Abstract
Ech hydrogenase (Ech) from the methanogenic archaeon Methanosarcina barkeri catalyzes the reversible reduction of ferredoxin by H2 and is a member of a distinct group of membrane-bound [NiFe] hydrogenases with sequence similarity to energy-conserving NADH:quinone oxidoreductase (complex I). To elucidate the physiological role(s) of Ech a mutant lacking this enzyme was constructed. The mutant was unable to grow on methanol/H2/CO2, H2/CO2, or acetate as carbon and energy sources but showed wild-type growth rates with methanol as sole substrate. Addition of pyruvate to the growth medium restored growth on methanol/H2/CO2 but not on H2/CO2 or acetate. Results obtained from growth experiments, cell suspension experiments, and enzyme activity measurements in cell extracts provide compelling evidence for essential functions of Ech and a 2[4Fe-4S] ferredoxin in the metabolism of M. barkeri. The following conclusions were made. (i) In acetoclastic methanogenesis, Ech catalyzes H2 formation from reduced ferredoxin, generated by the oxidation of the carbonyl group of acetate to CO2. (ii) Under autotrophic growth conditions, the enzyme catalyzes the energetically unfavorable reduction of ferredoxin by H2, most probably driven by reversed electron transport, and the reduced ferredoxin thus generated functions as low potential electron donor for the synthesis of pyruvate in an anabolic pathway. (iii) Reduced ferredoxin in addition provides the reducing equivalents for the first step of methanogenesis from H2/CO2, the reduction of CO2 to formylmethanofuran. Thus, in vivo genetic analysis has led to the identification of the electron donor of this key initial step of methanogenesis.
The coupling of thermodynamically unfavorable redox reactions to the consumption of a membrane ion gradient, often referred to as reverse electron transport, is poorly understood. This is particularly so in strict anaerobes, such as the methanogenic archaea, where it is thought to play a central role in metabolism. However, as the name implies, reverse electron transport is, at least in some cases, closely related to the well studied process of energy-conserving electron transport. Recently, a novel family of multisubunit membrane-bound [NiFe] hydrogenases, which are closely related to proton-pumping NADH:quinone oxidoreductases (complex I in electron transport chain of mitochondria), has been recognized (1, 2). This family includes hydrogenases 3 and 4 from Escherichia coli (3, 4), CO-induced hydrogenase from Rhodospirillum rubrum (5, 6), Eha and Ehb from Methanothermobacter species (1), and Ech hydrogenase (Ech) from Methanosarcina barkeri Fusaro (7). Various members of this family have been proposed to function in energy-conserving electron transport, reverse electron transport, or both.
From a biochemical perspective, the most thoroughly studied member of the family is the recently identified Ech found in the methanogenic archaeon M. barkeri (8). This enzyme catalyzes the reversible reduction of ferredoxin by H2 and is the only family member that has been purified to homogeneity. The enzyme is an integral membrane complex, which, when purified, is composed of four hydrophilic and two hydrophobic subunits, corresponding to the products of the identified echABCDEF operon (7). Biochemical studies indicate that a 2[4Fe-4S] ferredoxin is the physiological substrate for Ech, but the physiological function of Ech has yet to be established. RNA hybridization and immunoblotting experiments indicate that the ech operon is expressed at similar levels under all conditions examined, suggesting only that it is required under many growth conditions (7, 8).
Three possible functions have been proposed for Ech. First, because the enzyme is an integral membrane protein related to complex I, it has been suggested that Ech catalyzes an energy-conserving reaction. The oxidation of the carbonyl group of acetyl-CoA to CO2 and H2, a partial reaction in the production of methane from acetate (the acetoclastic pathway), has been proposed as the Ech-dependent coupling site (8). This reaction is coupled to the generation of a proton motive force in M. barkeri (9). Moreover, in vitro oxidation of CO to CO2 and H2 can be reconstituted by using purified Ech, ferredoxin, and acetyl-CoA decarbonylase/synthetase (ACDS) (8). A second possible role for Ech involves the first step of methanogenesis from H2/CO2, the reduction of CO2 to formyl-methanofuran (CHO-MFR). Because of the low midpoint potential of the CO2 + methanofuran/CHO-methanofuran couple (E0′ = ≈−500 mV), this reaction becomes endergonic with H2 as electron donor (E0′ = −414 mV) (10). This reaction is even more endergonic at the low hydrogen partial pressures prevailing in the natural habitats of methanogens. In freshwater sediments, for example, the H2 partial pressure is on the order of 5 Pa corresponding to an E′ for the 2H+/H2 couple of −286 mV. Therefore, this unfavorable reaction requires an additional input of energy to proceed. Cell suspension experiments have provided evidence that reduction of CO2 to CHO-MFR is driven by an electrochemical proton or sodium ion gradient (11–13). Ech and the related hydrogenases Eha and Ehb from Methanothermobacter species have been proposed to function as ion pumps in this reaction providing the low potential reducing equivalents necessary for the reduction of CO2 to CHO-MFR (1, 8). A third possible function of Ech involves the biosynthesis of acetate and pyruvate. In M. barkeri, carbon is assimilated into cell mass via the stepwise synthesis of acetyl-CoA and pyruvate, both of which are required for further biosynthetic reactions (14). The synthesis of acetyl-CoA by the ACDS complex and the reductive carboxylation of acetyl-CoA to pyruvate by pyruvate:ferredoxin oxidoreductase are endergonic with hydrogen as electron donor. Reduced ferredoxin (E0′ = −420 mV) was identified as the direct electron donor of these reduction steps (15, 16). Because Ech in vitro catalyzes ferredoxin reduction by H2 at high rates, it has been suggested that Ech provides the cell with reduced ferredoxin in vivo (8).
In vivo testing of these putative functional roles for Ech has not previously been possible because of a lack of methods for genetic analysis of Methanosarcina species; however, recent advances in the field now make such a study feasible (17–20). In this report, we describe construction and characterization of an M. barkeri mutant lacking Ech. These studies confirm each of the roles proposed above and strongly support the functioning of Ech in both energy-conserving electron transport and the coupling of thermodynamically unfavorable reactions to reverse electron transport. The data also indicate that ferredoxin is the long-sought in vivo electron donor for the first step in methanogenesis from H2 and CO2 and highlight the central role for Ech and ferredoxin in the metabolism of this methanogenic archaeon.
Materials and Methods
Strains, Media, and Growth Conditions.
M. barkeri Fusaro (DSM 804) was grown in single cell morphology (21) at 35°C in high salt (HS) broth medium (22). Growth of M. barkeri on agar-solidified media was as described (19). Carbon and energy sources were added to HS media as follows: methanol, 125 mM; sodium acetate, 80 mM; H2/CO2, 80:20 mixture added to gas phase at 1–200 kPa over ambient pressure. When required, sodium pyruvate was added at 100 mM from sterile, anaerobic stock solutions. Puromycin was added to media at 2 μg/ml for selection of Methanosarcina strains carrying the pac gene. To score for hpt mutations, 8-aza-2,6-diaminopurine (8-ADP) was added to media at 20 μg/ml; Hpt+ strains are incapable of growth on media containing 8-ADP, whereas Hpt− strains are able to grow.
DNA Methods and Plasmid Constructions.
Standard methods were used throughout for isolation and manipulation of plasmid DNA (23). Genomic DNA isolation from M. barkeri and DNA hybridizations were performed as described (18, 19, 22). All plasmid constructions are described in the supporting information, which is published on the PNAS web site, www.pnas.org. Complete DNA sequences of all plasmids used in the study are available on request.
Construction and Complementation of M. barkeri Ech1.
The M. barkeri Δech1∷pac-ori-aph mutant (hereafter designated Ech1) was constructed by transformation of M. barkeri Fusaro with NotI-cut pCK22. Transformants were selected on HS agar with methanol, acetate, and puromycin. M. barkeri Ech1 was complemented by transformation with AscI-cut pJK78 with selection on HS-agar with puromycin under H2/CO2 gas phase. Liposome-mediated transformation of M. barkeri and homologous recombination-mediated gene replacement experiments were as described (18, 19).
Cell Suspension Experiments.
Cells were collected from late exponential growth phase cultures (OD578 = 1) by centrifugation at 5,000 × g for 20 min, washed twice in either HS medium (no substrate) or 50 mM Mops (pH 7.0), 2 mM DTT, 400 mM NaCl, 54 mM MgCl2, and then resuspended in the same buffer. Assays were performed at 37°C in sealed 10-ml anaerobic vials with gas phase pressure at 200 kPa. Assay mixtures were held on ice and started by transfer to 37°C. Gas phase samples were withdrawn at various time points and assayed for CH4, CO, CO2, or H2 by gas chromatography. Total cell protein was determined by using the method of Bradford (24) after an aliquot of the cells was lysed by sonication.
Preparation of Cell Extracts and Membrane Fractions.
Late exponential-phase cultures were harvested by centrifugation at 5,000 × g for 20 min and lysed by resuspension in 50 mM Mops, pH 7.0/2 mM DTT, to which a few crystals of DNase I were added. Complete lysis was ensured by sonication, using a Bandelin (Berlin) Sonopuls HD 200 sonifier at a power of 200 W for 7 × 5 min. Intact cells and debris were removed by centrifugation at 10,000 × g for 30 min. Membrane fractions were prepared by centrifugation of cell extracts at 150,000 × g for 2 h and resuspension of the membrane pellets in 50 mM Mops, pH 7.0/2 mM DTT.
Formation of CO2 and H2 from CHO-MFR by Membrane Fractions.
Assays were conducted at 37°C in sealed, anaerobic vials containing 550 μg of membrane protein, 30 nmol of ferredoxin, and 400 nmol of CHO-MFR in 1 ml of 50 mM Mops, pH 7.0/2 mM DTT under a gas phase of 100% N2 at 200 kPa. H2, CO2, and CH4 were measured by gas chromatography. M. barkeri ferredoxin was prepared as described (8).
Determination of Enzyme Activities.
All enzyme activities were measured in anaerobic gas-tight cuvettes at 37°C. In hydrogenase assays, the gas phase was 100% H2 at 130 kPa. All other assays were performed under 100% N2 at 130 kPa. Ferredoxin-dependent hydrogenase (Ech) was assayed as described (8). F420 reducing hydrogenase (Frh) was assayed by following the reduction of F420 with H2 (25). CHO-MFR dehydrogenase (Fmd) was assayed by following the reduction of benzyl viologen with CHO-MFR (26). In the enzyme assays described below, tetrahydromethanopterin (H4MPT) isolated from Methanothermobacter marburgensis and its derivatives were used instead of the structurally similar compound tetrahydrosarcinapterin (H4SPT) found in M. barkeri. The two substrates are interchangeable in enzyme assays. Methylene-H4SPT dehydrogenase (Mtd) was assayed by following the formation of methenyl-H4MPT from methylene-H4MPT with F420 as electron acceptor (27). Methenyl-H4SPT cyclohydrolase (Mch) was assayed according to ref. 28 by following the conversion of methenyl-H4MPT to formyl-H4MPT. CHO-methanofuran:H4SPT formyltransferase (Ftr) was assayed by following the formation of formyl-H4MPT from H4MPT and CHO-MFR as described (29). Additional details of enzyme assays are provided in the supporting information.
Results
Construction and Verification of an M. barkeri Δech Mutant.
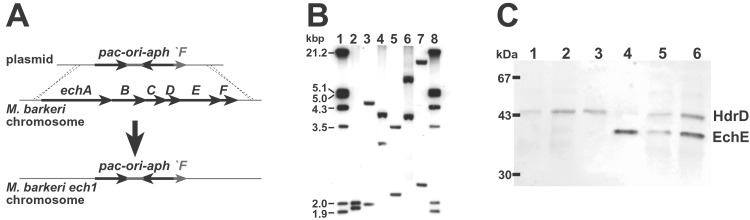
An M. barkeri mutant with a deletion of the echABCDEF operon, encoding Ech, was constructed by homologous recombination-mediated gene replacement as shown in Fig. 1A. To do this, the regions up- and downstream of the ech operon (about 500 bp each) were cloned into a plasmid flanking the pac-ori-aph cassette, which encodes resistance to puromycin in M. barkeri (18). The plasmid was then linearized and used to transform M. barkeri with selection for puromycin resistance. Several transformants were obtained and screened by DNA hybridization for ones that carried the Δech1∷pac-ori-aph mutation (Fig. 1B). Finally, Western blotting was used to show that the EchE subunit of the hydrogenase was undetectable in cell extracts (Fig. 1C). One mutant, designated M. barkeri Ech1, met these criteria and was used for all subsequent experiments.
Figure 1.
Construction and verification of an M. barkeri ΔechABCDEF mutant. (A) Ech1 was constructed by transformation of M. barkeri to PurR with linearized plasmid carrying the Δech1∷pac-ori-aph mutation. Recombination (dotted lines) between homologous sequences on the plasmid and chromosome resulted in replacement of the ech operon with the mutation. A small fragment of echF remains in the mutant. (B) Mutant (Ech1) and wild-type (WT) chromosome structure was verified by DNA hybridization as described. The predicted sizes of hybridizing bands (base pairs) are shown in parentheses: lanes 1 and 8, DNA markers; lane 2, Ech1-HindIII (1938, 2018); lane 3, WT-HindIII (2011, 4540); lane 4, Ech1-EcoRV (3076, 3984); lane 5, WT- EcoRV (2132, 3551); lane 6, Ech1-EcoRI (3912, >4000); lane 7, WT-EcoRI (2283, >6526). (C) EchE and HdrD were detected by Western blot as described (8). Lane 1, 5 μg of Ech1 cell extract; lane 2, 5 μg of Ech1 membrane protein; lane 3, 0.1 μg of purified Hdr (control); lane 4, 0.1 μg of purified Ech (control); lane 5, 5 μg of WT cell extract; lane 6, 5 μg of WT membrane protein. The molecular mass of subunit EchE is about 39 kDa, the molecular mass of HdrD is 43 kDa. The migration of molecular mass standards is shown on the left.
Phenotypic Characterization of M. barkeri Ech1.
M. barkeri is capable of growth on a variety of substrates including methanol, acetate, and H2/CO2. It also grows on a combination of methanol and H2/CO2 via a pathway that is distinct from the use of either substrate alone. We examined the capacity of M. barkeri Ech1 to use each of these growth substrates (Table 1). Although the mutant was isolated on media containing both methanol and acetate, it is capable of growth on methanol alone. As predicted, the mutant is unable to grow on either acetate or H2/CO2. Surprisingly, it also fails to grow on methanol/H2/CO2. We considered two possibilities for these phenotypes. First, that the Δech1 mutation blocks methanogenesis and second, that it blocks a required biosynthetic pathway.
Table 1.
Growth of M. barkeri strains in various media
| Me
|
H2/CO2
|
Me/H2/CO2
|
Ac
|
|||||
|---|---|---|---|---|---|---|---|---|
| −pyr | +pyr | −pyr | +pyr | −pyr | +pyr | −pyr | +pyr | |
| Wild type | + | + | + | + | + | + | + | + |
| Δech1∷pac-ori-aph | + | + | − | − | − | + | − | − |
| Δech1∷pac-ori-aph hpt∷echABCDEF | + | ND | + | ND | + | ND | + | ND |
Growth was scored in HS broth with the indicated substrates; Me, methanol; Ac, acetate; pyr, sodium pyruvate. Positive cultures (+), except acetate, typically grew within 2 days; Ac cultures grew within 4 weeks. ND, not done.
To assess the role of Ech in methanogenesis we quantified the amount of methane made from various substrates by dense suspensions (about 109/ml) of methanol-grown cells incubated overnight in the presence of pseudomonic acid [to prevent growth and protein synthesis (19)]. More dilute suspensions were used to measure the rates and stoichiometry of product formation (Table 2). The rate of methanogenesis from methanol was essentially identical for wild-type and Ech1 cell suspensions. Both strains completely converted methanol to methane and CO2 in the expected 3 (±0.3) to 1 stoichiometry. Wild-type cell suspensions converted about 80% of H2/CO2 to methane; however, Ech1 produced no methane, indicating that Ech is required for production of methane from H2/CO2. Interestingly, both wild-type and Ech1 cells rapidly produced methane from the combination of methanol/H2/CO2. Assuming that no CO2 was reduced to methane by Ech1 (as suggested by the inability of the mutant to make methane from H2/CO2), 99% of the input methanol was converted to methane, which agrees with the expected stoichiometry for this substrate combination. These data show that the inability of Ech1 to grow on methanol/H2/CO2 is not the result of a loss of methanogenesis. We also measured reduction of formaldehyde to methane by using H2, which occurred at identical rates in both wild-type and Ech1 cells. Formaldehyde enters the CO2 reduction pathway at the level of methylene-H4SPT (30). Therefore, the block in CO2 reduction lies between CO2 and methylene-H4SPT. Finally, acetate was not converted to CH4 and CO2 at detectable levels by cell suspensions of either wild-type or Ech1 grown on methanol. Thus, we measured the oxidation of CO to CO2 and H2, which represents the oxidative half of acetoclastic methanogenesis. The CO oxidation rate was 9-fold lower in Ech1 than in the wild type, suggesting that the inability to grow on acetate is caused by a defect in the oxidative segment of the acetoclastic pathway.
Table 2.
Reactions catalyzed by cell suspensions of M. barkeri strains
| Reaction | M. barkeri (nmol min−1 mg−1) | M. barkeri Ech1 (nmol min−1 mg−1) |
|---|---|---|
| 4 CH3OH → 3 CH4 + CO2 + 2 H2O* | 174 (±23) | 176 (±15) |
| CH3OH + H2 → CH4 + H2O† | 440 (±125) | 520 (±160) |
| CH2O + 2 H2 → CH4 + H2O‡ | 97 (±29) | 113 (±57) |
| CO2 + 4 H2 → CH4 + 2 H2O§ | 78 (±9) | ≪1 |
| CO + H2O → CO2 + H2¶ | 26 (±7) | 3 (±1) |
Assays contained 125 mM methanol and cell suspensions at protein concentrations of 5–20 mg/ml. CH4 production was assayed.
The gas phase was 100% H2. Assays contained 125 mM methanol and cell suspensions at protein concentrations of 5–20 mg/ml. CH4 production was assayed.
The gas phase was 100% H2. Assays contained 5 mM formaldehyde and suspended cells at a protein concentration of 6 mg/ml. CH4 production was assayed.
The gas phase was 80% H2/20% CO2. The assays contained cell suspensions at a protein concentration of 13 mg/ml. CH4 production was assayed.
To prevent methanogenesis, cultures were incubated with 20 mM bromoethanesulfonic acid (BES) for 1 h before harvesting. Cell suspensions also contained 20 mM BES. The gas phase was 10% CO/90% N2. The assays contained cell suspensions at protein concentrations of 5–10 mg/ml. Consumption of CO and production H2 and CH4 was assayed.
The production of methane from methanol/H2/CO2 by Ech1 demands an explanation, such as a block in a required biosynthetic pathway, for its lack of growth on this substrate combination. Thus, we attempted to grow the mutant on methanol/H2/CO2 media supplemented with various compounds that might bypass the supposed biosynthetic block. Initially, we showed that growth of the mutant was restored by a combination of casamino acids (0.1%), yeast extract (0.1%), acetate (10 mM), and pyruvate (10 mM), but, after many trials, we found that high concentrations of pyruvate (100 mM) alone were able to restore growth (Table 1). No combination of supplements lacking pyruvate restored growth on methanol/H2/CO2; however, the observation that additional compounds reduced the level of pyruvate needed indicates that they partially fulfill the biosynthetic requirement, probably by supplying compounds that are derived from pyruvate. Pyruvate did not restore growth on any other substrate, indicating that a defect in biosynthesis was not solely responsible for lack of growth on these compounds (Table 1). These data clearly show that, in addition to its role in methanogenesis, Ech has a required role in the biosynthesis of pyruvate.
Complementation of Δech1 Mutation.
To verify that the observed phenotypes were caused by deletion of the ech operon we attempted to complement the mutation by insertion of the echABCDEF genes into a second site in the Ech1 chromosome. We previously showed that the hpt locus, encoding hypoxanthine-phosphoribosyl transferase, is a permissive site for insertion of extraneous DNA in the close relative Methanosarcina acetivorans (M. Pritchett and W.W.M., unpublished data). We constructed a plasmid in which the ech operon was inserted into the M. barkeri hpt locus and then recombined this hpt∷echABCDEF allele into the Ech1 chromosome. DNA hybridization confirmed that the resulting transformants carried both the Δech1∷pac-ori-aph and hpt∷echABCDEF alleles (data not shown). The strains were resistant to 8-aza-2,6-diaminopurine, as expected for hpt mutants, and able to grow on all substrates used by the wild type (Table 1), indicating that the Ech1 growth phenotypes are solely the result of the Δech1 mutation.
Biochemical Analysis of Methanogenesis from H2/CO2.
To define more precisely the blocked step in the reduction of CO2 to methane, we assayed each of the known enzymatic steps between CO2 and methylene-H4SPT in extracts of mutant and wild-type cells (Table 3). As expected, ferredoxin-dependent hydrogenase activity was completely undetectable in cell extracts of Ech1, indicating that no other enzymes with this activity are present in M. barkeri. However, all other enzyme activities were present at similar levels in both Ech1 and wild-type cell extracts, leaving open the question of why Ech1 is incapable of CO2 reduction. With a single exception, each enzyme was assayed by using the natural substrate and electron-carrying cofactor. The exception is CHO-methanofuran dehydrogenase (Fmd), for which the in vivo electron donor/acceptor has never been established. Typically, as in Table 3, the enzyme is assayed by using viologen dyes as artificial electron carriers. We considered the possibility that ferredoxin was the in vivo electron carrier for Fmd and that Ech mediates the transfer of electrons between H2 and CHO-methanofuran. Both Fmd and Ech are present in the membrane fraction, therefore we assayed the oxidation of CHO-methanofuran to H2 and CO2 in membrane preparations. As shown in Fig. 2, fresh membrane preparations of wild-type cells were able to oxidize CHO-methanofuran to H2 and CO2, but only in the presence of ferredoxin. This activity is extremely labile and was not observed in preparations more than a few hours old, but was clearly reproducible in nine different measurements using three independent membrane preparations (specific activity 210 ± 60 nmol min−1 mg−1). Ech1 membranes never showed this activity despite using very fresh preparations at high protein concentrations. Because the system is so labile, this negative result must be interpreted with caution. Nevertheless, the data strongly support the conclusion that ferredoxin is the in vivo electron carrier for Fmd and suggest that the lack of growth by Ech1 on H2/CO2 is the result of an inability to produce the reduced ferredoxin needed for the Fmd-catalyzed reduction of CO2.
Table 3.
Reactions catalyzed by cell extracts
| Reaction | M. barkeri (nmol min−1 mg−1) | M. barkeri Ech1 (nmol min−1 mg−1) |
|---|---|---|
| Ferredoxin-dependent hydrogenase | 130 (±10) | ≪1 |
| CHO-MFR dehydrogenase | 1350 (±50) | 1700 (±100) |
| CHO-methanofuran:H4SPT formyltransferase | 6300 (±880) | 5780 (±960) |
| Methenyl-H4SPT cyclohydrolase | 1350 (±210) | 1200 (±100) |
| Methylene-H4SPT dehydrogenase | 3800 (±540) | 3100 (±170) |
| F420-reducing hydrogenase | 3700 (±260) | 4600 (±330) |
Reactions were assayed as described in Materials and Methods. Details are provided in the supporting information.
Figure 2.
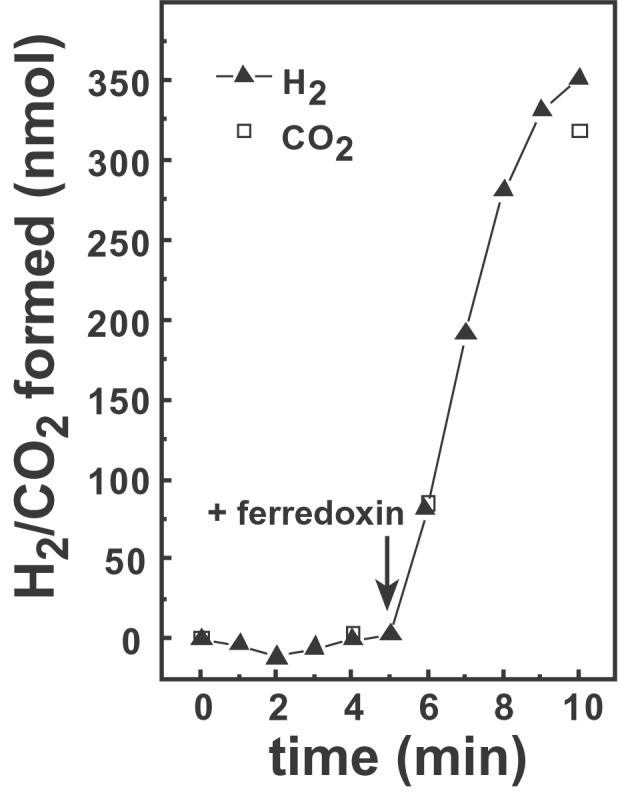
Ferredoxin-dependent formation of H2 and CO2 from CHO-MFR. Freshly prepared membrane fractions of methanol-grown M. barkeri were assayed for H2 and CO2 production as described. Ferredoxin was added at the time point indicated. The rate of H2 and CO2 formation was 200 nmol min−1 (mg membrane protein)−1.
Discussion
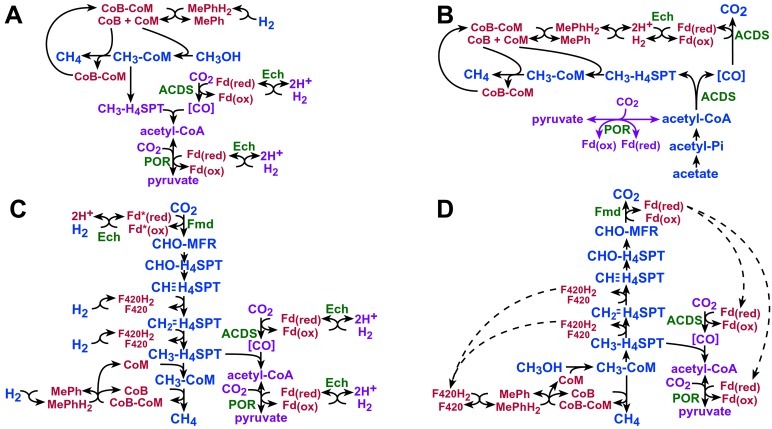
The data presented here indicate that Ech plays multiple roles in the metabolism of M. barkeri through its ability to catalyze the reversible reduction of ferredoxin by H2. These include central, but distinct, roles in methanogenesis from acetate and from H2/CO2, as well as a separate role in the biosynthesis of pyruvate (and probably acetate). Importantly, Ech is believed to couple ferredoxin-dependent hydrogenase activity to translocation of protons (or sodium ions) across the cell membrane, suggesting that Ech acts as a site of energy conservation under certain growth conditions and couples energetically unfavorable reactions to reverse electron transport under others. A model depicting the proposed roles of Ech under various growth conditions is shown in Fig. 3.
Figure 3.
Proposed functional roles for Ech and ferredoxin during growth of M. barkeri on various substrates. Models for growth on methanol plus H2/CO2 (A), acetate (B), H2/CO2 (C), and methanol (D) are shown. The methanogenic pathway is shown in blue, the energy-conserving electron transport chain in red, and biosynthetic pathway in purple. The steps catalyzed by Ech and ferredoxin-dependent enzymes are labeled in green. See text for detailed discussion.
During growth of M. barkeri on methanol/H2/CO2, Ech probably plays a strictly biosynthetic role. Methanogenesis using these substrates involves an initial transfer of the methyl group from methanol to coenzyme M (CoM-SH, mercaptoethanesulfonic acid), followed by reduction to methane with coenzyme B (CoB-SH, 7-mercaptoheptonylthreonine phosphate) as the electron donor (31). The products of these reactions are methane and the heterodisulfide of CoM-SH and CoB-SH (CoM-S-S-CoB). Energy is conserved as a transmembrane ion gradient in the subsequent electron transport chain leading from H2 to CoM-S-S-CoB, which also regenerates the reduced CoM-SH and CoB-SH needed for subsequent cycles. As shown in Fig. 3A, there is no role for Ech in this methanogenic pathway, consistent with the observation that cell suspensions of Ech1 produce methane from methanol/H2/CO2. However, Ech1 will not grow on methanol/H2/CO2 unless supplemented with pyruvate, suggesting that Ech is involved in pyruvate biosynthesis. As described above, M. barkeri assimilates carbon via the stepwise, ferredoxin-dependent synthesis of acetyl-CoA and pyruvate catalyzed by ACDS and pyruvate:ferredoxin oxidoreductase (POR). The inability of Ech1 to synthesize pyruvate during growth on methanol/H2/CO2 strongly suggests that Ech is responsible for producing the reduced ferredoxin used by ACDS and POR. The lack of an acetate requirement under these conditions is probably a result of the reversibility of POR (16). Thus, pyruvate can also serve as a source of acetyl-CoA. Importantly, the synthesis of both acetyl-CoA and pyruvate is endergonic at the H2 partial pressures found in natural habitats (1–10 Pa). The possibility that Ech couples reverse electron transport to the reduction of ferredoxin with H2 provides an attractive explanation for synthesis of these compounds under energetically unfavorable conditions.
In the acetoclastic pathway (32, 33), acetate is converted to acetyl-CoA and subsequently split into CH3-H4SPT and enzyme-bound CO by means of the ACDS enzyme. Enzyme-bound CO is oxidized to CO2 via ACDS with concomitant production of reduced ferredoxin, whereas CH3-H4SPT is sequentially reduced to methane with the production of CoM-S-S-CoB (Fig. 3B). CoM-S-S-CoB is reduced to free CoM-SH and CoB-SH as the terminal step in an energy-conserving electron transport chain. As suggested (8), the electrons from reduced ferredoxin may flow through H2 via the action of Ech, then through methanophenazine via the Vho/Vht hydrogenases, and finally to CoM-S-S-CoB via heterodisulfide reductase (34). The explanation that Ech1 fails to grow on acetate solely because of a biosynthetic defect is unlikely because we were unable to restore growth by pyruvate supplementation. Therefore, Ech probably plays a direct role in methanogenesis from acetate. Consistent with this idea, cell suspensions of Ech1 catalyzed the oxidative half of the acetoclastic pathway (conversion of CO to CO2 and H2) at a significantly lower rate than the wild type. Importantly, the conversion of CO to CO2 and H2 catalyzed jointly by ACDS and Ech is predicted to be a site of energy conservation in acetoclastic methanogensis. This reaction has been shown to be coupled to the generation of a proton-motive force (9) and is consistent with the putative ion-translocating activity of Ech.
Ech probably plays both biosynthetic and methanogenic roles during growth on H2/CO2 (Fig. 3C). The conversion of CO2 to methane involves, besides group transfer reactions, four H2-dependent reduction steps: the initial reduction of CO2 to CHO-MFR catalyzed by Fmd, the reduction of methenyl- to methylene-H4SPT (methylene-H4SPT dehydrogenase, Mtd), the reduction of methylene- to methyl-H4SPT (methylene-H4SPT reductase, Mer), and finally the reduction of methyl-CoM to methane (methyl-CoM reductase, Mcr) (35). As in the pathways described above, the latter reaction is not a direct reduction of methyl-CoM via H2, but rather involves an energy-conserving electron transport chain ending with the reduction of CoM-S-S-CoB. The cell suspension experiments clearly indicate that Ech is required for reduction of CO2 to methane with H2, specifically at a step between CO2 and methylene-H4SPT. However, each of the enzymes involved in these steps was present at wild-type levels in Ech1, leading us to believe that Ech was responsible for supplying electrons to Fmd for the reduction of CO2 to CHO-MFR. Although Fmd has been the subject of study for nearly 20 years, the in vivo electron donor/acceptor has never been rigorously established (for discussion see refs. 31 and 35). The reaction is highly endergonic in the reductive direction and cannot be measured in vitro, although the reverse reaction is easily measured by using artificial electron acceptors. CO2 reduction is thought to proceed in at least two steps: (i) the reduction of an electron carrier by H2 and (ii) the subsequent reduction of CO2 to CHO-MFR with the reduced electron carrier. The hydrogenase that provides the reducing equivalents in the first step has not previously been identified; however, it has been suggested that a reduced ferredoxin is the electron donor for the second step (31). Our data show that ferredoxin is indeed capable of serving as the electron acceptor for oxidation of CHO-methanofuran to CO2 and H2, and that Ech is required for this reaction when ferredoxin is limiting (Fig. 2). These data imply that the reverse reaction, i.e., the endergonic reduction of CO2 to CHO-MFR, also uses ferredoxin as the electron carrier, and that Ech is responsible for production of reduced ferredoxin. The putative ability of Ech to couple consumption of an ion gradient to ferredoxin reduction would allow this thermodynamically unfavorable reaction to proceed. Although growth on H2/CO2 was not restored by addition of pyruvate, it seems likely that Ech is also required for biosynthesis during growth on these substrates, because H2 is the only available reductant for biosynthesis of pyruvate and acetate under this growth condition. Thus, the biosynthetic situation is analogous to growth on methanol/H2/CO2.
Last among the growth substrates, an explanation is required for the dispensability of Ech during growth on methanol (Fig. 3D). In the methylotrophic pathway, methanol is oxidized to CO2 essentially by running the H2/CO2 pathway in reverse. The reducing equivalents thus generated are used for reducing three additional molecules to CH4 via the same steps used in the methanol/H2/CO2 pathway, except that the energy-conserving electron transport chain ending with reduction of CoM-S-S-CoB begins with reduced coenzyme F420 derived from the oxidation of methyl-H4SPT and methylene-H4SPT (31). Additional reducing equivalents in the form of reduced ferredoxin are also produced by Fmd in this pathway. Note that the Fmd reaction is exergonic in the oxidative direction and, therefore, does not require reverse electron transport. Thus, there is no Ech requirement for the Fmd reaction in the oxidative direction. Moreover, the reduced ferredoxin produced by Fmd should be more than sufficient to supply the requirement for biosynthesis of acetyl-CoA and pyruvate, and, thus, there is no biosynthetic requirement for Ech during growth on methanol.
Two additional conclusions can be drawn from the dispensability of Ech for growth on methanol. First, the finding that Ech is required for biosynthesis during growth on methanol/H2/CO2, but not during growth on methanol alone, suggests that the oxidative branch of the methylotrophic pathway is strictly repressed in the presence of hydrogen, thus precluding the use of the Fmd reaction for provision of reduced ferredoxin. This conclusion is consistent with previous data showing that many of the enzymes in the oxidative branch of the methylotrophic pathway are down-regulated by addition of hydrogen (36). Second, during growth on methanol there must be an additional ferredoxin oxidoreductase to replace Ech. Our results indicate that reduced ferredoxin generated in the oxidation of CHO-MFR is oxidized by Ech to form H2, which is then used for the reduction of CoM-S-S-CoB via the Vho/Vht hydrogenases. Because ferredoxin-dependent hydrogenase activity is undetectable in the mutant, methanogenesis and growth should cease once the ferredoxin pool is reduced. This growth cessation is not observed. Hence, oxidation of the ferredoxin in the mutant has to be carried out by an alternative reaction that does not produce H2 as intermediate. It seems possible that ACDS and POR could fulfill this role producing acetate and pyruvate as byproducts of growth. However, because the growth yield of mutant and wild type are similar (data not shown), we do not believe that one-third of the reducing equivalents are channeled into acetate or pyruvate synthesis. Further, because suspensions of wild-type and mutant cells catalyzed the conversion of methanol to CO2 and CH4 with the expected stoichiometry, it is likely that all of the reducing equivalents generated in the oxidative branch are channeled into the heterodisulfide reductase reaction. The nature of this putative link in electron transport between ferredoxin and CoM-S-S-CoB remains unclear at this time.
The reported results have general implications for the metabolism of methanogens. Hydrogenases related to Ech are encoded by the genomes of methanogens belonging to different phylogenetic groups. Examples are the two membrane-bound [NiFe] hydrogenases Eha and Ehb, found in the obligate hydrogenotrophs Methanothermobacter thermoauotrophicus and M. marburgensis (1). These enzymes have been proposed to catalyze the H2-dependent reduction of a low potential electron carrier in a reaction driven by reverse electron transport. This electron carrier is assumed to donate electrons to Fmd in the methanogenic pathway and to ACDS and POR in anabolic reactions. A similar set of membrane-bound hydrogenases is also encoded by the genome of Methanococcus jannaschii (1, 37).
Finally, the results presented here highlight the utility of Methanosarcina species for genetic analysis of methanogenesis. Most methanogens have only a single methanogenic pathway, which is believed to be essential for viability because all known methanogens are obligate methanogens. This belief is supported by the inability to construct mutants lacking a key hydrogenase in the hydrogenotrophic methanogen Methanococcus voltae (38). In contrast, Methanosarcina species are metabolically diverse, being able to use a number of substrates for methanogenesis via four distinct methanogenic pathways (see above). Thus, it should be possible to genetically block methanogenesis via one (or two) pathway(s) while leaving the mutant capable of growth via the remaining pathway(s). Our finding that ech mutants are viable on some, but not other, substrates validates this hypothesis. Further, by using a combined approach that includes in vivo genetics, a long-standing question of methanogenesis has been solved. Perhaps even more important, the insights gained regarding the physiology of M. barkeri go well beyond methanogenesis, suggesting that future genetic studies in Methanosarcina species will be extremely fruitful.
Supplementary Material
Acknowledgments
This work was supported by the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, National Science Foundation Grant MCB-987459, National Institutes of Health Grant T32-GM07283, and by a Searle Scholars Award from the Chicago Community Trust (to W.W.M.).
Abbreviations
- Ech
Ech hydrogenase
- ACDS
acetyl-CoA decarbonylase/synthetase
- CHO-MFR
to formyl-methanofuran
- HS
thiol moiety
- H4MPT
tetrahydromethanopterin
- H4SPT
tetrahydrosarcinapterin
- Fmd
CHO-MFR dehydrogenase
- CoM
coenzyme M
- CoB
coenzyme B
- POR
pyruvate:ferredoxin oxidoreductase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tersteegen A, Hedderich R. Eur J Biochem. 1999;264:930–943. doi: 10.1046/j.1432-1327.1999.00692.x. [DOI] [PubMed] [Google Scholar]
- 2.Friedrich T, Scheide D. FEBS Lett. 2000;479:1–5. doi: 10.1016/s0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 3.Böhm R, Sauter M, Böck A. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 4.Andrews S C, Berks B C, McClay J, Ambler A, Quail M A, Golby P, Guest J R. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 5.Fox J D, Kerby R L, Roberts G P, Ludden P W. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox J D, He Y, Shelver D, Roberts G P, Ludden P W. J Bacteriol. 1996;178:6200–6208. doi: 10.1128/jb.178.21.6200-6208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Künkel A, Vorholt J A, Thauer R K, Hedderich R. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- 8.Meuer J, Bartoschek S, Koch J, Künkel A, Hedderich R. Eur J Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 9.Bott M, Thauer R K. Eur J Biochem. 1989;179:469–472. doi: 10.1111/j.1432-1033.1989.tb14576.x. [DOI] [PubMed] [Google Scholar]
- 10.Bertram P A, Thauer R K. Eur J Biochem. 1994;226:811–818. doi: 10.1111/j.1432-1033.1994.t01-1-00811.x. [DOI] [PubMed] [Google Scholar]
- 11.Kaesler B, Schönheit P. Eur J Biochem. 1989;184:223–232. doi: 10.1111/j.1432-1033.1989.tb15010.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaesler B, Schönheit P. Eur J Biochem. 1989;186:309–316. doi: 10.1111/j.1432-1033.1989.tb15210.x. [DOI] [PubMed] [Google Scholar]
- 13.Winner C, Gottschalk G. FEMS Microbiol Lett. 1989;65:259–264. [Google Scholar]
- 14.Simpson P G, Whitman W B. In: Methanogenesis. Ferry J G, editor. New York: Chapman & Hall; 1993. pp. 445–472. [Google Scholar]
- 15.Grahame D A, DeMoll E. Biochemistry. 1995;34:4617–4624. doi: 10.1021/bi00014a015. [DOI] [PubMed] [Google Scholar]
- 16.Bock A K, Kunow J, Glasemacher J, Schönheit P. Eur J Biochem. 1996;237:35–44. doi: 10.1111/j.1432-1033.1996.0035n.x. [DOI] [PubMed] [Google Scholar]
- 17.Metcalf W W. In: Methods in Microbiology: Genetic Methods for Diverse Prokaryotes. Smith M, Sockett L, editors. Vol. 29. London: Academic; 1999. , Chap. 10. [Google Scholar]
- 18.Zhang J K, White A K, Kuettner H C, Boccazzi P, Metcalf W W. J Bacteriol. 2002;184:1449–1454. doi: 10.1128/JB.184.5.1449-1454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boccazzi P, Zhang J K, Metcalf W W. J Bacteriol. 2000;182:2611–2618. doi: 10.1128/jb.182.9.2611-2618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf W W, Zhang J K, Apolinario E, Sowers K R, Wolfe R S. Proc Natl Acad Sci USA. 1997;94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sowers K R, Boone J, Gunsalus R P. Appl Environ Microbiol. 1993;59:3832–3839. doi: 10.1128/aem.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf W W, Zhang J K, Shi X, Wolfe R S. J Bacteriol. 1996;178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1992. [Google Scholar]
- 24.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Afting C, Hochheimer A, Thauer R K. Arch Microbiol. 1998;169:206–210. doi: 10.1007/s002030050562. [DOI] [PubMed] [Google Scholar]
- 26.Karrasch M, Börner G, Thauer R K. FEBS Lett. 1990;274:48–52. doi: 10.1016/0014-5793(90)81326-j. [DOI] [PubMed] [Google Scholar]
- 27.te Brommelstroet B W, Geerts W J, Keltjens J T, van der Drift C, Vogels G D. Biochim Biophys Acta. 1991;1079:293–302. doi: 10.1016/0167-4838(91)90072-8. [DOI] [PubMed] [Google Scholar]
- 28.DiMarco A A, Donnelly M I, Wolfe R S. J Bacteriol. 1986;168:1372–1377. doi: 10.1128/jb.168.3.1372-1377.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitung J, Thauer R K. FEBS Lett. 1990;275:226–230. doi: 10.1016/0014-5793(90)81477-6. [DOI] [PubMed] [Google Scholar]
- 30.Escalante-Semerena J C, Rinehart K L, Jr, Wolfe R S. J Biol Chem. 1984;259:9447–9455. [PubMed] [Google Scholar]
- 31.Keltjens J T, Vogels G D. In: Methanogenesis. Ferry J G, editor. New York: Chapman & Hall; 1993. pp. 253–303. [Google Scholar]
- 32.Ferry J G. Biofactors. 1997;6:25–35. doi: 10.1002/biof.5520060104. [DOI] [PubMed] [Google Scholar]
- 33.Ferry J G. J Bacteriol. 1992;174:5489–5495. doi: 10.1128/jb.174.17.5489-5495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ide T, Bäumer S, Deppenmeier U. J Bacteriol. 1999;181:4076–4080. doi: 10.1128/jb.181.13.4076-4080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thauer R K, Hedderich R, Fischer R. In: Methanogenesis. Ferry J G, editor. New York: Chapman & Hall; 1993. pp. 209–252. [Google Scholar]
- 36.Mukhopadhyay B, Purwantini E, Daniels L. Arch Microbiol. 1993;159:141–146. [Google Scholar]
- 37.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 38.Pfeiffer M, Bestgen H, Burger A, Klein A. Arch Microbiol. 1998;170:418–426. doi: 10.1007/s002030050662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.