mutations are perturbing, at least mildly, normal mitochondrial function. Reassuringly, there are reasons to believe that these mutations indeed may be nonneutral. First, it appears that the particular nucleotide positions involved in the 16,025–16,055 expanded mutations are highly conserved; according to GenBank, no polymorphisms have been detected in these positions in the human population (i.e., among >5,000 sequences of this region that are available, BLAST search). None are listed in MITOMAP either. Moreover, all the mutated nucleotides are conserved among the great apes. The absence of polymorphisms or interspecies differences is especially impressive given the high variability of these sequences in cardiomyocytes, which implies that these positions are potentially highly mutable. The fact that these mutations are never inherited strongly supports the involvement of these sequences in some vital mitochondrial function. Second, we note that all the mutations in question are distributed within a very compact area, reminiscent of a protein binding site or a secondary structure site. It is tempting to speculate that mutations of conserved sequences around base pairs 16,025–16,055 might disrupt a binding site of a (yet unknown) protein or secondary structure involved in the regulation of synthesis and/or maintenance of DNA and/or RNA in the mitochondrion, which ultimately could result in altered mitochondrial function and positive selection via the mechanism mentioned above (42). The known functional site closest to the hotspot is the transcription-associated sequence, TAS1, mapped to base pairs 16,081–16,141 (43).
The other possible explanation for the observed differences between the spectra would be that the 16,025–16,055 hotspot mutations are generated by a myocyte-specific mutagen. The two mechanisms are not necessarily mutually exclusive, although the second mechanism seems less plausible, because mutations in the 16,025–16,055 hotspot are quite diverse chemically, whereas the hotspot is extremely compact. This pattern is more suggestive of a binding site inactivation mechanism. In our previous studies of a different target sequence (base pairs 10,030–10,130; ref. 44), we detected no significant differences between the mtDNA mutational spectra in different tissues, which also weakens the possibility that mutagens differ significantly between various tissues. It has been proposed recently (47) that clonal expansion of mtDNA mutations in nonproliferating cells may be explained by random drift. Although this possibility cannot be excluded, overall, our spectral data seem to support the selective mechanism of clonal expansion in such cells.
Spectra of Large mtDNA Deletions.
Tissue specificity of mutational spectra is even more pronounced if large mtDNA deletions are considered in addition to point mutations. In this work, no deleted mtDNA were detected in buccal cells. As we have reported elsewhere (11, 46), in cardiomyocytes a large number of clonally expanded deletions were detected (up to 25% of cells in a heart contain expanded deletions). There is a hotspot for deletion breakpoints within the control region very close to the point mutation hotspot (around base pair 16,070). This hotspot apparently is identical to that described originally in autosomal dominant mitochondrial myopathy (47).
One of us (J.V.) has reported recently mutational spectra of the small intestine and the heart as determined by using a mouse model with chromosomally integrated lacZ reporter gene (48, 49). Interestingly, the nuclear DNA spectrum of the small intestine (proliferative tissue) was similar to the mtDNA spectrum of buccal cells in that it was comprised predominantly of point mutations. The nuclear spectrum of the heart was similar to the heart mtDNA spectrum in that it was comprised of both point mutations and large genome rearrangements.
Intracellular Homogeneity of mtDNA Supports Mitochondrial Theory of Aging.
The mitochondrial theory of aging (2) in its mutational form (3) proposes that the aging process is caused by the life-long accumulation of somatic mtDNA mutations. An obstacle of this theory has been the large copy number of mtDNA per cell, which would be expected to prevent low copy somatic mtDNA mutants from influencing the cellular phenotype if these mutants were distributed randomly among cells. It has been suggested therefore that mtDNA somatic mutations should expand in aging human tissues by either random segregation (3) or selection (4). Although for mtDNA deletions such a postulate was confirmed promptly (7, 8), the present communication reports clonal expansion of somatic point mutations in nontumor somatic tissues. Our finding implies that point mutations, in addition to deletions, are likely to be involved in the aging process. This is an important addition in support of the theory because point mutations appear to be much more abundant than deletions, and thus their impact (even though most of them are likely neutral) may be even greater, especially considering the possibility of positive selection for detrimental mutations (42).
Implications for the Observed High Frequency of Homoplasmic mtDNA Mutations in Tumors.
The demonstration that clonally expanded mtDNA mutations are abundant in normal epithelial cells provides for a simple interpretation of the recent discovery (20–25) that a majority of human tumors of epithelial origin contained one or more mtDNA somatic mutations at a level close to 100%. Indeed, a typical tumor originates from a single cell of a precancerous cell lineage, which in turn originates from a normal cell of the parental tissue. Thus an abundance of cells with clonally expanded somatic mutations in normal tissues leads directly to the abundance of expanded mutations within tumor precursor cells and, finally, tumors. Moreover, additional cell divisions associated with the progression of a normal epithelial cell into a tumor precursor would provide an additional opportunity for mutation as well as for expansion, such that one would expect the frequency of homoplasmic mutations in tumors to be somewhat higher than the frequency of clonal expansions in normal epithelial cells, which indeed seems to be the case (34).
Acknowledgments
We acknowledge Pamela Sheridan for buccal swabs, Hilary Coller (Fred Hutchinson Cancer Research Center, Seattle) and Bill Thilly (Massachusetts Institute of Technology, Cambridge, MA) for indispensable discussions. This work was supported in part by National Institutes of Health Grants AG19787, ES11343, AG13314, AG08812, AG18536, AG00251, AG17242, AG13319, and AG18388.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harman D. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Linnane A W, Marzuki S, Ozawa T, Tanaka M. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 4.Kadenbach B, Muller-Hocker J. Naturwissenschaften. 1990;77:221–225. doi: 10.1007/BF01138485. [DOI] [PubMed] [Google Scholar]
- 5.Attardi G, Yoneda M, Chomyn A. Biochim Biophys Acta. 1995;1271:241–248. doi: 10.1016/0925-4439(95)00034-2. [DOI] [PubMed] [Google Scholar]
- 6.Moraes C T, Schon E A. Methods Enzymol. 1996;264:522–540. doi: 10.1016/s0076-6879(96)64046-4. [DOI] [PubMed] [Google Scholar]
- 7.Mita S, Schmidt B, Schon E A, DiMauro S, Bonilla E. Proc Natl Acad Sci USA. 1989;86:9509–9513. doi: 10.1073/pnas.86.23.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller-Hocker J, Seibel P, Schneiderbanger K, Kadenbach B. Virchows Arch A Pathol Anat Histopathol. 1993;422:7–15. doi: 10.1007/BF01605127. [DOI] [PubMed] [Google Scholar]
- 9.Schwarze S R, Lee C M, Chung S S, Roecker E B, Weindruch R, Aiken J M. Mech Ageing Dev. 1995;83:91–101. doi: 10.1016/0047-6374(95)01611-3. [DOI] [PubMed] [Google Scholar]
- 10.Brierley E J, Johnson M A, Lightowlers R N, James O F, Turnbull D M. Ann Neurol. 1998;43:217–223. doi: 10.1002/ana.410430212. [DOI] [PubMed] [Google Scholar]
- 11.Khrapko K, Bodyak N, Thilly W G, van Orsouw N J, Zhang X, Coller H A, Perls T T, Upton M, Vijg J, Wei J Y. Nucleic Acids Res. 1999;27:2434–2441. doi: 10.1093/nar/27.11.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar D R, Moonie P A, Jacobs H T, Holt I J. Proc Natl Acad Sci USA. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenuth J P, Peterson A C, Fu K, Shoubridge E A. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 15.Marchington D R, Hartshorne G M, Barlow D, Poulton J. Am J Hum Genet. 1997;60:408–416. [PMC free article] [PubMed] [Google Scholar]
- 16.Jenuth J P, Peterson A C, Shoubridge E A. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 17.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski K E, Miller C A, Askanas V, Engel W K, Bhasin S, Attardi G. Proc Natl Acad Sci USA. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D, Mott J L, Chang S W, Denniger G, Feng Z, Zassenhaus H P. Genomics. 2000;69:151–161. doi: 10.1006/geno.2000.6333. [DOI] [PubMed] [Google Scholar]
- 20.Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, Guzman A, Oliva H, Sancho M. Electrophoresis. 1997;18:682–685. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 21.Polyak K, Li Y, Zhu H, Lengauer C, Willson J K, Markowitz S D, Trush M A, Kinzler K W, Vogelstein B. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 22.Habano W, Nakamura S, Sugai T. Oncogene. 1998;17:1931–1937. doi: 10.1038/sj.onc.1202112. [DOI] [PubMed] [Google Scholar]
- 23.Richard S M, Bailliet G, Paez G L, Bianchi M S, Peltomaki P, Bianchi N O. Cancer Res. 2000;60:4231–4237. [PubMed] [Google Scholar]
- 24.Fliss M S, Usadel H, Caballero O L, Wu L, Buta M R, Eleff S M, Jen J, Sidransky D. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 25.Habano W, Sugai T, Nakamura S I, Uesugi N, Yoshida T, Sasou S. Gastroenterology. 2000;118:835–841. doi: 10.1016/s0016-5085(00)70169-7. [DOI] [PubMed] [Google Scholar]
- 26.van Orsouw N J, Zhang X, Wei J Y, Johns D R, Vijg J. Genomics. 1998;52:27–36. doi: 10.1006/geno.1998.5410. [DOI] [PubMed] [Google Scholar]
- 27.Anderson S, Bankier A T, Barrell B G, Young I G. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 28.Hirano M, Shtilbans A, Mayeux R, Davidson M M, DiMauro S, Knowles J A, Schon E A. Proc Natl Acad Sci USA. 1997;94:14894–14899. doi: 10.1073/pnas.94.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace D C, Stugard C, Murdock D, Schurr T, Brown M D. Proc Natl Acad Sci USA. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li-Sucholeiki X C, Khrapko K, Andre P C, Marcelino L A, Karger B L, Thilly W G. Electrophoresis. 1999;20:1224–1232. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1224::AID-ELPS1224>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Hu G, Thilly W G. Gene. 1994;147:197–204. doi: 10.1016/0378-1119(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 32.Hu G, Thilly W G. Curr Genet. 1995;28:410–414. doi: 10.1007/BF00310808. [DOI] [PubMed] [Google Scholar]
- 33.Zischler H, Geisert H, von Haeseler A, Paabo S. Nature (London) 1995;378:489–492. doi: 10.1038/378489a0. [DOI] [PubMed] [Google Scholar]
- 34.Coller H, Khrapko K, Bodyak N, Nekhaeva E, Herrero-Jimenez P, Thilly W. Nat Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 35.Gross N J, Getz G S, Rabinowitz M. J Biol Chem. 1969;244:1552–1562. [PubMed] [Google Scholar]
- 36.Agresti A. Stat Sci. 1992;7:131–177. [Google Scholar]
- 37.Fisher R A. Statistical Methods for Research Workers. Edinburgh, U.K.: Oliver and Boyd; 1970. [Google Scholar]
- 38.Kunkel T A. J Biol Chem. 1986;261:13581–13587. [PubMed] [Google Scholar]
- 39.Hayashi J, Tagashira Y, Yoshida M C, Ajiro K, Sekiguchi T. Exp Cell Res. 1983;147:51–61. doi: 10.1016/0014-4827(83)90270-7. [DOI] [PubMed] [Google Scholar]
- 40.Preiss T, Lowerson S A, Weber K, Lightowlers R N. Trends Genet. 1995;11:211–212. doi: 10.1016/s0168-9525(00)89048-4. [DOI] [PubMed] [Google Scholar]
- 41.Jones J B, Song J J, Hempen P M, Parmigiani G, Hruban R H, Kern S E. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 42.de Grey A. BioEssays. 1997;19:161–166. doi: 10.1002/bies.950190211. [DOI] [PubMed] [Google Scholar]
- 43.Sbisa E, Tanzariello F, Reyes A, Pesole G, Saccone C. Gene. 1997;205:125–140. doi: 10.1016/s0378-1119(97)00404-6. [DOI] [PubMed] [Google Scholar]
- 44.Khrapko K, Coller H A, Andre P C, Li X-C, Hanekamp J S, Thilly W G. Proc Natl Acad Sci USA. 1997;94:13798–13803. doi: 10.1073/pnas.94.25.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elson J, Samuels D, Turnbull D, Chinnery P. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bodyak N D, Nekhaeva E, Wei J Y, Khrapko K. Hum Mol Genet. 2001;10:17–24. doi: 10.1093/hmg/10.1.17. [DOI] [PubMed] [Google Scholar]
- 47.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. Nature (London) 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 48.Dolle M E, Snyder W K, Gossen J A, Lohman P H, Vijg J. Proc Natl Acad Sci USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolle M E, Snyder W K, Dunson D B, Vijg J. Nucleic Acids Res. 2002;30:545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]