Abstract
During the developmental process of the Gram-negative soil bacterium Myxococcus xanthus, vegetatively growing rod cells differentiate to ultimately become metabolically quiescent and environmentally resistant myxospores encased within fruiting bodies. This program, initiated by nutrient deprivation, is propagated by both cell-autonomous and cell-nonautonomous signals. Our goal was to determine whether M. xanthus, like many other developmental systems, uses cell-cycle cues to regulate and control its developmental program. To address this question, the DNA replication cycle was used as a marker to monitor progression through the cell cycle in vegetative, stationary, and developing M. xanthus populations. Using flow cytometry, quantitative fluorescence microscopy, and FISH to establish the chromosome copy number of myxospores, it was determined that vegetatively growing cells contain one to two copies of the genome, but upon entry into stationary phase, the chromosome copy number drops to a single copy. Of particular interest, fruiting body-derived myxospores contain a specific two-chromosome DNA complement with both origin and terminus regions localized to the periphery of the myxospore. We speculate that this duplication of genetic information in the myxospore would help assure viability during germination by providing a second copy of each gene. The results of this study imply that not only is DNA replication tightly regulated during the developmental process of M. xanthus, but that there are also regulatory mechanisms to ensure that all myxospores acquire two copies of the chromosome.
Keywords: development, chromosome copy number, myxospore
Preservation of the genetic integrity of an organism is fundamental for the continuation of the species. Throughout evolution, cells have developed mechanisms for ensuring the proper replication and segregation of genetic material from mother to daughter cells. Metabolically active and dividing cells have derived a variety of mechanisms to monitor, evaluate, and when necessary, repair damaged genetic material. One strategy used by a diverse group of microbes to protect their genetic information in response to environmental stress is the formation of a metabolically dormant state: the spore.
It is apparent that the regulatory mechanisms leading to the packaging and storing of a proper, specific amount of genetic material are as fundamental to spore formation as is the creation of physical barriers such as a thick spore coat. For example, in the budding yeast Saccharomyces, specific haploid spores are formed from meiotic division. In some prokaryotes, such as Bacillus subtilis, haploid spores are formed from a specific asymmetric cell division event (1, 2), whereas in Streptomyces spores are formed during the septation of the terminal coiled aerial hyphae, leading to what are believed to be unigenomic spores (3, 4). Although these organisms are diverse in terms of spore formation machinery, a common thread is that all of these systems use mechanisms that tightly regulate their DNA replication during the sporulation process, emphasizing the importance of this regulatory step in nature.
Myxococcus xanthus is a Gram-negative soil bacterium and, as such, must constantly face the pressures of the soil environment: predators, competition, and environmental stress. When confronted with the inevitable nutrient-limiting conditions, the population survives via the initiation of a multicellular developmental program that culminates in the formation of a dome-shaped fruiting body inside which the vegetative cells have differentiated into metabolically quiescent myxospores (for reviews see refs. 5-8). Interestingly, early work showed that when chemically induced, such as by the addition of 0.5 M glycerol (9), each and every M. xanthus cell has the capacity to differentiate into a spore, and this differentiation does not require a cell-division event (10). This finding is in contrast to sporulation in B. subtilis, which does requires a cell-division event to produce an endospore (2). In such a disparate system, does M. xanthus exert cell-cycle control over its developmental program, even though cell division does not appear to be a prerequisite for sporulation? By adding M. xanthus to the list of developmental systems in which cell-cycle regulation is being studied, we seek to gain a broader understanding of cell cycles during sporulation in general and gain insight into how nature uses different mechanisms to solve similar problems.
In these studies, flow cytometry, fluorescence microscopy, and FISH were applied to quantitate the chromosome copy of number of vegetative cells, developmental fruiting body-derived myxospores, and chemically induced myxospores. By using chromosome copy number as a marker of the cell cycle, we conclude that M. xanthus controls its developmental process with cell-cycle cues such that each fruiting body-derived myxospore has two copies of the chromosome.
Methods
Bacterial Growth and Media. Strain DK101 of M. xanthus was grown in casitone/Tris/yeast extract (CTTYE) medium and on CTTYE containing 1.5% agar (11). Growth of cultures was done at 33°C. Chloramphenicol was used as needed at a concentration of 25 μg/ml as reported (12). Neither increasing the chloramphenicol concentration nor increasing the duration of exposure increased the inhibitory effect of the drug. Strain PB2 of B. subtilis was grown on a modified Schaeffer's medium (designated 2× SG) and 2× SG containing 1.7% agar (13). Sporulation of B. subtilis was induced by growth for 24 h in 2× SG.
Development. M. xanthus was grown in CTTYE liquid media until a reading of 100 Klett units. Cells were concentrated 10 to 1 and spotted as 20-μl spots onto TPM agar (10 mM Tris, pH 7.6/8 mM MgSO4/1 mM KH2PO4 containing 1.5% agar). Plates were incubated at 33°C for 5 days before harvesting. Development was also initiated on clone fruiting agar (14) and in the submerged liquid culture buffer system (15).
Purification of Myxospores. Myxospores were purified from other cells in the fruiting body by using an adaptation of previous protocols (16, 17). Briefly, developmental cells were scraped from TPM agar with a spatula and resuspended in 1 ml of 10 mM sodium phosphate, pH 7.2. This resuspension was then applied to a sucrose step gradient with steps of 60%, 30%, 15%, and 5% sucrose in 10 mM sodium phosphate, pH 7.2. Samples were centrifuged at 400 × g for 15 min in a HB-4 rotor. The 5% sucrose fraction contains rods, and the 30-60% sucrose fractions contain myxospores. Samples are typically 90-95% pure populations when observed under the light microscope.
Germination of M. xanthus Myxospores. Germination of myxospores was done as described (18). Briefly, myxospores were allowed to germinate for 24 h at 33°C in 1% casitone/Tris liquid supplemented with 1 mM CaCl2. Rifampicin was used as needed at a concentration of 10 μg/ml. Progress was monitored by phase-contrast microscopy for conversion of phase-bright spores to phase dark.
Fixation and Staining for Flow Cytometry. Samples were fixed by resuspension in 500 μl of PME buffer (50 mM Pipes, pH 6.9/5 mM EGTA/1 mM MgSO4) containing 4% paraformaldehyde at room temperature for 10 min. Samples were then washed with 90% methanol for 20 min at -20°C and treated overnight at 37°C with 100 μg/ml RNase in 200 mM Tris, pH 7.6. Staining was done at 4°C for 30 min in 25 μg/ml propidium iodide in 200 mM Tris, pH 7.6. Flow cytometric analyses were done on a Becton Dickinson FACScalibur. Each experimental run consisted of an analysis of 20,000-25,000 individual cells. The position of the fluorescence intensity peak for B. subtilis endospores used as a standard for these experiments was further confirmed by comparison with endospores germinated in the presence of DNA replication inhibitors and comparison with a vegetatively growing population of B. subtilis.
Fixation and Staining for Fluorescence Microscopy. Samples were fixed as with flow cytometry. Staining was done in mounting medium containing 1 μl/ml DAPI with 100 mM Tris (pH 9.2), 50% glycerol, and 1 mg/ml o-phenylenediamime. Fields containing both B. subtilis and M. xanthus cells in focus were taken with a Nikon Eclipse E600 microscope, and the fluorescence intensity of each cell was quantified with the image pro software package (Media Cybernetics, Silver Spring, MD).
FISH. Myxospores were affixed to slides and prepared for FISH by using an adaptation of previously published protocols (19, 20). To remove the spore coat for enhanced probe entry, myxospores were additionally treated in 1 M NaCl in 10 mM sodium phosphate, pH 7.2 for 90 min (17) before fixation. Probe fragments were created by PCR amplification of a 20-kbp region centered around the estimated origin and terminus regions of M. xanthus as determined by regions of discontinuity in the GC base skew of the genome (The Institute for Genomic Research/Monsanto, personal communication) and conjugated to Cy3 and Cy5, respectively (19). After hybridization (20), cells were visualized on an Olympus IX70 Deltavision microscope. The parfocality of the various filter sets used in this experiment was checked by localization of a set of Tetraspeck Fluorescent Microspheres (Invitrogen).
Results
Quantitation of DNA Content of Vegetative and Stationary-Phase Cells. Flow cytometric methods were used to monitor and quantify DNA content of M. xanthus populations at different growth states and during development, using the DNA content of cells as a marker for cell-cycle status. The DNA content of M. xanthus as a population was examined during exponential growth and stationary phase by fixing cells in paraformaldehyde and then staining the DNA with the DNA/RNA-binding dye propidium iodide. The fluorescence intensity, which is proportional to the DNA content, was then quantified by flow cytometry.
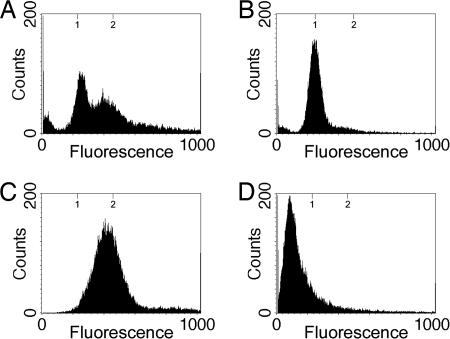
In exponentially growing cells, this technique provides a snapshot of the distribution of the numbers of cells that occupy the several stages of the cell cycle. These cells are growing asynchronously with respect to the cell cycle, and we would predict that flow cytometry would reveal a broad distribution of DNA contents, from 1n to 2n (where n is the copy number of chromosomes in the cell before DNA replication). The profile for exponentially growing M. xanthus cells in rich media is presented in Fig. 1A. As predicted, the flow cytometric data of an exponentially growing population were consistent with an asynchronous population of cells in various stages of the cell cycle and with previous studies that demonstrated a population of vegetatively growing M. xanthus cells contains an average of 1.7 chromosomes per cell (22). By this method, the cells' DNA contents range from 200 to 500 fluorescence units (FU) with a strong peak centered at 250 FU. About 40% of the cells are within the 250-FU peak, representing a 1n DNA content, whereas the remaining 60% of the cells are distributed between 250 and 500 FU, representing cells with chromosomes in varying stages of replication.
Fig. 1.
Flow-cytometric profiles of vegetative and stationary-phase M. xanthus cells. Shown are the flow-cytometric profiles of vegetative cells taken at Klett 100 units (A), 2-day stationary-phase cells taken at Klett 310 units (B), vegetative cells grown in the presence of 25 μg/ml chloramphenicol for 8 h (C), and B. subtilis spores used as a standard for quantification (D). The y axis represents cell number, and the x axis represents arbitrary FU ranging from 0 to 1,000 in a linear scale. Tick marks at the top of the graph represent the expected positions for the one and two copies of the chromosome based on the fluorescence of B. subtilis spores. The broad peak for the cells treated with chloramphenicol may be attributed to an incomplete inhibition of DNA initiation similar to what has been previously observed (12, 21). Increased concentrations of chloramphenicol and increased durations of exposure yielded identical results.
The DNA content of stationary-phase M. xanthus cells was also examined (Fig. 1B). In Escherichia coli, chromosome number changes at growth transitions: during exponential growth in rich medium cells have a distribution of two to eight chromosomes per cell, but after cells enter stationary phase the chromosome number diminishes to one to two chromosomes per cell (23, 24). As M. xanthus enters stationary phase there is also a shift to a lower chromosome copy number as the DNA profile compresses into a single 1n peak at 250 FU (Fig. 1B).
To determine the genome complement of cells after all rounds of DNA replication had been completed, we exposed exponentially growing cells to chloramphenicol before flow cytometry. Previous studies have suggested that chloramphenicol inhibits protein synthesis but not DNA replication so that cells complete DNA replication but do not divide, the net result being arrest at the completion of chromosome replication (12, 25). In the presence of 25 μg/ml chloramphenicol, exponentially grown M. xanthus cells arrested with cells having a single peak slightly <2n (500 FU) (Fig. 1C), demonstrating the position of the 2n peak where M. xanthus has doubled its DNA content. Taken together, these studies establish that exponentially growing vegetative cells are an asynchronous population.
Although these experiments demonstrate that exponentially growing cells double their DNA content, it does not establish the absolute number of chromosomes, n, present before DNA replication. If cells contain two copies of the chromosome before DNA replication (in which case n would be two), cells would be going from a two-chromosome state to a four-chromosome state. To quantify the number of chromosomes present in cells before replication, we standardized our fluorescence intensities to a known standard: B. subtilis spores. Because of the mechanism for the production of B. subtilis spores, each spore will have a single 4.2-Mbp completed chromosome (26-28) and, thus, will be uniform with respect to DNA content. B. subtilis spores were purified, fixed, stained with propidium iodide, and then subjected to flow cytometry in a manner identical to vegetative M. xanthus cells (Fig. 1D). B. subtilis spores show a strong single peak centered ≈120 FU. Because the M. xanthus chromosome is 9.1 Mbp (The Institute for Genomic Research/Monsanto, personal communication) or ≈2.2 times the size of the 4.2-Mbp B. subtilis chromosome (29), we conclude that under these growth conditions, M. xanthus cells contain a single copy of the chromosome before replication and progression through the cell cycle such that in this case n is one.
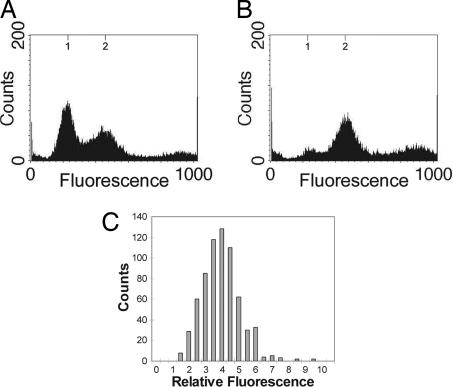
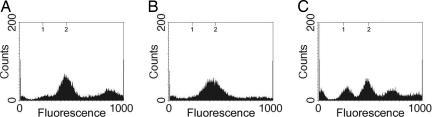
Quantitation of DNA Content of Myxospores. The DNA contents of cells isolated from both the fruiting bodies and purified myxospores were quantified by flow cytometry. Flow-cytometric analysis from all cells associated with fruiting bodies show two peaks at ≈250 and 500 FU, corresponding to the expected positions for one and two copies of chromosome (Fig. 2A). This population consists of a mixture of myxospores and rod-shaped cells that are found associated with fruiting bodies (16). When myxospores were purified from 5-day-old fruiting bodies, flow cytometry revealed that they display a single peak at ≈500 FU (Fig. 2B). This single peak demonstrates the homogeneity of myxospores with respect to DNA content and their possession of two copies of the chromosome.
Fig. 2.
Quantitation of DNA content in fruiting body-derived myxospores. (A and B) Shown are the flow cytometric profiles of 5-day-old fruiting bodies (A) purified into myxospores (B). Axes are as in Fig. 1. (C) A histogram of the fluorescence measurements for myxospores (n = 679). The y axis represents cell number, and the x axis represents the fluorescence intensity of DAPI in each myxospore relative to B. subtilis spores in the same field.
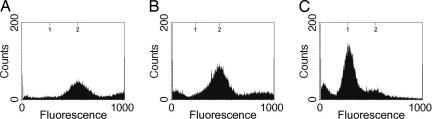
The thick spore coat of the myxospore could potentially interfere with the entry of the various dyes used in this study, leading to inaccurate fluorescence quantification, even though we consistently observed that all myxospores were stained under the fluorescence microscope and that increasing or decreasing dye concentrations did not significantly change the spectrum profiles observed, only the background (data not shown). However, to specifically eliminate the possibility of artifacts caused by dye uptake, myxospores were partially germinated, allowing for spore-coat breakdown, which would theoretically facilitate dye entry. Partial germination was achieved in the presence of the RNA polymerase inhibitor, rifampicin, which allows breakdown of the spore coat while inhibiting the spores from germinating fully back into vegetative cells (30). As the spore coat was broken down, myxospores could be observed to go from phase bright to phase dark when observed by phase-contrast microscopy. Flow-cytometric analysis demonstrated that germinating spores treated with rifampicin appeared to remain in the two-chromosome state (Fig. 3B), similar to purified myxospores (Fig. 3A), whereas untreated myxospores, which were allowed to fully germinate, progressed to the one-chromosome state (Fig. 3C). These experiments indicate that the quantification data obtained are not an artifact of spore impermeability.
Fig. 3.
Flow-cytometric profiles of myxospores germinating in the presence of rifampicin. Shown are the flow-cytometric profiles of 5-day-old fruiting body-derived myxospores (A) and myxospores germinated for 24 h in the presence of 50 μg/ml rifampicin (B) or in its absence (C). Axes are as in Fig. 1.
These data are supported by an alternate method of measuring DNA content, quantitative fluorescence microscopy (31). Purified 5-day-old myxospores were stained with the DNA-binding fluorescent dye DAPI. The fluorescence intensity of each cell in each field was quantified as described in Methods, and fluorescence intensities were measured relative to DAPI-labeled B. subtilis spores (whose DNA contents are known). These studies support the findings of the flow-cytometric analysis: the DNA content of myxospores is uniform, with myxospores having approximately four times the DNA content of the B. subtilis spores (Fig. 2C).
To explore DNA replication during sporulation further, FISH was performed on purified myxospores with probes against the replication origin region of the M. xanthus chromosome. In this analysis, fluorescent foci were observed when origin-labeled probe bound to this specific region of the chromosome. The number of foci within a cell should correspond to the number of chromosomes. After observation of >1,400 origin-labeled myxospores, ≈70% of the labeled myxospores were determined to contain exactly two distinct fluorescent foci with the remaining 30% consisting of 16% of myxospores with one focus and 14% of myxospores with three or more foci.
The observation of a single focus can be explained by the orientation of the myxospore during microscopy; using deconvolution microscopy, we have been able to separate a single focus into two distinct foci lying on top of each other but in different planes as can be observed when a 3D model is constructed from optical sections (Movie 1, which is published as supporting information on the PNAS web site). The quantitation of three or more foci usually corresponded to myxospores with what appeared to be numerous small, irregularly shaped foci obviously exterior to the spore and possibly representing nonspecific binding to the spore coat.
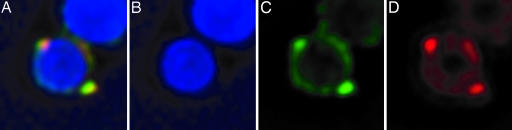
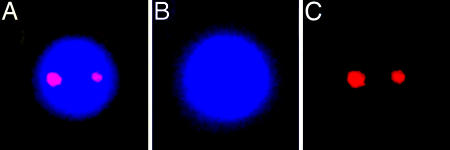
Interestingly, when simultaneous labeling was done with both origin and terminus probes, the fluorescent foci were observed to always be associated with the periphery of the myxospore's chromosomal region (Fig. 4). Furthermore, it appears that the origin and terminus regions colocalize to common areas in the periphery, although it is still unclear whether these regions are in the exact same position. When another site on the chromosome was labeled, the sdeK locus located ≈1.2 Mbp away from the origin (The Institute for Genomic Research/Monsanto, personal communication), it was possible to observe the two foci in the interior of the myxospore (Fig. 5), thus demonstrating that the observation of the terminus and origin peripheral labeling was not an artifact of the protocols.
Fig. 4.
FISH of myxospores against the origin and terminus. Shown are fluorescence micrographs depicting localization of DNA (DAPI) (B), the origin region of the chromosome (Cy3) (C), the terminus region of the chromosome (Cy5) (D), and a merge of B-D (A). (Magnification: ×7,500.)
Fig. 5.
FISH of myxospores against the sdeK locus. Shown are fluorescence micrographs depicting a myxospore labeled with DAPI (B) and with a Cy3-conjugated sdeK probe (C), and a merge of B and C (A). (Magnification: ×7,000.)
Two Mechanisms of Spore Induction. The DNA contents of myxospores induced by different methods of starvation were examined to determine whether the method of nutrient limitation regulates the DNA content of myxospores. Development was examined in clone fruiting agar (14) (which allows for very slight growth before cells initiate development) and in a submerged liquid culture buffer system (15). These results were compared with starvation on TPM.
In all three cases where myxospores were produced by fruiting body development (TPM, clone fruiting, and submerged culture), the myxospores produced at the end of development had virtually identical flow-cytometric profiles: a peak centered ≈500 FU, the two-chromosome peak (Fig. 6 A and B). These data suggest that the regulation of chromosome number in myxospores is not a phenomenon isolated to an abrupt starvation, such as what occurs on TPM media, but is observed in all tested forms of myxospore development so it represents a process inherent to M. xanthus development in general.
Fig. 6.
Flow-cytometric profiles of myxospores induced by various conditions. Shown are the flow-cytometric profiles of myxospores developed in clone fruiting medium (A), myxospores developed under submerged culture (B), and Klett 100-unit cells treated for 24 h with 0.5 M glycerol (C). All cells in the glycerol spore population were converted to phase-bright spores, and no vegetative cells were observed. Axes are as in Fig. 1.
In addition to nutrient limitation, M. xanthus cells can be chemically induced to form spores independently of fruiting body formation by using a variety of chemical agents, including glycerol and dimethyl sulfoxide (9). This chemical treatment, here 0.5 M glycerol, causes almost 100% of vegetatively growing M. xanthus cells to differentiate into spores that are physiologically and morphologically distinct from fruiting body-derived myxospores (32). Spores formed by chemical induction showed a very different flow-cytometric profile than fruiting body-derived myxospores. Whereas fruiting body-derived myxospores showed a specific two-chromosome DNA content, glycerol-induced spores showed a chromosome number distribution with one, two, or even more copies of chromosomes (Fig. 6C).
Discussion
In our findings, we have observed that fruiting bodies have a distribution of chromosome numbers from one to two chromosomes, but purified myxospores possess two copies of the chromosome. In a field of developing M. xanthus cells, it can be noted that there are not only fruiting bodies with myxospores, but that there are also peripheral rod cells remaining between them (16). When these rod-shaped cells are separated from myxospores, we have observed that these cells primarily have a one-chromosome complement similar to stationary-phase cells. We conclude that these rod-shaped cells are responsible for the one n chromosome peak observed in the fruiting body DNA content distribution previously described (Fig. 2 A). Interestingly, in previous studies peripheral rod cells have been shown to have their expression of C signal-dependent genes reduced compared with aggregating cells. This reduction in gene expression can be related to the lower levels of C signal in this population (33). One possible hypothesis is that the one-chromosome DNA complement of these rod-shaped cells is also a consequence of the low levels of C signal.
FISH demonstrates that there are not only two chromosomes in each myxospore, but that the origin regions of the M. xanthus chromosome are associated with the peripheral regions of the myxospore and appear to commonly be at opposite ends. This localization strongly suggests that there is a polarizing mechanism active during myxospore differentiation to separate the replicated chromosomes in advance of cell division, which occurs upon germination. This notion is supported by the observation that the terminus is also associated with the periphery of the myxospore. By contrast, a probe labeling another site in the chromosome, the sdeK locus, appears internal to the myxospore. Observations have been made in Caulobacter crescentus where it has been shown that, in newly divided cells, the chromosome is extended across the length of the cell with the origin near one pole and the terminus near the opposite; however, after chromosome replication, origins are located to opposite poles with termini at midcell. Regions of the chromosome are moved to specific locations after replication (34). The configuration of M. xanthus myxospores containing two replicated chromosomes with one origin and one terminus at each pole appears to be unique, but there are the obvious similarities that each organism needs to have specific localizations within the cell for specific regions of the chromosome. Furthermore, the proximity of the fluorescent foci to the cell periphery in all M. xanthus spores implies that the termini (and not just the origins as in previously observed systems) have close associations with the membrane of the cell and that the origin and terminus can possibly share a common association point with the membrane. With M. xanthus having such a large genome size compared with other bacteria, perhaps association of both the origin and terminus with the cell periphery is crucial for proper separation of replicated chromosomes. Related to this observation is that examination of the M. xanthus genome reveals four homologues of the ParA ATPase, which suggests multiple chromosome-partitioning functions in M. xanthus, possibly for the differential localization of the origin and the terminus.
Quantification of the chromosome copy number of M. xanthus myxospores revealed that spores specifically have two copies of the chromosome. In contrast, spores produced by chemical induction, which do not go through fruiting-body formation, exhibit a range of chromosome copy number consistent with previous findings on glycerol-derived spores (22, 35, 36). Although it has been shown that there is a common mechanism to induce the actual shape change in both fruiting body-derived and chemically derived spores (37), it appears that DNA replication is not controlled identically in these two sporulation methods. These results predict that there are specific regulatory mechanisms coupling DNA replication to sporulation during the developmental process of fruiting-body formation and not in chemical induction of sporulation.
As to the question “why are there two chromosomes in the myxospore?” we propose that such an arrangement enhances myxospore survival and the ability to germinate in the soil environment. It has been observed that the larger, more voluminous, bacilli such as Bacillus cereus and Bacillus megaterium contain two copies of the chromosome, whereas the spores of B. subtilis contain only one (26, 28), and furthermore, it has been found that these larger bacilli have increased resistance to ionizing radiation over the smaller bacilli (28). Similarly, cultures of E. coli grown on minimal media, with one to two chromosomes per cell, were found to be more sensitive to radiation than cultures grown in rich media, with two to eight chromosomes per cell (38), and Saccharomyces cerevisiae haploid strains are more radiation-sensitive than are diploid and tetraploid strains (39). Finally, the highly DNA damage-resistant Deinococcus radiodurans has been observed to exhibit recA-dependent interchromosomal recombination between the four copies of its genome as its second step for recovery after exposure to extreme doses of ionizing radiation (40, 41). Therefore, it is not surprising then that it has been shown that myxospores are significantly more UV-resistant than vegetative cells (42). We hypothesize that in the case of genetic damage from the environment in myxospores that have not been able to germinate because of lack of nutrient, the probability at least one good copy of an essential gene is increased, and an old myxospore would retain a complete set of essential genes and be able to germinate. During germination and before cell division, DNA repair could copy the undamaged allele, which would act as a template.
The next challenge is to uncover the mechanisms that regulate DNA replication and cell division during myxospore development. Virtually nothing is known about how the DNA replication and cell division machinery are regulated during development in M. xanthus or how development modulates the replication process, but it is possible to glean hints from other more well studied systems. In bacteria, a central player in DNA replication is the replication initiation protein, DnaA. In E. coli, cells in stationary phase have their chromosome number diminished to only one to two chromosomes per cell (23, 24). It has been shown that the expression of DnaA is inhibited as cells enter stationary phase as a direct result of the stringent response (43). Interestingly, previous studies have shown that the stringent response is also essential for the initial phases of development in M. xanthus (44). However, there appear to be fundamental differences in dnaA regulation during M. xanthus development as compared with the paradigm of E. coli. Recent DNA microarray studies of M. xanthus development have suggested that dnaA expression is actually activated 3- to 4-fold during development (45) instead of being inhibited as in the standard E. coli stringent response model. This contrast points to the possibility of the sporulation process in M. xanthus having altered previously established cellular pathways, such as the stringent response, to suit its own needs.
In B. subtilis, DnaA regulates Sda expression, preventing sporulation from occurring before the completion of DNA replication (46). Coincidentally, we found that when DNA replication inhibitors hydroxyurea, nalidixic acid, and novobiocin are added at the beginning of development in submerged culture, developmental arrest ensues in the late aggregation stage of development; whereas addition of these same inhibitors past the aggregation stage (12 h after initiation of development) results in normal development and sporulation (unpublished data). These observations might suggest that M. xanthus, like B. subtilis, has a regulatory circuit that prevents progression of development until DNA replication functions are fully completed.
Of particular interest is that several developmental regulatory pathways are known to converge at the aggregation stage of development, including the C-signal (47) and SdeK (48) pathways and activation of the devTRS operon (49). The molecular mechanism of DNA replication coupling to development is undetermined, but it is possible that DNA replication is under the control of one or more of these regulatory pathways.
Supplementary Material
Acknowledgments
We thank W. Burkholder, C. Gross, and D. Kaiser for critically reading the manuscript and providing helpful discussions and suggestions; D. Lagarias for technical assistance with flow cytometry; B. Liu, M. Paddy, and K. Shiozaki for assistance with fluorescence and deconvolution microscopy; S. Burgess for assistance with FISH; and the Monsanto Company and The Institute for Genomic Research for privileged access to the M. xanthus genome sequence before publication. This work was supported in part by Public Health Service Grant GM54592 (to M.S.) from the National Institutes of Health.
Author contributions: L.T. and M.S. designed research; L.T. performed research; L.T. and M.S. analyzed data; and L.T. and M.S. wrote the paper.
Abbreviations: FU, fluorescence units; TPM, Tris/MgSO4/KH2PO4.
References
- 1.Sonenshein, A. L. (2000) in Prokaryotic Development, eds. Brun, Y. V. & Shimkets, L. J. (Am. Soc. Microbiol., Washington, DC), pp. 133-150.
- 2.Errington, J. (1991) Mol. Microbiol. 5, 785-789. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F. (2000) in Prokaryotic Development, eds. Brun, Y. V. & Shimkets, L. J. (Am. Soc. Microbiol., Washington, DC), pp. 33-48.
- 4.Chater, K. F. & Losick, R. (1997) in Bacteria as Multicellular Organisms, eds. Shapiro, J. A. & Dworkin, M. (Oxford Univ. Press, New York), pp. 149-182.
- 5.Shimkets, L. J. & Dworkin, M. (1997) in Bacteria as Multicellular Organisms, eds. Shapiro, J. A. & Dworkin, M. (Oxford Univ. Press, New York), pp. 220-244.
- 6.Dworkin, M. (1996) Microbiol. Rev. 60, 70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser, D. & Losick, R. (1993) Cell 73, 873-885. [DOI] [PubMed] [Google Scholar]
- 8.Shimkets, L. J. (1990) Microbiol. Rev. 54, 473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler, W. & Dworkin, M. (1966) J. Bacteriol. 91, 1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dworkin, M. & Gibson, S. M. (1964) Science 146, 243-244. [DOI] [PubMed] [Google Scholar]
- 11.Bretscher, A. P. & Kaiser, D. (1978) J. Bacteriol. 133, 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimchi, A. & Rosenberg, E. (1976) J. Bacteriol. 128, 69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leighton, T. J. & Doi, R. H. (1971) J. Biol. Chem. 246, 3189-3195. [PubMed] [Google Scholar]
- 14.Hagen, D. C., Bretscher, A. P. & Kaiser, D. (1978) Dev. Biol. 64, 284-296. [DOI] [PubMed] [Google Scholar]
- 15.Kuner, J. M. & Kaiser, D. (1982) J. Bacteriol. 151, 458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connor, K. A. & Zusman, D. R. (1991) J. Bacteriol. 173, 3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye, M., Inouye, S. & Zusman, D. R. (1979) Proc. Natl. Acad. Sci. USA 76, 209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elías, M. & Murillo, F. J. (1991) Gen. Microbiol. 137, 381-388. [Google Scholar]
- 19.Peoples, T. L., Dean, E., Gonzalez, O., Lambourne, L. & Burgess, S. M. (2002) Genes Dev. 16, 1682-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, R. B. & Shapiro, L. (1999) Proc. Natl. Acad. Sci. USA 96, 10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steen, H. B. & Boye, E. (1980) Cytometry 1, 32-36. [DOI] [PubMed] [Google Scholar]
- 22.Zusman, D. & Rosenberg, E. (1970) J. Mol. Biol. 49, 609-619. [DOI] [PubMed] [Google Scholar]
- 23.Åkerlund, T., Nordstrom, K. & Bernander, R. (1995) J. Bacteriol. 177, 6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boye, E. & Løbner-Olesen, A. (1991) Res. Microbiol. 142, 131-135. [DOI] [PubMed] [Google Scholar]
- 25.Zusman, D., Krotoski, D. M. & Cumsky, M. (1978) J. Bacteriol. 133, 122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser, P. M. & Karamata, D. (1992) Biochimie 74, 723-733. [DOI] [PubMed] [Google Scholar]
- 27.Oishi, M., Yoshikawa, H. & Sueoka, N. (1964) Nature 204, 1069-1073. [DOI] [PubMed] [Google Scholar]
- 28.Woese, C. R. (1958) J. Bacteriol. 75, 5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessières, P., Bolotin, A., Borchert, S., et al. (1997) Nature 390, 249-256. [DOI] [PubMed] [Google Scholar]
- 30.Juengst, F. W., Jr. & Dworkin, M. (1973) J. Bacteriol. 113, 786-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vischer, N. O., Huls, P. G., Ghauharali, R. I., Brakenhoff, G. J., Nanninga, N. & Woldringh, C. L. (1999) J. Microsc. 196, 61-68. [PubMed] [Google Scholar]
- 32.Komano, T., Inouye, S. & Inouye, M. (1980) J. Bacteriol. 144, 1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julien, B., Kaiser, A. D. & Garza, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viollier, P. H., Thanbichler, M., McGrath, P. T., West, L., Meewan, M., McAdams, H. H. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zusman, D. & Rosenberg, E. (1968) J. Bacteriol. 96, 981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg, E., Katarski, M. & Gottlieb, P. (1967) J. Bacteriol. 93, 1402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licking, E., Gorski, L. & Kaiser, D. (2000) J. Bacteriol. 182, 3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasin, F. & Hutchinson, F. (1977) J. Mol. Biol. 116, 81-98. [DOI] [PubMed] [Google Scholar]
- 39.Mortimer, R. K. (1958) Radiat. Res. 66, 158-169. [Google Scholar]
- 40.Levin-Zaidman, S., Englander, J., Shimoni, E., Sharma, A. K., Minton, K. W. & Minsky, A. (2003) Science 299, 254-256. [DOI] [PubMed] [Google Scholar]
- 41.Daly, M. J. & Minton, K. W. (1995) J. Bacteriol. 177, 5495-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudo, S. Z. & Dworkin, M. (1969) J. Bacteriol. 98, 883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiaramello, A. E. & Zyskind, J. W. (1990) J. Bacteriol. 172, 2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer, M. & Kaiser, D. (1995) Genes Dev. 9, 1633-1644. [DOI] [PubMed] [Google Scholar]
- 45.Jakobsen, J. S., Jelsbak, L., Welch, R. D., Cummings, C., Goldman, B., Stark, E., Slater, S. & Kaiser, D. (2004) J. Bacteriol. 186, 4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkholder, W. F., Kurtser, I. & Grossman, A. D. (2001) Cell 104, 269-279. [DOI] [PubMed] [Google Scholar]
- 47.Kroos, L. & Kaiser, D. (1987) Genes Dev. 1, 840-854. [DOI] [PubMed] [Google Scholar]
- 48.Pollack, J. S. & Singer, M. (2001) J. Bacteriol. 183, 3589-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thöny-Meyer, L. & Kaiser, D. (1993) J. Bacteriol. 175, 7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.