Abstract
Hedgehog pathway activation is required for proliferation of cerebellar granule cell neuron precursors during development and is etiologic in certain cerebellar tumors. To identify genes expressed specifically in granule cell neuron precursors, we used oligonucleotide microarrays to analyze regulation of 13,179 genes/expressed sequence tags in heterogeneous primary cultures of neonatal mouse cerebellum that respond to the mitogen Sonic hedgehog. In conjunction, we applied experiment-specific noise models to render a gene-by-gene robust indication of up-regulation in Sonic hedgehog-treated cultures. Twelve genes so identified were tested, and 10 (83%) showed appropriate expression in the external granular layer (EGL) of the postnatal day (PN) 7 cerebellum and down-regulation by PN 15, as verified by in situ hybridization. Whole-organ profiling of the developing cerebellum was carried out from PN 1 to 30 to generate a database of temporal gene regulation profiles (TRPs). From the database an algorithm was developed to capture the TRP typical of EGL-specific genes. The “TRP-EGL” accurately predicted expression in vivo of an additional 18 genes/expressed sequence tags with a sensitivity of 80% and a specificity of 88%. We then compared the positive predictive value of our analytical procedure with other widely used methods, as verified by the TRP-EGL in silico. These findings suggest that replicate experiments and incorporation of noise models increase analytical specificity. They further show that genome-wide methods are an effective means to identify stage-specific gene expression in the developing granule cell lineage.
Keywords: Sonic hedgehog|proliferation|expression profiling|oligonucleotide microarray| noise
During early central nervous system development, multipotent progenitors are instructed by extrinsic signals from organizing centers such as the floorplate and midbrain–hindbrain isthmus (1). The transcriptional response to such signals is complex, depending on intrinsic restrictions on cellular competence to proliferate or develop into specific classes of neurons or glia. For such reasons, central nervous system developmental interactions are difficult to model by using homogeneous cell lines. Recent work indicates that expression profiling can identify genes expressed in the central nervous system (2–7) as confirmed by in situ hybridization (ISH; refs. 2 and 3). However, the application of gene-expression profiling to model systems in developmental biology poses unique technical and analytical challenges. Analysis of gene expression in particular cells within a heterogeneous tissue often is hampered by the inability to purify cells that maintain the biological characteristics of populations in situ. Additionally, the analytic algorithms used for identification of genes may fail to account for noise in the experimental data (8, 9).
As a model system, the rodent neonatal cerebellum is suitable for study of these issues, because it is relatively simple anatomically and is comprised predominantly of granule cell neurons. Granule cell neuron precursors (GCNPs) are generated by the embryonic hindbrain and migrate dorsally to form the outer layer of the cerebellum, or external granule layer (EGL; Fig. 1A; ref. 10). Cerebellar growth is largely postnatal, and the secreted glycoprotein Sonic hedgehog (Shh) is necessary for GCNP proliferation in vivo. Primary cultures of neonatal cerebellum have been used to study granule cell development, and Shh has been shown to cause proliferation and prevent differentiation of GCNP in vitro (11–14). We have incubated postnatal day (PN) 5 GCNP cultures in serum-containing medium for 12–16 h without Shh proteins, which allows most immature granule cells to leave the cell cycle (14). Approximately 20% of remaining GCNPs continue to express the immature marker, Math1 (15), and proliferate after subsequent Shh treatment. Thus, these cultures are highly heterogeneous, comprising populations of neurons and glia that are postmitotic or at various phases of the cell cycle.
Figure 1.
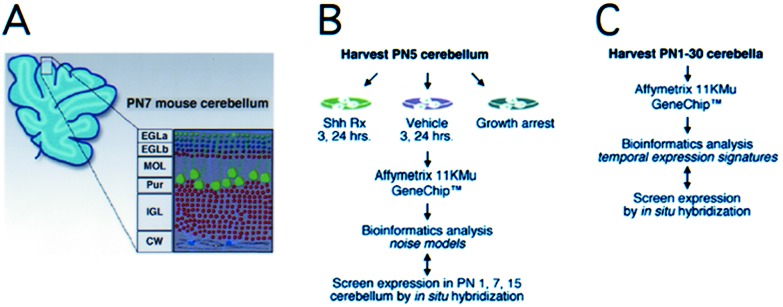
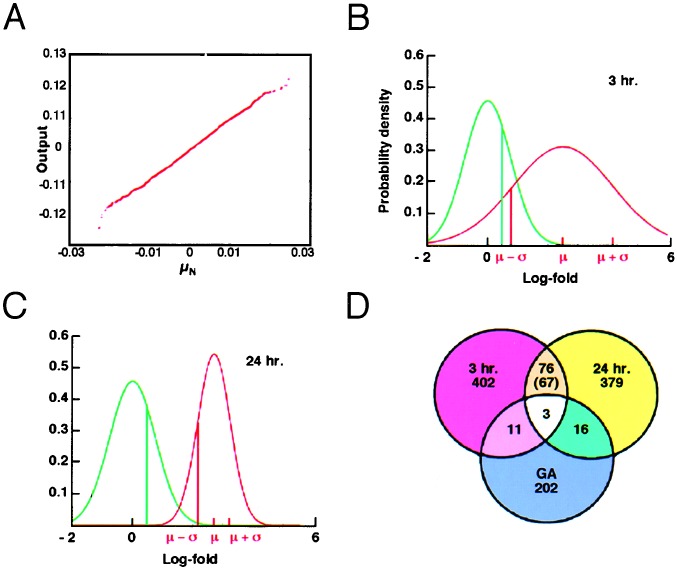
Structure of PN 7 mouse cerebellum and experimental approach. (A) The layered architecture of the cerebellum in a parasagittal section. The EGL comprises actively proliferating granule cells (EGLa) and those that recently have left the cell cycle (EGLb). The Purkinje cell (Pur), molecular (MOL), internal granule cell (IGL), and cerebellar white matter (CW) layers are indicated. (B) Schematic diagram of experimental strategy used to identify 76 genes/EST significantly up-regulated by Shh under various culture conditions and verification by ISH. (C) Whole cerebella during PN1–30 were analyzed by GeneChips to generate a database of temporal gene regulation used to validate results of analytical techniques in silico.
Here we show that it is possible to prospectively identify genes expressed specifically in GCNP in vivo with high specificity via microarray analysis of primary cerebellar cultures treated with Shh. Our results indicate that patterns of temporal regulation in silico, derived from whole-organ profiling during cerebellar development, can be used to recognize as well as validate GCNP-specific gene expression. We then use this information to compare various analytic procedures in terms of the relative false-positive rate. Incorporation of noise models augmented analytical specificity and identification of genes with temporal-spatial restriction to GCNP in the developing cerebellum.
Materials and Methods
Primary Cerebellar Cultures and Oligonucleotide Microarrays.
Detailed procedures for preparation of primary cerebellar culture of neonatal Swiss–Webster B PN 4–5 mice and treatment with Shh and growth-arrest conditions are described (14). Approximately 1.5 × 107 cells or 3–5 pooled cerebella from PNs 1, 3, 5, 7, 10, 15, 21, and 30 were collected. From all conditions, 20 μg of total RNA purified by CsCl2 density gradient or Trizol was prepared for hybridization to Affymetrix (Santa Clara, CA) Mu11K GENECHIP; chips were scanned, and data capture was carried out as described (16).
Average of Ratios (AR) Analysis Method.
Affymetrix software uses the average difference (Avg Diff) as a quantitative indicator of expression of a transcript and the absolute call to assess the reliability of transcript measurement (absolute call values: absent, marginal, or present). Our experiments used the Mu11K ensemble of 13,179 probe sets for expressed sequence tags (ESTs) and controls, which we refer to as the Chip data set. For these, the Avg Diff values were normalized by a linear regression technique (17–19) using a 3-h vehicle data set as its normalizing reference. Our normalization method assumes that the overall distribution of expression values is similar. Our analyses showed a maximum of 493 genes differentially expressed between two states (3 h untreated vs. treated) or less than 4% of the total number of genes measured (Fig. 2). Because the scatter of genes is not affected by the linear transformation that the normalization provides, we believe that the assumption is sound.
Figure 2.
Genes up-regulated under experimental conditions. (A) Distribution of measurement error. The y axis is the output of Gaussian random number generator N (0, 0.0065), and the x axis is μN from 5,000 permutations τ of condition labels. The linear relationship between X and Y indicates Gaussian distribution of measurement error. (B and C) Example of the log-fold distribution caused by noise (FDN; green line) and log-fold distribution of Shh-treated vs. vehicle-treated cells (red line) at 3 (B) and 24 (C) h for the gene cdc20. Areas underneath the curves indicate respective probabilities. μ, mean; σ, SD. (D) Total number of genes/EST significantly up-regulated at 3 (red) and 24 (yellow) h and under conditions of growth arrest (blue).
An FDN representing the probabilistic distribution of logarithmic fold changes was calculated from the pair of most highly correlated 3-h vehicle data sets (two identical targets (aliquots) that had been hybridized onto separate microarrays). Let τ = (τ1, τ2, . . . , τ13,179), where τi takes values 1 or 2, and be a permutation of the Avg Diff values of the 3-h replicate vehicle experiments. For each τ, we compute a mean and SD as follows in Eqs. 1 and 2.
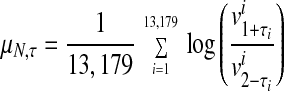 |
1 |
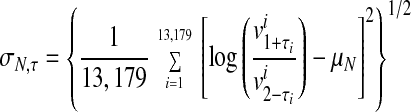 |
2 |
Here
v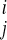 is the Avg Diff value for gene i under vehicle condition
j. (we define
s
is the Avg Diff value for gene i under vehicle condition
j. (we define
s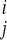 similarly for the Shh-treated condition). For 5,000 different
permutations of τ, we found the μN to be
Gaussian-distributed ≈0.0065 (1.0065-fold; Fig. 2) and the average
and median of σN to be 0.7608 (2.1400-fold).
All folds were transformed logarithmically for numerical symmetry
around zero. For each EST, we calculated a statistic for its fold
change from vehicle to Shh-treated conditions as follows in Eqs.
3 and 4,
similarly for the Shh-treated condition). For 5,000 different
permutations of τ, we found the μN to be
Gaussian-distributed ≈0.0065 (1.0065-fold; Fig. 2) and the average
and median of σN to be 0.7608 (2.1400-fold).
All folds were transformed logarithmically for numerical symmetry
around zero. For each EST, we calculated a statistic for its fold
change from vehicle to Shh-treated conditions as follows in Eqs.
3 and 4,
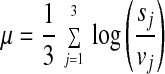 |
3 |
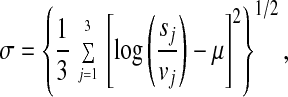 |
4 |
where vj and sj denote Avg Diff values for the EST in the jth vehicle and Shh-treated conditions, respectively. μ is invariant under the order of taking folds, and there are an equal number of replicate experimental data sets, three, in each of the vehicle and Shh-treated conditions. We call this the fold method based on the statistic of ratios. A gene is considered to be up-regulated significantly from vehicle to Shh-treated condition if (i) the fold statistic average was greater than one SD above the FDN (i.e., μ > σN), and (ii) μ − σ > 0.24 (i.e., ≥1.27-fold increase at the lower end).The initial selection of parameter values was arbitrary with the intention to modify them based on empirical testing of their ability to identify genes expressed in the developing cerebellum. These initial parameter values were found to yield a success rate of 83% (Table 1) of the genes identified, and therefore no further modifications were carried out. A similar rationale explains the use of a Pearson correlation value of >0.75 in the temporal gene regulation profile (TRP)-EGL algorithm (below).
Table 1.
Expression of genes/ESTs in vivo in the neonatal mouse cerebellum
| Genes | Accession No. | PN 1
|
PN 7
|
PN 15
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EGL | EGLa | EGLb | PK | IGL | EGLa | EGLb | PK | IGL | ||
| Training set: EGL expression—positive* | ||||||||||
| CycD1 | W08016 | +++ | +++ | − | − | − | − | − | − | − |
| STK1 | D21099 | +++ | ++ | ++ | − | − | − | − | − | − |
| Cdc20 | AA00468 | +++ | +++ | +++ | − | − | + | + | + | + |
| CKS2 | AA11263 | ++ | ++ | − | − | − | − | − | − | − |
| AYK1 | U80932 | +++ | ++ | − | − | − | + | + | − | − |
| IPL1 | AA050055 | + | ++ | ++ | − | − | − | − | − | − |
| SET | W78604 | +++ | +++ | +++ | − | − | + | + | + | + |
| BM28 | AF004105 | +++ | +++ | − | − | − | − | − | − | − |
| RanBP1 | X56045 | +++ | +++ | − | − | − | − | − | − | − |
| DP1 | X72310 | ++ | +++ | ++ | − | − | − | − | − | ++ |
| Training set: EGL expression—negative* | ||||||||||
| Stra13 | Y07836 | − | − | − | − | − | − | − | − | ++ |
| EST69† | C76791 | − | − | +/− | ++ | + | − | − | − | − |
| Test set: EGL expression—positive‡ | ||||||||||
| E2F1 | L21973 | + | ++ | ++ | − | − | − | − | − | ++ |
| RNM2 | M14233 | ++ | +++ | +++ | − | − | − | − | − | − |
| CycB2 | X66032 | +++ | ++ | ++ | − | − | − | − | − | − |
| N-myc | X03919 | +++ | +++ | +++ | − | − | − | − | − | − |
| CycA2 | Z26580 | +++ | +++ | +++ | − | − | − | − | − | − |
| CycB1 | AA426917 | ++ | +++ | +++ | − | − | − | − | − | − |
| mMIS5 | AA689977 | +++ | +++ | +++ | − | − | − | − | − | − |
| EIF4A | AA166088 | ++ | +++ | +++ | + | + | − | − | − | − |
| HMG1 | Z11997 | ++ | +++ | +++ | − | − | − | − | − | − |
| PAL31 | W48027 | ++ | +++ | +++ | + | + | − | − | − | − |
| Test set: EGL expression—negative‡ | ||||||||||
| AA84 | AA688784 | − | + | + | − | − | − | − | − | − |
| AA96 | AA267296 | − | + | + | − | − | − | − | − | − |
| FKBP-13† | AA072278 | − | +/− | +/− | − | − | − | − | − | − |
| BC73 | AA16900 | − | +/− | +/− | + | − | − | − | − | − |
| RCC | AA051583 | − | + | + | − | − | − | − | − | − |
| AA88 | AA275288 | − | + | + | − | − | − | − | − | − |
| QKI5A | AA174970 | − | − | − | +++ | +++ | − | − | − | − |
| FIN-14 | U42386 | − | − | − | ++ | ++ | − | − | − | − |
PK, Purkinje cell; IGL, internal granule layer; EGLa, actively proliferating granule cells; EGLb, granule cells that recently have left the cell cycle.
A training set of 12 genes was tested for EGL-specific expression in PN 1, 7, and 15 mouse cerebella by ISH. Ten of these (CycD1, STK1, CDC20, CKS2, AYK1, IPL1, SET, BM28, RanBP1, and DP1) showed expression in the PN 7 EGL in contrast to Stra13 and EST69.
Entries identified by the TRP-EGL that failed to show expression in the EGL in vivo (false positives). False negatives, not identified by the TRP-EGL but having EGL-specific expression, are underlined.
Test data set expression in vivo. Of an additional 18 genes tested, 10 were expressed in the PN 7 EGL, and 8 genes were not expressed specifically in the EGL. The relative intensity of expression is shown as strong (+++), moderate (++), weak (+) or absent (−).
Permutation Testing.
Permutation analysis was carried out as described (19) whereby the condition labels of the data were permuted within their respective 3- and 24-h experiment groups. The relabeled data were analyzed for the number of significant genes, and the procedure was repeated with different label permutations. Results of 10,000 permutations for the intersection of genes up-regulated across conditions in both the 3- and 24-h yielded a false-discovery rate (FDR; ref. 19) for our AR method of 15.2%. In contrast, an FDR of 60.5% was obtained for the conventional ratio of the averages (RA) technique (3, 20, 21) where the fold is calculated as follows in Eq. 5.
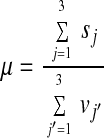 |
5 |
Generation of TRP.
Duplicated microarray data sets were obtained from eight postnatal time points: PN 1, 3, 5, 7, 10, 15, 21, and 30. Each data set was normalized by linear regression (17) against a PN 1 set. A gene possessed an “EGL-specific temporal pattern” if in its temporal profile: (i) the average of Avg Diffs for PN 3–7 was significantly greater (Student t test, with P < 0.05) than the average of Avg Diffs for PN 15–30, (ii) data for the gene contained at least one “P” call, (iii) Ave Diffs of at least one entry was >200, and finally (iv) that the profile had a maximum Pearson correlation greater than 0.75 with respect to the patterns: exp[−0.2(t − 3)2], exp[−0.2(t − 5)2], or exp[−0.2(t − 7)2].
ISH.
Neonatal cerebella (PN 1, 7, and 15) from perfused Swiss–Webster B mice and all genes/ESTs in Table 1 were analyzed as described (ref. 22; detailed protocol available on request).
Results
We asked whether stimulation of primary cerebellar cultures with Shh could be used to identify genes expressed in the GCNP component. For the oligonucleotide microarray experiments, RNA samples were harvested from primary PN 4–5 cerebellar cultures after 3- and-24 h treatment with Shh or vehicle (control), corresponding to the timing of G1 cyclin gene up-regulation and maximal levels of proliferation, respectively (14). Further, to distinguish Shh-regulated genes expressed within postmitotic granule neurons or glia, we generated expression profiles from cultures that were growth-arrested and then treated with Shh (14). Fig. 1B illustrates the overall organization of experiments (data available at www.chip.org/∼kho/sonic.txt).
The analytic method was developed to measure the likelihood that a reported fold change in expression was caused by a genuine physiological response as opposed to being an artifact of microarray measurement error or noise. To account for biological sample variability, all experiments were performed in duplicate (interexperimental noise). In addition, an analysis was performed to account for inter-Chip variability. In sum, three separate hybridizations/analyses were performed for each of the 3- and 24-h vehicle and Shh-treated samples. As illustrated in Fig. 2, a gene was regarded as significantly up-regulated when (i) its fold statistic average μ was one SD (σN) above the FDN average of 0.0 and (ii) μ − σ > 0.24 (see Materials and Methods). In other words, we selected for genes with up-regulation above a chosen threshold with low variance and therefore a higher likelihood of fold reproducibility across replicates.
By using this procedure, of 13,179 potential genes/ESTs, 493 (4.4%) and 475 (4.3%) genes showed significant up-regulation after 3 and 24 h of Shh treatment, respectively (Fig. 2B). Only 79 genes (0.7%) were up-regulated at both time points (Fig. 2B). Permutation analysis indicated that these findings were highly unlikely to have been obtained by chance alone. Of the 79 genes identified, many were involved in cell cycle regulation, chromatin remodeling, and DNA/RNA synthesis. For instance, 10 genes (14%) encoded protein components of cell cycle machinery, 12 (17%) were nuclear-chromosomal proteins implicated in DNA replication, and others (e.g., DNA topoisomerase II, tRNA ligase, and endonuclease 1) are associated with ribosomal assembly. These findings suggested that the majority of genes up-regulated by Shh at both 3 and 24 h were associated with the GCNP component of the cultures. Interestingly, within these primary cerebellar cultures, proliferating GCNPs account for only ≈20% of total cells (14). Under culture conditions of growth arrest, a distinct and largely nonoverlapping set of genes was up-regulated by Shh (Fig. 2D), suggesting differences in the transcriptional response between GCNP and postmitotic granule cells. Three genes also identified in conditions of growth arrest were subtracted to form the final list of 76 genes specifically up-regulated in Shh-treated cerebellar cultures (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org).
Validation of Candidate Genes/ESTs at Key Developmental Time Points.
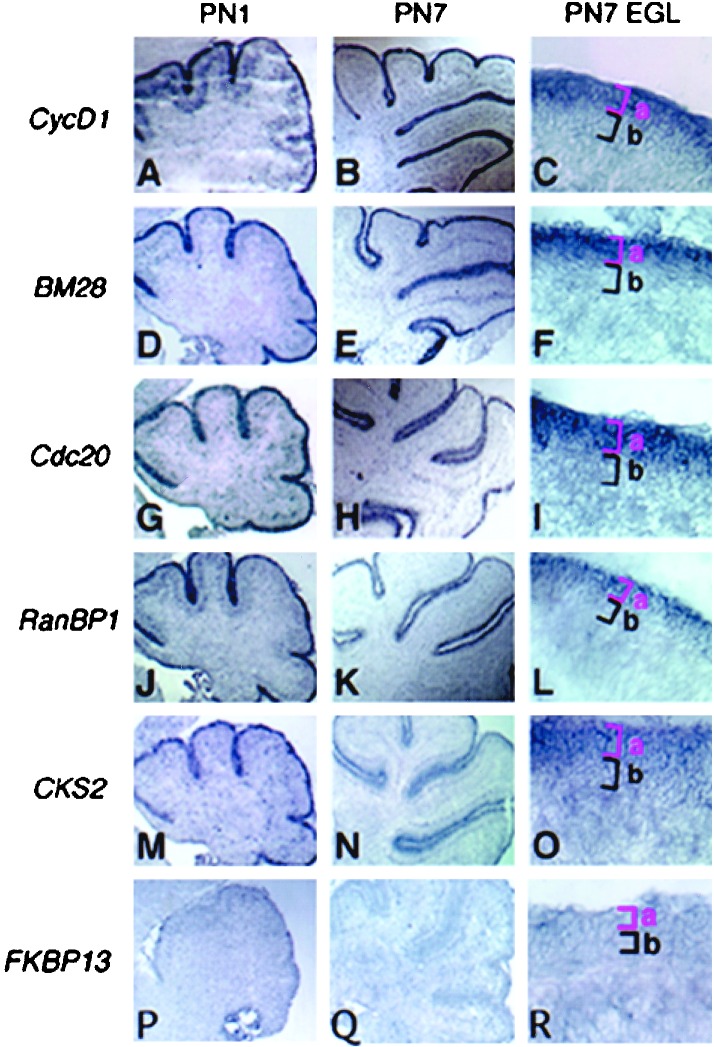
To determine whether some or all of the 76 candidates were expressed in the EGL, we chose a “training set” of 12 randomly selected genes/ESTs and submitted them to ISH on cryosections of PN 1, 7, and 15 neonatal cerebellum. These stages correspond to periods of GCNP proliferation (PN 1 and 7) and differentiation (PN 15 and later stages) (23). In particular, PN 7 most closely approximates the temporal stage of cerebellar development reflected in the cultures of PN 5 granule cells after ≈40 h in vitro. Strikingly, we observed that 10 of 12 (83%) training-set genes showed specific expression in the PN 1–7 EGL (Fig. 3 and Table 1).
Figure 3.
Expression of candidate genes/ESTs in vivo in the neonatal mouse cerebellum. Representative results of ISH showing the gene-expression pattern of identified genes in the PN 1 and 7 postnatal mouse cerebellum at low power magnification (×40) are shown. (C, F, I, L, O, and R) The right-hand column shows high-power magnification (×400) images of the EGL and delineates the actively proliferating granule cells (from those that recently have left the cell cycle) (Fig. 1A). The gene-expression pattern for the group I genes cycD1 (A–C), BM28 (D–F), cdc20 (G–I), RanBP1 (J–L), and CKS2 (M–O) indicated mRNA transcripts in the EGLa. In contrast, genes such as FKBP13 (P–R) did not show expression in PN 7 EGL.
Whole-Organ Temporal Expression Profiling.
We observed that 83% of genes in the training set were expressed in PN 1–7 cerebellum and down-regulated significantly by PN 15 (Table 1), which is consistent with normal temporal regulation of GCNP proliferation. These findings suggested the concept of a TRP, characteristic of genes expressed within the GCNP compartment of the EGL. If so, it was possible that such expression signatures could be identified via microarray analysis.
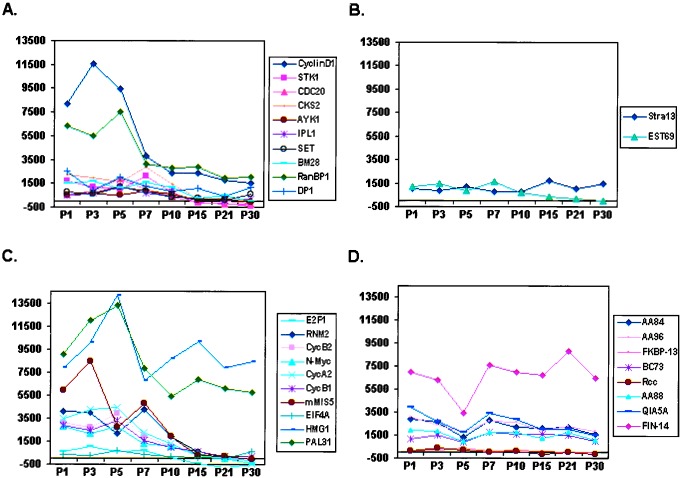
We submitted duplicate samples of whole developing mouse cerebellum from PN 1–30 for analysis (Fig. 1C). As shown (Fig. 4A), 10 of 12 genes with validated expression in the EGL were highly expressed at PN 3–7 relative to levels at PN 15–30. Of the remaining 2 of 12 genes, Stra13 showed neither this temporal pattern (Fig. 4B) nor EGL expression (Table 1). Although EST69 did show the temporal pattern characteristic of EGL-specific genes, its expression was not confirmed in situ (Table 1). The failure to detect EST69 expression may be caused by technical problems such as faulty probe design or low endogenous expression levels. In any case, these findings suggest a high correspondence between genes expressed in cerebellar granule precursors, as detected by ISH and the temporal sequence of regulation typified by genes shown in Fig. 4A, as determined by whole-organ expression profiling.
Figure 4.
Temporal analysis of candidate genes/EST expression by whole-organ expression profiling of mouse neonatal cerebellum. (A) Graphical representation of temporal regulation of the 10 of 12 genes from the training set with confirmed expression in the cerebellar EGL (Table 1). Note that they show a similar pattern comprising relatively high levels of expression at PN 3–7 relative to PN 15–30. (B) Temporal analysis of the remaining 2 of 12 genes (Table 1) from the training set. (C and D) Pattern of temporal regulation of 18 additional genes tested (“test set,” Table 1). Note that 8 of 10 genes (C) displayed a similar pattern to genes in A. Conversely, HMG1 exhibited a late peak at PN 15. Seven of nine genes (D) showed a nonspecific temporal pattern of regulation. It should be noted that the failure to identify expression of EST69 and FKBP13 could be because of technical reasons such as faulty probe design or low endogenous levels of expression in vivo. x axis, duplicate samples of mouse cerebella harvested at PNs 1, 3, 5, 7, 10, 15, 21, and 30 were analyzed; y axis, absolute value of Avg Diff as reported by Affymetrix software (scale set at 2,000).
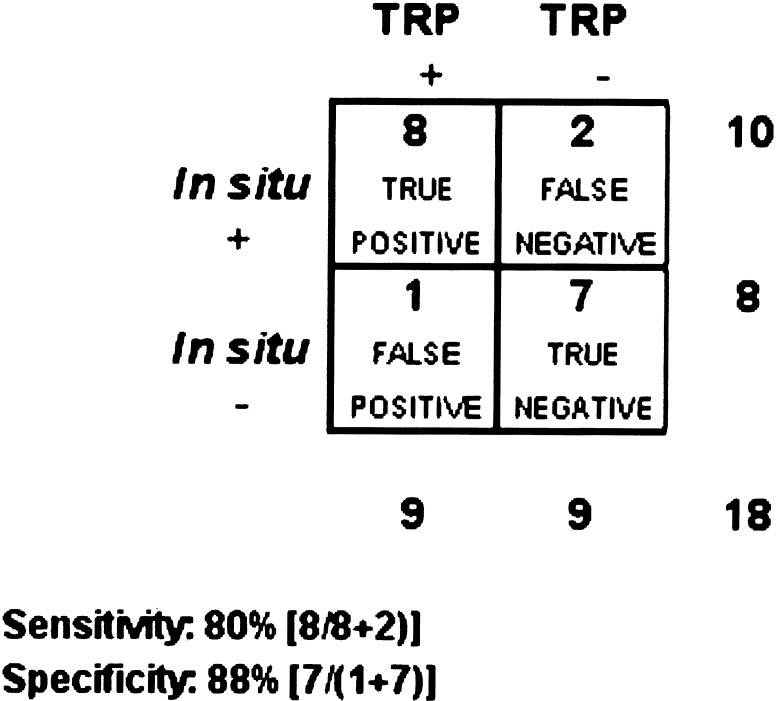
We formalized our knowledge-based approach to analyze additional genes prospectively. The TRP-EGL was designed to identify genes with a temporal pattern of expression typical of true-positive genes in the training set. We applied the TRP-EGL to the list of remaining candidates from Table 3 and chose nine that met TRP-EGL criteria (positives) and nine that did not meet criteria (negatives). These 18 genes comprised the test data set, which was validated by ISH (Table 1). As shown (Fig. 5), the TRP-EGL predicted the in vivo pattern of expression with a high sensitivity of 80% and specificity of 88%.
Figure 5.
Sensitivity and specificity of using temporal gene-expression pattern in silico to predict expression of candidate genes/EST in vivo. As shown, the TRP results in an overall sensitivity of 80% and specificity of 88%. Assuming the stochastic independence of the in situ confirmation outcome and the temporal gene regulatory pattern, a χ2 test of the contingency table gives a probability (P < 0.01) that the association between confirmation and pattern is insignificant.
Application of Noise Models Increases Analytical Specificity.
These findings suggested that the TRP-EGL database in silico provided an independent means of identifying and/or validating genes expressed in GCNPs in vivo. As shown in Table 2, the yield of genes meeting TRP-EGL criteria was improved significantly with biological replication of experiments. Of the list of 76 genes common to both 3- and 24-h treatment data sets, 52% met TRP-EGL criteria vs. only 17 and 23% of genes identified from 3- or 24-h time points alone, respectively (Table 2). We note that the observed rate of test genes expressed in the EGL by ISH (10 of 18, 56%) was quite close to the expected rate based on validation by TRP-EGL. These findings are further evidence that the temporal gene-expression database provides an independent and accurate means of validating gene expression in silico.
Table 2.
Validation of analytical methods by temporal regulation profile
| Analytical Methods | A Total numbers of genes/ESTs | B TRP-EGL | C (+) Predictive value, % |
|---|---|---|---|
| 3 h (AR method) | 493 | 84 | 17 |
| 3 h (RA method) | 890 | 135 | 15 |
| 3 h (SAM, Δ = 0.22 54.6% FDR) | 494 | 67 | 14 |
| 24 h (AR method) | 475 | 111 | 23 |
| 24 h (RA method) | 1214 | 211 | 17 |
| 24 h (SAM, Δ = 0.17 49.5% FDR) | 476 | 51 | 11 |
| (3 + 24) h (AR method) | 79 | 41 | 52 |
| (3 + 24) h (RA method) | 195 | 72 | 37 |
| (3 + 24) h (SAM) | 14 | 1 | 7 |
Column A shows the compilation of total genes identified using the AR method at 3-, 24-, and 3 + 24-h time points. All results were screened against the database of temporal gene regulation profiles and TRP+ genes with appropriate temporal pattern are shown in column B. Column C shows the positive predictive value (i.e., the percentage of validated genes). Note that the percentage of validated genes increases by 2.2–3-fold when genes common to the 3- and 24-h data sets are selected. Moreover, compared to the alternative RA and SAM methods, our custom AR procedure yielded a 1.4-fold and >7-fold increase in positive predictive value, respectively.
Using this resource, we compared our custom analytic (AR) procedure with others in current use including fold significance (RA) methods (19–21) and significance analysis of microarrays (SAM; ref. 19). As shown (Table 2), although the RA method yields a higher overall number of genes, the positive predictive value is decreased slightly as compared with the AR method at 3 or 24 h. However, the AR method yields a 1.4-fold increase in identification of authenticated genes when the 3- and 24-h data sets are combined. For SAM, we performed two tests. First, to keep the same sensitivity, we ran SAM with Δ3 h = 0.21670 and Δ24 h = 0.17392 (FDRs 54.6 and 49.5%, respectively), which generated gene lists of similar size to the AR list. As shown (Table 2), 494 and 476 genes were identified by SAM as significantly up-regulated in the 3- and 24-h data sets, and of these, 67 (14%) and 51 (11%) genes were validated by TRP-EGL. However, only 14 genes from these lists overlapped, one of which was confirmed. In a second analysis using SAM for Δ values (Δ3 h = 0.91 and Δ24 h = 1.31) that resulted in an FDR of 10.3 and 10.8%, only 31 and 26 genes were identified as up-regulated, and none of them overlapped (data not shown).
Thus, our custom AR method yields a 7-fold increase in positive predictive value compared with SAM. We conclude that our procedure, which incorporates experiment-specific noise models, yields an increased specificity in the analysis of 3- and 24-h data sets. In practical terms, the method results in an enhanced ability to identify genes with authentic expression in the developing GCNP compartment.
Discussion
Functional genomic approaches attempting to elucidate mechanisms of normal development have been faced with the challenge of extracting useful information from data that is contaminated by many sources of noise. Primary or explant cultures are inherently heterogeneous, and components of such cultures may react differently to developmentally relevant signaling molecules as reflected in distinct transcriptional responses. The surprising finding that Shh affects predominantly a single cell type (GCNP) may be a feature unique to the cerebellar granule culture system. It is possible that this is a special case, albeit an important one, because it does make the point that with the right biological system a strong and specific stimulus for a specific cell type can be discriminated.
The number of genes/ESTs we observed to be up-regulated at both 3- and 24-h time points were an order of magnitude less than those found to be implicated in cell cycle regulation in yeast (24). However, our studies used complex heterogeneous cultures with only a small fraction of cells responding to Shh. It therefore is not surprising that many genes that may show significant up-regulation in synchronous, homogeneous (unicellular) cultures may have been masked in our studies. Further, the up-regulation of genes as detected by microarray may come from two sources, namely (i) the up-regulation of genes in a portion of the cells, e.g., a particular cell type, or (ii) the increased proportion of cell types that express this gene. In the 3-h Shh vs. control data, the first effect may dominate, because the GCNPs have not yet begun to proliferate in Shh-treated cultures. Thus, the up-regulation of the genes is likely reflective of the cellular response to Shh. Conversely, in the 24-h Shh vs. control data the second effect may play a larger part. This may explain why only a small number of the “significant” genes found in 3- and 24-h Shh experiments overlapped. We observed that the intersection of these two differentially expressed gene sets enhanced the likelihood of identifying genes with GCNP-specific expression. Interestingly, we have extended our analysis to another type of cell in the heterogeneous GCNP cultures. To identify genes expressed in postmitotic granule cells, we analyzed the vehicle-treated cultures at 3 and 24 h (i.e., conditions that promote cell-cycle exit). We found over 300 genes that were correlated to expression of the mature granule cell-specific marker, Zic1, including En2, LIM 1, P27, GIRK1, and ID3, all known markers of the cerebellar internal granule cell. Thus, with the proper experimental design it may be possible to dissect out later stage-specific granule cell markers.
As has been observed (3), the heterogeneous nature of tissues during development and the precise spatial restrictions on gene-expression domains dictates confirmation of gene expression by ISH. Our studies differ from other recent expression profiling studies in that we have emphasized achievement of higher specificity. Such methodology should benefit biological investigation by identifying genes with a relatively low false-positive rate that are likely to have an appropriate pattern of expression in vivo. Although our studies used three hybridizations (two interexperiment and two intra-experiment) for each data point, enhanced specificity and sensitivity would be possible with larger numbers of replicates. Nonetheless, for any given number of replicates, our technique seems to improve specificity for a given level of sensitivity compared with the standard ratio calculation. By calculating the FDN from AR rather than the typically used RA, we captured how the fold ratio was affected by measurement variation. The AR allowed us to keep the false-negative rate relatively low as evidenced by the results of the ISH and temporal gene-expression studies.
The database of postnatal cerebellar gene expression seems to provide a comprehensive source of information that can be used to predict genes expressed in GCNPs as well as other developing cerebellar cellular populations. However, because we have focused on confirmation of gene-expression patterns in candidates regulated by Shh, we cannot rule out that bias may have been a factor leading to the high sensitivity and specificity of our knowledge-based TRP-EGL approach. The design of the TRP-EGL is based on the known developmental behavior of granule cell precursors, a largely transient population present in the first postnatal week of life. Indeed, the TRP-EGL is highly effective at identification of many other known EGL-specific genes including Math1 (15) and HES1 (25). Future work will be required to test the utility of using solely TRP-EGL as a means of identifying genes with tissue-specific expression.
Hedgehog Pathway Effects During Development and Tumorigenesis.
We have used expression profiling with oligonucleotide microarrays to identify genes expressed in immature GCNPs maintained by Shh. Scrutiny of 40 of 76 validated genes from the combined 3- and 24-h data sets (Table 2) revealed that they comprise ≈90% proliferation-associated genes. This is consistent with the fact that Shh is a mitogen with common effects on cell-cycle progression at these time points (14). In addition, we observed nonproliferation-associated genes in cultures treated with Shh for 3 or 24 h. For instance, transcription factors comprised ≈10% of total genes identified at individual 3- and 24-h time points; these included the transcription factor Pax2, a paired-box protein with known roles in mid-hindbrain and cerebellar development (26), and Math1, a basic helix–loop–helix protein that is essential for granule cell development (15), and the Shh transcriptional target, Gli2 (12). Identification of transcription factors and signaling molecules in the 3- and 24-h data sets may provide new markers and candidate regulators of granule cell development.
Inappropriate activation of the Shh signaling pathway can promote development of medulloblastoma, a cerebellar tumor (27). Previous work indicates that Shh promotes continued cell-cycle progression in proliferating neural precursor cells by maintaining expression of G1 cyclins (14). It is of interest that our analysis identified a number of genes closely associated with cell-cycle regulation that were induced as quickly as 3 h. Interestingly, N-myc expression (Table 3; A.M.K. and D.H.R., unpublished observations) has been found recently in human medulloblastoma associated with Hedgehog pathway activation (28). Up-regulation of proliferation-associated genes has been linked to various cancer cell lines (29) and human malignancy (30). Thus, further analysis of the identified genes/ESTs may shed further light on Hedgehog pathway activation and tumorigenesis.
Supplementary Material
Acknowledgments
We are especially grateful to Drs. Todd Golub and Eric Lander for advice and support, Dr. Atul Butte for informative discussions, and Michelle Gaasenbeek, Christine Ladd, and Sovann Kaing for technical assistance. Q.Z and A.M.K. thank the American Brain Tumor Association for postdoctoral fellowships. A.K. acknowledges the Dana-Mahoney Neuro-Oncology Program for support. These studies were funded by National Institute of Neurological Disorders and Stroke Grants R21 NS41764-01 and RO1 NS4051 (to D.H.R.), the Edward Mallinkrodt, Jr. Foundation, the National Multiple Sclerosis Society and the Claudia Adams Barr Foundation. D.H.R is a Kimmel Scholar.
Abbreviations
- ISH
in situ hybridization
- GCNP
granule cell neuron precursor
- EGL
external granule layer
- Shh
Sonic hedgehog
- PN
postnatal day
- AR
average of ratios
- RA
ratio of the averages
- Avg Diff
average difference
- EST
expressed sequence tag
- FDN
fold distribution caused by noise
- TRP
temporal gene regulation profile
- FDR
false-discovery rate
- SAM
significance analysis of microarrays
References
- 1.Edlund T, Jessell T M. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 2.Geschwind D H, Ou J, Easterday M C, Dougherty J D, Jackson R L, Chen Z, Antoine H, Terskikh A, Weissman I L, Nelson S F, Kornblum H I. Neuron. 2001;29:325–339. doi: 10.1016/s0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 3.Zirlinger M, Kreiman G, Anderson D J. Proc Natl Acad Sci USA. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dent G W, O'Dell D M, Eberwine J H. Physiol Behav. 2001;73:841–847. doi: 10.1016/s0031-9384(01)00521-2. [DOI] [PubMed] [Google Scholar]
- 5.Sandberg R, Yasuda R, Pankratz D G, Carter T A, Del Rio J A, Wodicka L, Mayford M, Lockhart D J, Barlow C. Proc Natl Acad Sci USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terskikh A V, Easterday M C, Li L, Hood L, Kornblum H I, Geschwind D H, Weissman I L. Proc Natl Acad Sci USA. 2001;98:7934–7939. doi: 10.1073/pnas.131200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mody M, Cao Y, Cui Z, Tay K Y, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien J Z. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M Q. Genome Res. 1999;9:681–688. [PubMed] [Google Scholar]
- 9.Mirnics K. Nat Rev Neurosci. 2001;2:444–447. doi: 10.1038/35077587. [DOI] [PubMed] [Google Scholar]
- 10.Wingate R J, Hatten M E. Development (Cambridge, UK) 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- 11.Wechsler-Reya R J, Scott M P. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 12.Dahmane N, Ruiz-i-Altaba A. Development (Cambridge, UK) 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 13.Wallace V A. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 14.Kenney A M, Rowitch D H. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, Zoghbi H Y. Nature (London) 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 16.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang C H, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov J P, et al. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butte, A. J., Ye, J., Haring, H. U., Stumvoll, M., White, M. F. & Kohane, I. S. (2001) Pac. Symp. Biocomput., 6–17. [DOI] [PubMed]
- 18.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ly D H, Lockhart D J, Lerner R A, Schultz P G. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 21.Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner M L, Kallioniemi A. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Q R, Yuk D, Alberta J A, Zhu Z, Pawlitzky I, Chan J, McMahon A P, Stiles C D, Rowitch D H. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 23.Altman J. J Comp Neurol. 1972;145:353–397. doi: 10.1002/cne.901450305. [DOI] [PubMed] [Google Scholar]
- 24.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solecki D J, Liu X L, Tomoda T, Fang Y, Hatten M E. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 26.Urbanek P, Fetka I, Meisler M H, Busslinger M. Proc Natl Acad Sci USA. 1997;94:5703–5708. doi: 10.1073/pnas.94.11.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corcoran R B, Scott M P. J Neurooncol. 2001;53:307–318. doi: 10.1023/a:1012260318979. [DOI] [PubMed] [Google Scholar]
- 28.Pomeroy S L, Tamayo P, Gaasenbeek M, Sturla L M, Angelo M, McLaughlin M E, Kim J Y, Goumnerova L C, Black P M, Lau C, et al. Nature (London) 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 29.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van de Rijn M, Waltham M, et al. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 30.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.